Abstract
Yellow spot (syn. tan spot), caused by Pyrenophora tritici-repentis, is an important foliar disease of wheat in Australia that causes losses exceeding 50 % when conditions are favourable for disease development. Although good progress has been made internationally to understand yellow spot resistance, relatively few resistance genes have been identified and mapped in Australian germplasm and only one (tsn1 on chromosome 5BL) is in general and known use in Australian breeding programs. Although tsn1 is an important yellow spot resistance gene, it doesn’t explain the full spectrum of resistance and there is a significant opportunity to enhance expression of yellow spot resistance through identification of resistance factors other than tsn1. Six doubled haploid (DH) mapping populations (five of which were fixed for tsn1) were screened for yellow spot resistance at the seedling/tillering and adult plant stages at the Department of Agriculture and Food, Western Australia (DAFWA) and the Department of Environment and Primary Industries Victoria (DEPIVic) from 2009 to 2012. Four of the above populations were screened at the Department of Agriculture, Fisheries and Forestry Queensland (DAFFQ). Frequency distribution of individuals within each population for various levels of yellow spot resistance was continuous indicating that resistance is conditioned by several genes with partial effects. A few lines within each population consistently showed high levels of resistance probably resulting from a combination of several genes with additive effects. Nine new loci for yellow spot resistance were mapped by the Australian Wheat and Barley Molecular Marker Program (AWBMMP) at the University of Adelaide on chromosomes 1AS, 2AS, 5AS, 5AL, 4B, 6BS, 7BL, 2D and 7D in five of the six populations phenotyped so far. High LOD scores of 9–11 have been obtained for some of the QTL with a percentage disease reduction of 24–42 %. Efforts are now focused on identifying additional yellow spot resistance genes using newly developed populations and rapid phenotyping methods and developing a series of fixed lines, each carrying yellow spot resistance genes from various sources, in elite Australian backgrounds. These materials will provide proof-of-concept for achieving better resistance by pyramiding resistance genes, and they should be directly useful as parents for wheat breeding.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
Pyrenophora tritici-repentis is a necrotophic fungus that causes yellow spot of wheat. The disease is also known as yellow leaf spot or tan spot. There has been an increased prominence of this disease in recent years due to implementation of new farm practices such as minimum tillage, stubble retention, increased wheat-after-wheat cropping and use of susceptible varieties. In Australia, Shipton (1968), Rees et al. (1982), Rees and Platz (1983), and Bhathal et al. (2003) measured 20–50% losses in grain yield of bread wheat from this disease. In Western Australia this disease often occurs as a complex with Stagonospora nodorum blotch.
P. tritici-repentis is known to produce at least three host specific toxins (HSTs) that interact with specific host sensitivity genes to cause necrosis and/or extensive chlorosis (Ali et al. 1999; Lamari et al. 1995, 2005; Strelkov et al. 2002). These are Ptr ToxA, for which sensitivity is conditioned by the gene Tsn1 on chromosome arm 5BL (Anderson et al. 1999; Faris et al. 1996); Ptr ToxB, for which sensitivity is conditioned by the gene Tsc2 on chromosome arm 2BS (Friesen and Faris 2004), and Ptr ToxC, for which sensitivity is conditioned by the gene Tsc1 on chromosome arm 1AS (Effertz et al. 2002). Isolates of the pathogen can be classified into eight theoretical races based on the presence or absence of each of the three toxins. All eight of these combinations have been found among North American isolates. Recently, however, ToxA and ToxB gene-deficient isolates from Arkansas have been found not to fit this race classification system (Ali et al. 2010). These isolates may contain different toxin compounds that produce ToxA-like symptoms on some cultivars of wheat.
Improved resistance to the disease is the most economical and effective method of control and is therefore a key target for Australian wheat breeding programs. Resistance can be both qualitative (Gamba and Lamari 1998; Lamari and Bernier 1989) and quantitative (Elias et al. 1989; Friesen and Faris 2004). Genes such as tsn1, tsn2, tsn3, tsn4, tsn5, tsn6, and tsn-syn1 and Tsn-syn2, are known to affect toxin sensitivity and to confer race-specific resistance to necrosis (Anderson et al. 1999; Singh et al. 2006, 2008; Tadesse et al. 2006a, b, 2010) and the recessive genes tsc1 and tsc2 condition resistance to chlorosis (Abeysekara et al. 2010; Effertz et al. 2002; Friesen and Faris 2004). Non-race-specific quantitative trait loci have been mapped on almost all chromosomes of wheat (Chu et al. 2008, 2010; Faris and Friesen 2005; Faris et al. 2012; Li et al. 2011; Singh et al. 2012; Sun et al. 2010; Patel et al. 2013). Incorporating both qualitative and quantitative resistance is essential to develop cultivars with high levels of durable resistance.
Although good progress has been made internationally to understand yellow spot resistance, only one gene (tsn1 on chromosome 5BL) is in general and known use in Australian breeding programs. While tsn1, is an important YS resistance gene, it doesn’t explain the full spectrum of resistance in the Australian germplasm. Furthermore, Faris et al. (2012) have shown that the amount of variation explained by tsn1 can vary considerably (5–30 %) with different isolates and suggest possible variability in ToxA gene regulation amongst isolates.
The Department of Agriculture and Food Western Australia (DAFWA) has been conducting research for the past 3 years on genetic enhancement of resistance to the disease and improved germplasm development under a national project involving several collaborating organisations and co-funded by the Grains Research and Development Corporation. The project aims to provide improved genetic solutions to yellow spot management by: (i) development of improved phenotyping methods, (ii) identification of new resistance genes, (iii) improved germplasm and associated genetic knowledge that enhance resistance expression and diversity and (iv) improved parental stocks which can be readily utilised within breeding programs.
Phenotyping methods have been improved in three areas. Firstly, enhanced spore production techniques have been developed based on a modified method of Evans et al. (1993) which allow the production and storage of large amounts of yellow spot inoculum. This has greatly improved inoculation of a large number of plots both in the field and glasshouse. Secondly improved field phenotyping methods have been developed based on Shankar et al. (2008) which overcome the effect of maturity and height on disease expression allowing the assessment of true adult plant resistance. This basically involves inoculating individual plots at heading and rating at a specific thermal time after inoculation. Thirdly, a rapid assay for evaluating adult plant resistance has been developed under controlled environmental conditions under which the same plant can be assessed at both seedling and adult stages. Under this method plants head within 5–8 weeks under controlled environment as compared to 11–16 weeks in the field. Moderate to good correlations (r = 0.7–0.8) are obtained between assessments made under controlled environmental conditions and field conditions.
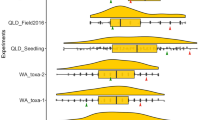
Twelve doubled haploid mapping populations have been developed or made available to the project. Of these, six populations (five of which were fixed for tsn1) were phenotyped for yellow spot resistance from 2009 to 2012 at the seedling/tillering and adult plant stages at DAFWA and the Department of Environment and Primary Industries Victoria (DEPIVic). Four of the above populations were phenotyped at the seedling/tillering and adult plant stages at the Department of Agriculture, Fisheries and Forestry Queensland (DAFFQ). Frequency distribution of individuals within each population for various levels of yellow spot resistance was continuous indicating that resistance is conditioned by several genes with partial effects. A few lines within each population consistently showed high levels of resistance probably resulting from a combination of several genes with additive effects.
Genetic maps were developed by the Australian Wheat and Barley Molecular Marker Program (AWBMMP) at the University of Adelaide using DArT, SSR and SNP markers. Nine major QTL other than tsn1 have been mapped on chromosomes 1AS, 2AS, 5AS, 5AL, 4B, 6BS, 7BL, 2D and 7D in five of the six populations phenotyped so far. Of these, QTL on 2A and 6B appear commonly between two populations but it is yet to be determined if they are at the same chromosomal positions. Statistical analyses have been completed on two of the populations and good correlations (r = 0.6–0.8) and high heritability estimates (H2 = 0.7–0.9) were obtained for yellow spot severity at various growth stages, environments and sites. High LOD scores of 9–11 have been obtained for the resistance QTL detected in these populations with a percentage disease reduction of 24–42 %.
Germplasm development is being carried out collaboratively by DAFWA and AWBMMP. The strategy involves pyramiding at least three resistance loci from different sources into elite Australian backgrounds. In a first set of crosses QTL on 2A and 5BL detected in one population are being pyramided with QTL on 1A detected in another population. Resistant parents were selected using both phenotypic and genotypic data and a cross was made in the spring of 2011. Although all the 100 F1s produced turned out to be grass clumps 194 F2 seeds were produced off these grass clumps by growing them at a constant 26 °C temperature and gibberellic acid treatment. The F2s were then grown out in single seed trays and leaf material sent to AWBMMP for marker-assisted selection. F2 and F3 segregation indicated that the grass clump trait was controlled by two to three complementary genes.
Currently 16 F4 lines, homozygous at the three resistance loci as well as fixed at the Vrn-A1 locus have been selected. Fourteen of these are mid to late maturing while two are early maturing. In addition 28 other F4 lines, also homozygous at the three resistance loci but segregating for either grass clumps or at the Vrn-A1 locus, have been selected. These are being further selected against the grass clump trait and are being fixed for the Vrn-A1 locus. All F4/F5 selections will be grown out for a bulk F5/F6 harvest and phenotyped for proof of concept. These are potential parental stocks enriched for resistance in wheat breeding.
This year crosses are being made to pyramid up to five resistance loci in both short season and long season backgrounds by crossing lines with three stacked genes with parental selections involving two additional genes from other available populations.
A spin off research activity under gene pyramiding for yellow spot resistance is developing sister lines with different resistance QTL combinations. For this, individuals which are heterozygous at the three resistance loci are being selected in each generation while allowing the rest of the genome to become homozygous. The aim is to obtain different QTL combinations within sister lines so that effects of individual loci and loci in various combinations can be compared within a similar background. Currently 25 F4 lines heterozygous at the three resistance loci have been selected and will be further selected in F5 for various QTL combinations.
In another activity within the project, genetic variability within the Australian germplasm is being combined by developing a large multiparent advanced generation intercross (MAGIC) population from a complex cross involving different sources of yellow spot resistance. The four parents include GBA Ruby, H45, King Rock and Magenta. The four-way cross was completed last spring and 583 F1s produced. Two F2 seeds from each of these F1s are being progressed through SSD to F5. Four way crossing is continuing this year with the aim to produce a total of around 2,000 F5 lines. This population will provide a unique opportunity to maximise genetic recombination, generate potential parental stocks enriched for yellow spot resistance and create opportunities for fine mapping resistance loci.
Another major activity within this project is identification of elite yellow spot resistant lines amongst material acquired from CIMMYT and ICARDA and amongst various mapping populations. Of over 1,000 lines screened so far 42 lines representing targeted yellow spot resistant material were tested under a ring test at various growth stages across Australia and 19 of these have been identified as having good levels of broad spectrum yellow spot resistance.
New resistance genes, germplasm and screening methodologies developed under this project are important resources that can be used by breeders for rapid development of varieties with high levels of resistance at both seedling and adult plant stages that is effective in various environments and across various national sites. Newly developed resistant varieties will greatly reduce the estimated annual losses of $212 M per annum caused by this disease in Australia (Murray and Brennan 2009).
References
Abeysekara S, Friesen TL, Liu Z et al (2010) Marker development and saturation mapping of the tan spot Ptr ToxB sensitivity locus Tsc2 in hexaploid wheat. Plant Genome 3:179–189
Ali S, Francl LJ, de Wolf ED (1999) First report of Pyrenophora tritici-repentis race 5 from North America. Plant Dis 83:591
Ali S, Gurung S, Adhikari TB (2010) Identification and characterization of novel isolates of Pyrenophora tritici-repentis from Arkansas. Plant Dis 94:229–235
Anderson JA, Evertz RJ, Faris JD et al (1999) Genetic analysis of sensitivity to Pyrenophora tritici-repentis necrosis-inducing toxin in durum and common wheat. Phytopathology 89:293–297
Bhathal JS, Loughman R, Speijers J (2003) Yield reduction in wheat in relation to leaf disease from yellow (tan) spot and septoria nodorum blotch. Eur J Plant Pathol 109:435–443
Chu CG, Friesen TL, Xu SS, Faris JD (2008) Identification of novel tan spot resistance loci beyond the known host-selective toxin insensitivity genes in wheat. Theor Appl Genet 117:873–881
Chu CG, Chao S, Friesen TL et al (2010) Identification of novel tan spot resistance QTLs using an SSR-based linkage map of tetraploid wheat. Mol Breed 25:327–338
Effertz RJ, Meinhardt SW, Anderson JA et al (2002) Identification of a chlorosis-inducing toxin from Pyrenophora tritici-repentis and the chromosomal location of an insensitivity locus in wheat. Phytopathology 92:527–533
Elias E, Cantrell RG, Horsford RM Jr (1989) Heritability of resistance to tan spot in durum wheat and its association with other agronomic traits. Crop Sci 29:299–304
Evans CK, Hunger RM, Siegerist WC (1993) Enhanced production of Pyrenophora tritici repentis conidial suspensions. Plant Dis 77:981–984
Faris JD, Friesen TL (2005) Identification of quantitative trait loci for race-nonspecific resistance to tan spot in wheat. Theor Appl Genet 111:386–392
Faris JD, Anderson JA, Francl LJ, Jordahl JG (1996) Chromosomal location of a gene conditioning insensitivity in wheat to a necrotic inducing culture filtrate from Pyrenophora tritici-repentis. Phytopathology 86:459–463
Faris JD, Abeysekara NS, McClean PE et al (2012) Tan spot susceptibility governed by the Tsn1 locus and race-nonspecific resistance quantitative trait loci in a population derived from the wheat lines Salamouni and Katepwa. Mol Breed 30:1669–1678
Friesen TL, Faris JD (2004) Molecular mapping of resistance to Prenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor Appl Genet 109:464–471
Gamba FM, Lamari L (1998) Mendelian inheritance of resistance to tan spot (Pyrenophora tritici-repentis) in selected genotypes of durum wheat (Triticum turgidum). Can J Plant Pathol 20:408–414
Lamari L, Bernier CC (1989) Toxin of Pyrenophora tritici-repentis: host specificity, significance in disease and inheritance of host reaction. Phytopathology 79:740–744
Lamari L, Sayoud R, Boulif M, Bernier CC (1995) Identification of a new race in Pyrenophora tritici-repentis: implications for the current pathotype classification system. Can J Plant Pathol 17:312–318
Lamari L, Strelkov SE, Yahyoui A et al (2005) Virulence of Pyrenophora tritici-repentis in the countries of the Silk Road. Can J Plant Pathol 27:383–388
Li HB, Yan W, Liu GR et al (2011) Identification and validation of quantitative trait loci conferring tan spot resistance in the bread wheat variety Ernie. Theor Appl Genet 122:395–403
Murray GM, Brennan JP (2009) The current and potential costs from diseases of wheat in Australia. Report published by GRDC (Grains Research and Development Corporation). http://www.grdc.com.au/director/events/grdcpublications
Patel JS, Mamidi S, Bonman MJ, Adhikari TB (2013) Identification of QTL in spring wheat associated with resistance to a novel isolate of Pyrenophora tritici-repentis. Crop Sci 53:842–852
Rees RG, Platz GJ (1983) Effects of yellow spot on wheat: comparison of epidemics at different stages of crop development. Aust J Agric Res 34:39–46
Rees RG, Platz GJ, Mayer RJ (1982) Yield losses in wheat from yellow spot: comparison of estimates derived from single tillers and plots. Aust J Agric Res 33:899–908
Shankar M, Walker E, Golzar H et al (2008) Quantitative trait loci for seedling and adult plant resistance to Stagonospora nodorum in wheat. Phytopathology 98:886–893
Shipton WA (1968) The effect of septoria diseases on wheat. Aust J Exp Agr Anim Husb 8:89–93
Singh PK, Gonzalez-Hernandez JL, Mergoum M et al (2006) Identification and molecular mapping of a gene in tetraploid wheat conferring resistance to Pyrenophora tritici-repentis race 3. Phytopathology 96:885–889
Singh PK, Mergoum M, Gonzalez-Hernandez JL et al (2008) Genetic and molecular mapping of resistance to necrosis inducing race 5 of P. tritici-repentis in tetraploid wheat. Mol Breed 21:293–304
Singh S, Hernandez MV, Crossa J et al (2012) Multi-trait and multi-environment QTL analyses for resistance to wheat diseases. PLoS One 7:e38008
Strelkov SE, Lamari L, Sayoud R, Smith RB (2002) Comparative virulence of chlorosis-inducing races of Pyrenophora tritici-repentis. Can J Plant Pathol 24:29–35
Sun XC, Bockus W, Bai G (2010) Quantitative trait loci for resistance to Pyrenophora tritici-repentis race 1 in a Chinese wheat. Phytopathology 100:468–473
Tadesse W, Hsam SLK, Wenzel G, Zeller FJ (2006a) Identification and monosomic analysis of tan spot resistance genes in synthetic wheat lines (Triticum turgidum L. × Aegilops tauschii Coss.). Crop Sci 46:1212–1217
Tadesse W, Hsam SLK, Zeller FJ (2006b) Evaluation of common wheat (Triticum aestivum L.) cultivars for tan spot resistance and chromosomal location of a resistance gene in cultivar ‘Salamouni’. Plant Breed 125:318–322
Tadesse W, Schmolke M, Hsam SLK et al (2010) Chromosomal location and molecular mapping of a tan spot resistance gene in the winter wheat cultivar Red Chief. J Appl Genet 51:235–242
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 Crown Copyright as represented by the Western Australian Government
About this paper
Cite this paper
Shankar, M. et al. (2015). Germplasm Enhancement for Resistance to Pyrenophora tritici-repentis in Wheat. In: Ogihara, Y., Takumi, S., Handa, H. (eds) Advances in Wheat Genetics: From Genome to Field. Springer, Tokyo. https://doi.org/10.1007/978-4-431-55675-6_21
Download citation
DOI: https://doi.org/10.1007/978-4-431-55675-6_21
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-55674-9
Online ISBN: 978-4-431-55675-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)