Abstract
Dietary iron (Fe) deficiency affects 14% of the world population with significant health impacts. Biofortification is the process of increasing the density of vitamins and minerals in a crop, through conventional breeding, biotechnology approaches, or agronomic practices. This process has recently been shown to successfully alleviate micronutrient deficiency for populations with limited access to diverse diets in several countries (https://www.harvestplus.org/). The Fe breeding target in the HarvestPlus program was set based on average rice consumption to fulfil 30% of the Estimated Average Requirement of Fe in women and children. In this review, we present the reported transgenic approaches to increase grain Fe. Insertion of a single or multiple genes encoding iron storage protein, metal transporter, or enzyme involved in the biosynthesis of metal chelator in the rice genome was shown to be a viable approach to significantly increase grain-Fe density. The most successful approach to reach the Fe breeding target was by overexpression of multiple genes. Despite this success, a significant effort of 8–10 years needs to be dedicated from the proof of concept to varietal release. This includes large-scale plant transformation, event selection, collection of data for premarket safety assurance, securing biosafety permits for consumption and propagation, and collection of data for variety registration.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Nutritional deficiency is an important global health concern that affects approximately 1.86 billion people worldwide, and 61% of it is caused by dietary iron (Fe) deficiency (James et al. 2018). Iron deficiency anemia results in decreased work productivity, increased maternal mortality, increased child mortality, slowed child development, and increased susceptibility to infectious diseases (Stoltzfus 2001). Iron deficiency anemia is estimated to be responsible for 800,000 deaths/year (WHO 2002).
Although providing access to more diverse diets is the ideal solution to alleviate micronutrient deficiency , this may not be achieved in the near future in developing and less developed countries. Iron supplementation and industrial fortification have been shown to alleviate micronutrient deficiencies but require continuous significant budget allocation at the government or household level. Despite the potential to diminish iron deficiency in the population, this may not help people living in remote rural areas because of the lack of infrastructure, purchasing power, or access to markets and healthcare systems (Mayer et al. 2008). Biofortification, a process of increasing the concentration of micronutrient in the edible part of a crop through conventional plant breeding, transgenic methods, and agronomic practices, offers a feasible and cost-effective approach, complementing other efforts to reach rural populations (Bouis and Saltzman 2017).
2 Target Concentration for Fe in Polished Rice
The Estimated Average Requirement (EAR) of iron for non-pregnant, non-lactating women is 1460 mg/day, and for children 4–6 years old is 500 mg/day (WHO/FAO 2004). Current studies show that 90% of the iron remains in the grain after processing, while the nutritionist assumption is that 10% of the iron is bioavailable. With 400 g/day per capita rice consumption for an adult woman and 120 g/day for children 4–6 years old, the HarvestPlus program set 13 μg/g as the final target concentration for Fe in polished rice to achieve 30% of the EAR (Bouis et al. 2011).
3 Iron Biofortification via Conventional Plant Breeding
Initial screening of the germplasm collection at the International Rice Research Institute (IRRI) showed that the range in Fe concentration in brown rice among 1138 genotypes tested was 6.3–24.4 μg/g (Gregorio et al. 1999). However, the variation in Fe concentration in milled rice becomes narrow due to the high proportion of Fe lost during milling. A study on the iron concentration of brown and milled rice of six varieties collected from ten commercial rice mills in one province in Vietnam showed that the percentage of Fe loss due to milling ranged from 65% to 82% (Hoa and Lan 2004). Furthermore, the maximal iron concentration in milled rice was 8 μg/g over 11,337 genotypes from the International Center for Tropical Agriculture (CIAT) core collection (Martínez et al. 2010). At IRRI, the highest Fe concentration of 7.4 μg/g in polished rice was achieved by classical breeding (Virk et al. 2006, 2007).
To reach the Fe target of 13 μg/g in polished rice, transgenic approaches are potential options because of the low concentration of Fe found in the rice gene pool. Efforts have been made to study the physiology as well as the genetic basis and biochemical mechanisms involved in Fe uptake and translocation in crops and model plants. These studies have facilitated the detection of the limiting factors that could be manipulated to increase Fe concentration in rice grain.
4 Iron Uptake and Translocation
Iron is an important micronutrient required in various processes such as photosynthesis and respiration. Based on the strategy they use to uptake iron from the rhizosphere, higher plants can be categorized into three different groups (Connorton et al. 2017): (1) Strategy I plants (all dicotyledonous plants and non-graminaceous monocots) that rely on the reduction of ferric Fe(III) to ferrous Fe(II), (2) Strategy II plants (graminaceous monocots) that rely on the chelation strategy involving phytosiderophore secretion, and (3) a combination of both. Arabidopsis has been used as a model plant to study Strategy I plants. Some major genes responsible for iron uptake using this strategy have been identified: iron-regulated transporter 1 (IRT1) (Eide et al. 1996), ferric-chelate oxidase 2 (FRO2) (Robinson et al. 1999), and HC-ATPase (HA) genes (Kobayashi and Nishizawa 2012).
Rice has been used as a model plant to study Strategy II plants. Plants in this group that include the most important cereals in the world secrete phytosiderophore (PS) in the rhizospere. PS is a high-Fe-affinity organic molecule from the mugineic acid family (Bashir et al. 2017; Borrill et al. 2014; Kobayashi et al. 2005; Suzuki et al. 2006). Figure 1 presents the basic scheme for the genes involved in iron homeostasis in rice.
TOM1 transporter of mugineic acid family phytosiderophores 1, YS1 yellow stripe 1, YSL15 yellow stripe 1-like, IRT1 iron-regulated transporter, NRAMP1 natural resistance-associated macrophage protein 1, FRO ferric reductase oxidase, FRDL1 ferric reductase defective-like1, ENA efflux transporter of nicotianamine, VIT vacuolar iron transporter, FER ferritin, MA mugineic acid, DMA 2′-deoxymugineic acid, NA nicotianamine, SAM S-adenosyl-l-methionine, NAS nicotianamine synthase, NAAT nicotianamine aminotransferase, DMAS deoxymugineic acid synthase
Mugineic acid is synthesized through a conserved pathway that starts from S-adenosyl-l-methionine. It is then followed by sequential reactions catalyzed by nicotianamine synthase (NAS), nicotianamine aminotransferase (NAAT), and deoxymugineic acid synthase (DMAS) enzymes, producing 20-deoxymugineic acid (DMA), a precursor of different types of mugineic acids (Bashir et al. 2006; Higuchi et al. 1999; Kobayashi and Nishizawa 2012; Takahashi et al. 1999). In rice roots, secretion of DMA is influenced by the expression of the TOM1 geneencoding efflux transporter of DMA (Nozoye et al. 2011). YELLOW STRIPE 1 (YS1) and YELLOW STRIPE 1-like (YSL1) transporters are known for their role in facilitating the uptake of Fe–MA complexes into root cells (Curie et al. 2001; Inoue et al. 2009). After reduction by ascorbate, the Fe(III)–DMA complex is likely converted to Fe(II)–NA, and then excreted to the xylem. The Fe may create complexes primarily with citrate and some with DMA for further transport (Yoneyama et al. 2015).
Aside from using Strategy II, rice may be able to uptake Fe(II) directly, as indicated by the presence of a ferrous transporter (OsIRT1) in the genome. Other metal transporters involved in Strategy I and Strategy II were also identified such as natural resistance-associated macrophage protein (NRAMP) and ZIP (zinc-regulated transporter, IRT-like protein) family (Cailliatte et al. 2010; Guerinot 2000; Lanquar et al. 2005).
Fe translocation in higher plants is a complex process involving xylem loading/unloading, phloem loading/unloading, and reabsorption (Kim and Guerinot 2007). Different chelators such as citrate, nicotianamine (NA), and MAs play an essential role in symplast metal homeostasis (Garcia-Oliveira et al. 2018). FERRIC REDUCTASE DEFECTIVE-LIKE 1 (OsFRDL1) in rice (Fig. 1) is known to encode a citrate transporter involved in the transport of Fe-citrate complex (Inoue et al. 2004; Yokosho et al. 2009).
The rice YSL family encoding influx transporter consists of 18 members (Curie et al. 2009). The OsYSL2 transporter is a carrier of Fe(II)–NA and is involved in iron transport to sink tissues (Koike et al. 2004). OsYSL15 transports Fe(III)–DMA and is involved in Fe uptake in the root and long-distance Fe transport. The Fe transporter OsYSL18 may play a specific role in fertilization, as indicated by specific expression in the pollen and pollen tubes. OsYSL18 may also be involved in Fe transport in the phloem (Kobayashi and Nishizawa 2012). As indicated by their vascular tissue expression in rice, OsIRT1 and OsTOM1 may be involved in Fe translocation within the plant as well (Ishimaru et al. 2006; Nozoye et al. 2011).
5 Iron Biofortification via Genetic Engineering
Several efforts have been conducted to increase Fe concentration in rice grains. These studies can be categorized into different approaches: (1) overexpression of geneencoding iron storage protein, (2) overexpression of geneencoding enzyme involved in the biosynthesis of metal chelator, (3) overexpression of geneencoding metal transporter, and (4) a combination of two or three approaches (Tables 1 and 2).
Fe concentration of 38.1 μg/g in brown rice was achieved by endosperm-specific expression of a soybean Fe storage protein, SoyFerH1 (Goto et al. 1999). Similar approaches using the SoyferH1 gene driven by different promoters (OsGluB1, OsGluB4, OsGlb1, ZmUbi-1) in different backgrounds (Swarna, IR68144, BR29, IR64, M12) were reported (Slamet-Loedin et al. 2015). Stable Fe concentrations of 9.2 or 7.6 μg/g over several generations were obtained (Khalekuzzaman et al. 2006; Oliva et al. 2014). Overexpression of the OsFer2 gene was also studied and Fe concentration of 15.9 μg/g, vis-à-vis 7 μg/g in control variety Pusa-Sugandh II, was observed (Paul et al. 2012).
Another approach in improving grain-Fe concentration in rice is by increasing the expression of genes encoding enzymes involved in the biosynthesis of metal chelator. OsNAS1 overexpression resulted in Fe concentration of 19 μg/g in brown rice; however, the concentration decreased to only 5 μg/g after polishing (Zheng et al. 2010). Co-overexpression of OsNAS1 and HvNAAT genes in japonica rice resulted in Fe concentration of 18 μg/g in the polished grain (Banakar et al. 2017b). Fe concentration of 55 μg/g in the succeeding generation was observed; however, this unusually high Fe concentration suggests either low milling degree or Fe contamination (Díaz-Benito et al. 2018). Overexpression and activation tagging of OsNAS2, on the other hand, resulted in 19 μg/g and 10 μg/g Fe concentration in polished rice, respectively (Johnson et al. 2011; Lee et al. 2012). Meanwhile, activation tagging of OsNAS3 achieved 12 μg/g Fe in polished grain vis-à-vis 4 μg/g Fe in the wild type (Lee et al. 2009b).
Several studies reported increased Fe uptake and translocation by overexpression of genes encoding metal transporter, including OsYSL2 (Ishimaru et al. 2010), OsYSL15 (Lee et al. 2009a), and OsYSL9 (Senoura et al. 2017). OsYSL2 and OsYSL15 are responsible for the uptake of Fe(II)–NA and Fe(III)–DMA, respectively, whereas OsYSL9 is involved in the transport of both complexes. Although only a minimal Fe increase was detected in T1 brown rice of OsYSL9 and OsYSL15 OE lines, overexpression of OsYSL2 resulted in a fourfold increase in Fe concentration in T1 polished rice.
Recently, two studies reported significant Fe concentration increases in rice grains by overexpressing multiple genes. Wu et al. (2019) overexpressed the AtNAS1, Pvfer, and AtNRAMP3 genes, resulting in 13.65 μg/g Fe in polished grains under greenhouse conditions. Trijatmiko et al. (2016), on the other hand, reported an Fe concentration of 15 μg/g in polished grains under field conditions by overexpressing nicotianamine synthase (OsNAS2) and soybean ferritin (SoyferH-1) genes. This high-Fe rice event did not show a yield penalty in field trials in the Philippines and Colombia. The grain quality of the transgenic event was similar to that of the IR64 genotype background used for transformation. These two studies show the potential for further advanced development of a biofortified rice product with elevated Fe concentration.
6 Future Directions
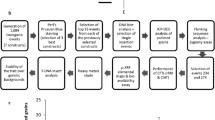
There is little prospect of achieving the target Fe concentration to reach 30% of the EAR via conventional plant breeding because of limited genetic variation in the Fe concentration in polished grains within the global rice germplasm collection. On the contrary, recent studies show that the target concentration can be achieved via genetic engineering. Under the current regulations in different countries, it usually takes 8-10 years from proof of concept to market release of genetically modified (GM) crops (Mumm 2013). The best performing events need to be selected from large-scale transformation. These events, aside from showing stable and acceptable trait expression, should have a simple integration of transgenes and have no disruption of endogenous genes with important phenotypic manifestation. Significant efforts need to be dedicated to collecting data for premarket safety assurance of the potential product, such as detailed molecular characterization of the event, safety of newly expressed proteins, novel protein expression and dietary exposure analysis, comparative nutritional analysis, and some environmental safety data collected from multi-location and multi-season field trials. After a biosafety permit for propagation of the event has been secured, developers need to follow similar procedures as in conventional breeding of a product for variety registration.
High Fe content is a consumer trait. To facilitate adoption by farmers, this micronutrient trait needs to be combined with agronomic traits. The most prospective agronomic trait for farmer adoption is higher yield. For this purpose, the possibility to incorporate the high-Fe trait into hybrid rice needs to be explored. The high-yield trait obtained through heterosis can be combined with nutritional traits. In addition, we observed that overexpression of some genes for Fe enhancement might cause unintended effects such as a yield decrease when the plants were in a homozygous condition. However, this is not detrimental when only one allele is present in hemizygous condition, and in some cases the micronutrient concentration can be retained in the hemizygous condition. In such a situation, hybrid rice can be a solution to achieve higher micronutrient concentration using a wider gene pool.
Recent developments in the regulation of genome-edited crops in different countries have attracted many scientists to work on genome editing. In the United States, certain categories of modified plants would be exempted from the regulations if the product can also be created through conventional breeding (APHIS 2020). In Argentina, a resolution on New Breeding Techniques (NBT) was passed in 2015, which rules that, if a transgene is not used or a transgene is used but is removed in the final product, it will not be classified as a GM product (Friedrichs et al. 2019). Precise genome editing technology that produces a double-stranded break in the genome, followed by the repair of this break that leads to a mutation or deletion, may result in a product that meets the non-GM regulatory classification.
Increased Fe concentration in polished grains was observed on the T-DNA insertion mutant of OsVIT2 (Bashir et al. 2013). The insertion of the T-DNA in the promoter region in this mutant led to the knockdown of the OsVIT2 gene (Bashir et al. 2013). Genome editing can be used to mutate the regulatory elements of genes involved in Fe homeostasis. This type of editing could result in altered expression of the genes and consequently enhanced Fe concentration in rice grains.
Although genome editing has great potential to ease the burden of regulatory requirements, genetic engineering will still be the primary tool to achieve the target Fe concentration. Overexpression of other genes involved in Fe homeostasis needs to be explored. Special attention needs to be given to the possible yield decrease in transgenic plants. Fine-tuning the expression of the genes by choosing a moderate constitutive promoter or tissue- and/or stage-specific promoter may need to be tested to avoid any yield penalty.
References
APHIS (2020) Movement of certain genetically engineered organisms. 85 FR 29790. Federal register document: 2020-10638. APHIS, Riverdale, MD
Aung MS, Masuda H, Kobayashi T, Nakanishi H, Yamakawa T, Nishizawa NK (2013) Iron biofortification of Myanmar rice. Front Plant Sci 4:158. https://doi.org/10.3389/fpls.2013.00158
Banakar R, Alvarez Fernández Á, Abadía J, Capell T, Christou P (2017a) The expression of heterologous Fe (III) phytosiderophore transporter HvYS1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport. Plant Biotechnol J 15(4):423–432. https://doi.org/10.1111/pbi.12637
Banakar R, Alvarez Fernandez A, Díaz-Benito P, Abadia J, Capell T, Christou P (2017b) Phytosiderophores determine thresholds for iron and zinc accumulation in biofortified rice endosperm while inhibiting the accumulation of cadmium. J Exp Bot 68(17):4983–4995. https://doi.org/10.1093/jxb/erx304
Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2006) Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J Biol Chem 281:32395. https://doi.org/10.1074/jbc.M604133200
Bashir K, Takahashi R, Akhtar S, Ishimaru Y, Nakanishi H, Nishizawa NK (2013) The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice 6:31. https://doi.org/10.1186/1939-8433-6-31
Bashir K, Nozoye T, Nagasaka S, Rasheed S, Miyauchi N, Seki M et al (2017) Paralogs and mutants show that one DMA synthase functions in iron homeostasis in rice. J Exp Bot 68:1785. https://doi.org/10.1093/jxb/erx065
Boonyaves K, Gruissem W, Bhullar NK (2016) NOD promoter-controlled AtIRT1 expression functions synergistically with NAS and FERRITIN genes to increase iron in rice grains. Plant Mol Biol 90(3):207–215. https://doi.org/10.1007/s11103-015-0404-0
Boonyaves K, Wu TY, Gruissem W, Bhullar NK (2017) Enhanced grain iron levels in iron-regulated metal transporter, nicotianamine synthase, and ferritin gene cassette. Front Plant Sci 8:130. https://doi.org/10.3389/fpls.2017.00130
Borrill P, Connorton JM, Balk J, Miller AJ, Sanders D, Uauy C (2014) Biofortification of wheat grain with iron and zinc: integrating novel genomic resources and knowledge from model crops. Front Plant Sci 5:53. https://doi.org/10.3389/fpls.2014.00053
Bouis HE, Saltzman A (2017) Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob Food Secur 12:49–58. https://doi.org/10.1016/J.GFS.2017.01.009
Bouis HE, Hotz C, McClafferty B, Meenakshi JV, Pfeiffer WH (2011) Biofortification: a new tool to reduce micronutrient malnutrition. Food Nutr Bull 32(1 Suppl 1):S31–S40. https://doi.org/10.1177/15648265110321S105
Cailliatte R, Schikora A, Briat JF, Mari S, Curie C (2010) High-affinity manganese uptake by the metal transporter nramp1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22:904. https://doi.org/10.1105/tpc.109.073023
Che J, Yokosho K, Yamaji N, Ma JF (2019) A vacuolar phytosiderophore transporter alters iron and zinc accumulation in polished rice grains. Plant Physiol 181(1):276–288. https://doi.org/10.1104/pp.19.00598
Connorton JM, Balk J, Rodríguez-Celma J (2017) Iron homeostasis in plants: a brief overview. Metallomics 9(7):813–823. https://doi.org/10.1039/C7MT00136C
Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346. https://doi.org/10.1038/35053080
Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M et al (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot 103:1. https://doi.org/10.1093/aob/mcn207
Díaz-Benito P, Banakar R, Rodríguez-Menéndez S, Capell T, Pereiro R, Christou P, Abadia J, Fernández B, Álvarez-Fernández A (2018) Iron and zinc in the embryo and endosperm of rice (Oryza sativa L.) seeds in contrasting 2′-deoxymugineic acid/nicotianamine scenarios. Front Plant Sci 9:1190. https://doi.org/10.3389/fpls.2018.01190
Drakakaki G, Christou P, Stöger E (2000) Constitutive expression of soybean ferritin cDNA in transgenic wheat and rice results in increased iron levels in vegetative tissues but not in seeds. Transgenic Res 9:445. https://doi.org/10.1023/A:1026534009483
Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci U S A 93:5624. https://doi.org/10.1073/pnas.93.11.5624
Friedrichs S, Takasu Y, Kearns P, Dagallier B, Oshima R, Schofield J, Moreddu C (2019) Meeting report of the OECD conference on “Genome Editing: Applications in Agriculture—Implications for Health, Environment and Regulation”. Transgenic Res 28(3–4):419–463. https://doi.org/10.1007/s11248-019-00154-1
Garcia-Oliveira AL, Chander S, Ortiz R, Menkir A, Gedil M (2018) Genetic basis and breeding perspectives of grain iron and zinc enrichment in cereals. Front Plant Sci 9:937. https://doi.org/10.3389/fpls.2018.00937
Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17(3):282–286. https://doi.org/10.1038/7029
Gregorio G, Senadhira D, Htut T (1999) Improving iron and zinc value of rice for human nutrition. Agric Dév 23:77–81
Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta Biomembr 1465:190. https://doi.org/10.1016/S0005-2736(00)00138-3
HarvestPlus (n.d.). https://www.harvestplus.org/. Accessed 14 Jul 2020
Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119(2):471–479. https://doi.org/10.1104/pp.119.2.471
Hoa TTC, Lan NTP (2004) Effect of milling technology on iron content in rice grains of some leading varieties in the Mekong delta. Omonrice 12:38–44
Inoue H, Mizuno D, Nakanishi H, Mori S, Takahashi M, Nishizawa NK (2004) A rice FRD3-like (OsFRDL1) gene is expressed in the cells involved in long-distance transport. Soil Sci Plant Nutr 50:1133. https://doi.org/10.1080/00380768.2004.10408586
Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K et al (2009) Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 284:3470. https://doi.org/10.1074/jbc.M806042200
Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T et al (2006) Rice plants take up iron as an Fe 3+ -phytosiderophore and as Fe 2+. Plant J 45(3):335–346. https://doi.org/10.1111/j.1365-313X.2005.02624.x
Ishimaru Y, Masuda H, Bashir K, Inoue H, Tsukamoto T, Takahashi M et al (2010) Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J 62(3):379–390. https://doi.org/10.1111/j.1365-313X.2010.04158.x
James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N et al (2018) Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1789. https://doi.org/10.1016/S0140-6736(18)32279-7
Johnson AAT, Kyriacou B, Callahan DL, Carruthers L, Stangoulis J, Lombi E, Tester M (2011) Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS One 6(9):e24476. https://doi.org/10.1371/journal.pone.0024476
Khalekuzzaman M, Datta K, Oliva N, Alam MF, Joarder OI, Datta SK (2006) Stable integration, expression and inheritance of the ferritin gene in transgenic elite indica rice cultivar BR29 with enhanced iron level in the endosperm. Indian J Biotechnol 5:26
Kim SA, Guerinot ML (2007) Mining iron: iron uptake and transport in plants. FEBS Lett 581:2273. https://doi.org/10.1016/j.febslet.2007.04.043
Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63:131. https://doi.org/10.1146/annurev-arplant-042811-105522
Kobayashi T, Suzuki M, Inoue H, Itai RN, Takahashi M, Nakanishi H et al (2005) Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J Exp Bot 56:1305. https://doi.org/10.1093/jxb/eri131
Kobayashi T, Ozu A, Kobayashi S, An G, Jeon JS, Nishizawa NK (2019) OsbHLH058 and OsbHLH059 transcription factors positively regulate iron deficiency responses in rice. Plant Mol Biol 101:471. https://doi.org/10.1007/s11103-019-00917-8
Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39(3):415–424. https://doi.org/10.1111/j.1365-313X.2004.02146.x
Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D et al (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 24:4041. https://doi.org/10.1038/sj.emboj.7600864
Lee S, An G (2009) Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ 32:408. https://doi.org/10.1111/j.1365-3040.2009.01935.x
Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, An G (2009a) Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol 150:786. https://doi.org/10.1104/pp.109.135418
Lee S, Jeon US, Lee SJ, Kim Y-K, Persson DP, Husted S et al (2009b) Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci U S A 106(51):22014–22019. https://doi.org/10.1073/PNAS.0910950106
Lee S, Kim YS, Jeon US, Kim YK, Schjoerring JK, An G (2012) Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol Cell 33(3):269–275. https://doi.org/10.1007/s10059-012-2231-3
Lucca P, Hurrell R, Potrykus I (2002) Fighting iron deficiency anemia with iron-rich rice. J Am Coll Nutr 21:184S–190S. https://doi.org/10.1080/07315724.2002.10719264
Martínez C, Borrero J, Taboada R, Viana J, Neves P, Narvaez L et al (2010) Rice cultivars with enhanced iron and zinc content to improve human nutrition. In: Presented at the 28th International Rice Research Conference, 8–12 November 2010. Hanoi, Vietnam
Masuda H, Usuda K, Kobayashi T, Ishimaru Y, Kakei Y, Takahashi M et al (2009) Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2(4):155–166. https://doi.org/10.1007/s12284-009-9031-1
Masuda H, Ishimaru Y, Aung MS, Kobayashi T, Kakei Y, Takahashi M et al (2012) Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci Rep 2:543. https://doi.org/10.1038/srep00543
Masuda H, Kobayashi T, Ishimaru Y, Takahashi M, Aung MS, Nakanishi H et al (2013) Iron-biofortification in rice by the introduction of three barley genes participated in mugineic acid biosynthesis with soybean ferritin gene. Front Plant Sci 4:132. https://doi.org/10.3389/fpls.2013.00132
Mayer JE, Pfeiffer WH, Beyer P (2008) Biofortified crops to alleviate micronutrient malnutrition. Curr Opin Plant Biol 11:166. https://doi.org/10.1016/j.pbi.2008.01.007
Mumm RH (2013) A look at product development with genetically modified crops: examples from maize. J Agric Food Chem 61:8254. https://doi.org/10.1021/jf400685y
Nozoye T, Nagasaka S, Kobayashi T, Takahashi M, Sato Y, Sato Y et al (2011) Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J Biol Chem 286:5446. https://doi.org/10.1074/jbc.M110.180026
Nozoye T, Nagasaka S, Kobayashi T, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK (2015) The phytosiderophore efflux transporter TOM2 is involved in metal transport in rice. J Biol Chem 290(46):27688–27699. https://doi.org/10.1074/jbc.M114.635193
Ogo Y, Itai RN, Kobayashi T, Aung MS, Nakanishi H, Nishizawa NK (2011) OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol Biol 75:593-605. https://doi.org/10.1007/s11103-011-9752-6
Oliva N, Chadha-Mohanty P, Poletti S, Abrigo E, Atienza G, Torrizo L et al (2014) Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol Breed 33:23. https://doi.org/10.1007/s11032-013-9931-z
Paul S, Ali N, Gayen D, Datta SK, Datta K (2012) Molecular breeding of Osfer 2 gene to increase iron nutrition in rice grain. GM Crops Food 3(4):310–316. https://doi.org/10.4161/gmcr.22104
Paul S, Ali N, Datta SK, Datta K (2014) Development of an iron-enriched high-yielding indica rice cultivar by introgression of a high-iron trait from transgenic iron-biofortified rice. Plant Foods Hum Nutr 69:203. https://doi.org/10.1007/s11130-014-0431-z
Qu LQ, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222:225. https://doi.org/10.1007/s00425-005-1530-8
Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397:694. https://doi.org/10.1038/17800
Senoura T, Sakashita E, Kobayashi T, Takahashi M, Aung MS, Masuda H et al (2017) The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol Biol 95(4–5):375–387. https://doi.org/10.1007/s11103-017-0656-y
Singh SP, Gruissem W, Bhullar NK (2017) Single genetic locus improvement of iron, zinc and β-carotene content in rice grains. Sci Rep 7(1):6883. https://doi.org/10.1038/s41598-017-07198-5
Slamet-Loedin IH, Johnson-Beebout SE, Impa S, Tsakirpaloglou N (2015) Enriching rice with Zn and Fe while minimizing Cd risk. Front Plant Sci 6:1–9. https://doi.org/10.3389/fpls.2015.00121
Stoltzfus RJ (2001) Iron-deficiency anemia: reexamining the nature and magnitude of the public health problem. J Nutr 131:697S–700S
Suzuki M, Takahashi M, Tsukamoto T, Watanabe S, Matsuhashi S, Yazaki J et al (2006) Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barley. Plant J 48:85. https://doi.org/10.1111/j.1365-313X.2006.02853.x
Suzuki M, Morikawa KC, Nakanishi H, Takahashi M, Saigusa M, Mori S, Nishizawa NK (2008) Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil. Soil Sci Plant Nutr 54:77. https://doi.org/10.1111/j.1747-0765.2007.00205.x
Takahashi M, Yamaguchi H, Nakanishi H, Shioiri T, Nishizawa NK, Mori S (1999) Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (strategy II) in graminaceous plants. Plant Physiol 121:947. https://doi.org/10.1104/pp.121.3.947
Trijatmiko KR, Dueñas C, Tsakirpaloglou N, Torrizo L, Arines FM, Adeva C et al (2016) Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci Rep 6:19792. https://doi.org/10.1038/srep19792
Vasconcelos M, Datta K, Oliva N, Khalekuzzaman M, Torrizo L, Krishnan S et al (2003) Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci 164:371. https://doi.org/10.1016/S0168-9452(02)00421-1
Virk P, Barry G, Das A, Lee J, Tan J (2006) Research status of micronutrient rice development in Asia. In: Proceedings of the International Symposium on Rice Biofortification: Improving Human Health Through Biofortified Rice, 15 September 2006, Suwon, Korea, pp 123–148
Virk P, Barry G, Bouis H (2007) Genetic enhancement for the nutritional quality of rice. In: Genetic enhancement for the nutritional quality of rice. Proceedings of the 26th International Rice Research Conference, 9–12 October 2006. Macmillan India Ltd, New Delhi, pp 279–285
WHO (2002) The world health report 2002, vol 16. World Health Organization, Geneva
WHO/FAO (2004) Vitamin and mineral requirements in human nutrition, 2nd edn. WHO, Geneva. https://doi.org/10.1128/AAC.03728-14
Wirth J, Poletti S, Aeschlimann B, Yakandawala N, Drosse B, Osorio S et al (2009) Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol J 7:631. https://doi.org/10.1111/j.1467-7652.2009.00430.x
Wu TY, Gruissem W, Bhullar NK (2018) Facilitated citrate-dependent iron translocation increases rice endosperm iron and zinc concentrations. Plant Sci 270:13. https://doi.org/10.1016/j.plantsci.2018.02.002
Wu TY, Gruissem W, Bhullar NK (2019) Targeting intracellular transport combined with efficient uptake and storage significantly increases grain iron and zinc levels in rice. Plant Biotechnol J 17(1):9–20. https://doi.org/10.1111/pbi.12943
Yokosho K, Yamaji N, Ueno D, Mitani N, Jian FM (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol 149:297. https://doi.org/10.1104/pp.108.128132
Yoneyama T, Ishikawa S, Fujimaki S, Yoneyama T, Ishikawa S, Fujimaki S (2015) Route and regulation of zinc, cadmium, and iron transport in rice plants (Oryza sativa L.) during vegetative growth and grain filling: metal transporters, metal speciation, grain Cd reduction and Zn and Fe biofortification. Int J Mol Sci 16(8):19111–19129. https://doi.org/10.3390/ijms160819111
Zhang Y, Xu YH, Yi HY, Gong JM (2012) Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J 72:400. https://doi.org/10.1111/j.1365-313X.2012.05088.x
Zhang C, Shinwari KI, Luo L, Zheng L (2018) OsYSL13 is involved in iron distribution in rice. Int J Mol Sci 19:3537. https://doi.org/10.3390/ijms19113537
Zheng L, Cheng Z, Ai C, Jiang X, Bei X, Zheng Y et al (2010) Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS One 5(4):e10190. https://doi.org/10.1371/journal.pone.0010190
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this chapter
Cite this chapter
Matres, J.M., Arcillas, E., Cueto-Reaño, M.F., Sallan-Gonzales, R., Trijatmiko, K.R., Slamet-Loedin, I. (2021). Biofortification of Rice Grains for Increased Iron Content. In: Ali, J., Wani, S.H. (eds) Rice Improvement. Springer, Cham. https://doi.org/10.1007/978-3-030-66530-2_14
Download citation
DOI: https://doi.org/10.1007/978-3-030-66530-2_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66529-6
Online ISBN: 978-3-030-66530-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)