Abstract
Labeled data is the current bottleneck of medical image research. Substantial efforts are made to generate segmentation masks to characterize a given organ. The community ends up with multiple label maps of individual structures in different cases, not suitable for current multi-organ segmentation frameworks. Our objective is to leverage segmentations from multiple organs in different cases to generate a robust multi-organ deep learning segmentation network. We propose a modified cost-function that takes into account only the voxels labeled in the image, ignoring unlabeled structures. We evaluate the proposed methodology in the context of pectoralis muscle and subcutaneous fat segmentation on chest CT scans. Six different structures are segmented from an axial slice centered on the transversal aorta. We compare the performance of a network trained on 3,000 images where only one structure has been annotated (PUNet) against six UNets (one per structure) and a multi-class UNet trained on 500 completely annotated images, showing equivalence between the three methods (Dice coefficients of 0.909, 0.906 and 0.909 respectively). We further propose a modification of the architecture by adding convolutions to the skip connections (CUNet). When trained with partially labeled images, it outperforms statistically significantly the other three methods (Dice 0.916, \(p\!<\!0.0001\)). We, therefore, show that (a) when keeping the number of organ annotation constant, training with partially labeled images is equivalent to training with wholly labeled data and (b) adding convolutions in the skip connections improves performance.
This study was supported by the NHLBI awards R01HL116931, R01HL116473, and R21HL140422. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Segmentation of structures of interest is one of the main tasks of medical image analysis, serving as a prior step to biomarker quantification. Deep learning has been used to solve many segmentation problems [1] in images ranging from computed tomography [2] to MRI [3] or even in multi-modality images with the same network, [4] for one or multiple-organs [5].
Current deep-learning segmentation algorithms are trained on a dataset where the structures of interest are annotated, producing a complete mask per case. Every voxel is given a label, as being either a structure or background. This enables to optimize cost functions such as the normalized cross entropy or the dice coefficient [6, 7].
While this learning methodology has achieved great performance in single and multi-structure detection, it is not scalable to complete multi-organ segmentation, since it would require an extensive dataset where all the voxels are annotated. The expenses incurred in the generation of such dataset are beyond the scope of the effort that the community can afford. However, through the organization of challenges and public datasets, a great wealth of annotated cases with one or few structures of interest are currently available. What if we could leverage these single-organ databases for the generation of multiple-organ segmentation algorithms?
In this manuscript, we address this issue and propose a principled methodology to train a multi-class deep-learning segmentation algorithm from partially labeled datasets. The proposed method encodes the labels in a one-hot schema and optimizes the average per-structure dice coefficient. The proposed custom loss function adapts to the labels being provided. One of the most popular segmentation network architectures is the UNet [8], consisting of an encoding path, a decoding path and a set of skip connections [9]. We will, therefore, perform our experiments with UNet-based networks. We further such architecture by adding convolutions in the skip connections. Such is done to allow for flexibility between the information used in the encoding and decoding paths of the UNet. Such UNet, labeled CUNet, shows statistically significant improved performance over the baseline UNet.
We illustrate the proposed methodology in the problem of pectoralis and subcutaneous fat segmentation. Those structures have been shown to be of clinical relevance in different diseases like Chronic Obstructive Pulmonary Disease and Lung Cancer [10, 11]. Prior work has attempted to segment this structures using atlas-based techniques [12] and standard UNets [13].
This work is closely related to the work of [14], where the authors use few 2D annotated axial slices to train networks able to segment the whole 3D structures using a weighted softmax cost function. In their work, unlabeled voxels are given a zero weight and therefore do not contribute to the computation of the error. Our works differs from [14] in the sense that we use a weighted cost function on the per-structure dice score. Our proposed cost function penalizes pixels that are not assigned to the right structure, even if the precise right structure of such pixel is unknown.
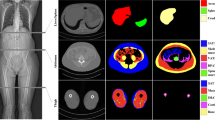
Left: Axial slice at the level of the transversal aorta zoomed at the pectoralis region. Middle: Reference standard. Right: Segmentation obtained with the proposed method. Color code: blue: right pectoralis major, green: right pectoralis minor, yellow: right subcutaneous fat, light blue: left pectoralis minor, magenta: left pectoralis minor, red: left subcutaneous fat. (Color figure online)
Schema of the proposed training methodology. The input to the network is an image, the segmentation mask where only one of the structures is segmented and the structure identifier. The output of the network is a segmentation of all the structures present on the image encoded in a one-hot schema. Each channel has information of only one structure or the background. Only the channel corresponding to the labeled structure is used to compute the loss metric. The structure of the network is, in this case, the proposed CUNet - a UNet with convolutions in the skip connections.
2 Materials and Methods
2.1 Data
CT scans were acquired from a large retrospective COPD observational study [15]. An expert identified the axial slice where pectoralis muscles were most visible at the level of the transversal aorta and segmented six different structures: left pectoralis major, left pectoralis minor, right pectoralis major, right pectoralis minor, left pectoralis subcutaneous fat and right pectoralis subcutaneous fat. The annotations were generated by applying intensity thresholds to the image and manually in-painting the structures of interest. Subcutaneous fat was defined as the layer of fat between lying between the margins of the major pectoralis muscle and the skin. Complete annotations (for the six structures) were generated for 2,000 cases, forming the completely annotated dataset. Partial annotations (only one structure per case) were generated for 3,000 cases, forming the partially annotated dataset.
2.2 Algorithm
Network:
The network structure of the proposed algorithm is the same as the UNet [8], but allowing for multi-class segmentation by adding a one-hot coding schema in the last layer, which has a softmax activation. We name such network a partial-UNet (PUNet). The output of the network is an image of the same dimensions as the original, but with N+1 channels, one per each of the N structures and an extra one for the background. We further modify such architecture by adding convolutions in the skip connections (CUNet). The schema of the modified network is depicted in Fig. 2. The input to the networks is the 512\(\,\times \,\)512 pixels CT axial slice, where the Hounsfield units (HU) have been clipped to the range \([-300, 500]\) and then normalized to the range \([-0.5, 0.5]\). The training set is formed by \(\{X_i, (Y_i, id_i)\}\), where \(X_i\) is the image, and \(Y_i\) is the segmentation mask associated with the structure identifier \(id_i\). The final per-pixel class is computed in a maximum likelihood fashion.
Cost Function: We use a cost-function that is the sum of the per-structure soft dice score for the structures that are present in the mini-batch. Thus, the loss function for a training point can be written as:
where \(\delta \) is a function equal to one if the structure is present in the masks of the minibatch and zero otherwise, dice stands for the Dice coefficient, \(\hat{Y}\) is the output of the softmax layer of the network, and n stands for the number of structures in the problem. Please note that \(\hat{Y}\) is a real-valued scored over all the voxels of the image. Therefore the cost function is an approximation of the real dice coefficient.
Baseline Algorithms: We compare the results of the UNet trained on with partial labels (PUNet) and the modified architecture trained with partial labels (CUNet) against (a) a multi-class u-net trained completely annotated images (UNet) using as cost function the per-class normalized dice score and (b) six per-organ u-nets (6xUNet) trained on the partially labeled dataset.
Training:
500 cases with complete annotations were used to train the baseline UNet, 3,000 cases with partial annotations were used to train the CUNet, the PUNet and six the per-structure UNets; 500 cases with complete annotations were used to validate the training, perform model selection and optimize meta-parameters and 1,000 cases with complete annotations were used only for testing and to report the results. We use the well-known ADAM optimizer to train the network with a learning rate fixed to 0.00005. The training is performed for a maximum of 30 epochs, and the validation loss is monitored. Training is stopped if the validation loss does not improve or decreases for five consecutive epochs.
2.3 Statistical Analysis
We use the Kruskal-Wallis statistical method to test if the per-method Dice score samples are coming from the same distributions. Upon rejection of the null hypothesis, we perform a non-parametric comparison for all pairs of methods using the Dunn method for joint ranking. Statistical analysis was performed with JMP Statistical Software (SAS Institute Inc.).
3 Results
The UNet trained with partial labels (PUNet) obtained a Dice score of 0.909, similar to that of the six per-class UNets (0.907) and the UNet trained with complete annotation (0.909). The modified architecture, (CUNet) achieved an overall average dice score of 0.916, improving over the other methods. The per-structure analysis can be found in Table 1. The Kruskal-Wallis test showed differences between the CUNet and the other methods for the average dice (\(p\!<\!0.0001\)). PUNet, 6xUNets and UNet average dice scores did not reach significance between them, indicating an equivalent behavior between such methods.
Figure 3 displays box-plots of the performance of the method per structure. There is an evident presence of outliers for all the structures. Some selected outliers are displayed in Fig. 4. We performed a post-hoc difference analysis between each method pair for each structure using the Dunn’s non-parametric test. The modified architecture, CUNet, mean dice score was greater than the traditional UNet for all structures analyzed (\(p\!<\!0.0001\)). The CUNet did not show significant differences with the 6xUNets for the left major and right minor pectoralis and performed worse for the left subcutaneous fat and left minor pectoralis structures (\(p\!<\!0.01\)). However, CUNet performed on average better than 6xUNets (\(p\!<\!0.0001\)).
Training time ranged from \(\approx 3\) min/epoch for the UNet trained with complete labels to \(\approx 18\) min/epoch for the other methods, since they need to circle through six times the number of raw training images. At test time, all methods analyzed an image in \(\approx 1s\), while the 6xUNet needed 6s. All times measured in a 1080Ti GPU.
Boxplots of the dice scores obtained with the different methdos. Left: all dice scores per method. Horizontal bars with stars denote statistical significance. Only the CUNet is statistically significantly different to the other methods. Right: per structure boxplot. Statsitical significance bars have been removed for clarity.
Some challenging cases of the database. Each column is a different case. From top to bottom: reference standard, jointly trained UNet, individually trained UNet, UNet trained with partial labels (PUNet) and the UNet with convolutions in the skip connections trained with partial labels (CUNet). We use the same color schema as in Fig. 1. (Color figure online)
Three extra segmentations of cases with moderate DICE score in at least one structure. The color conventions follows that of Fig. 4. Best viewed in color.
4 Discussion
We have presented a training methodology and a cost-function that enable the generation of multi-class deep learning segmentation algorithms from partially labeled images. We further the results by proposing a modification of the network architecture. Our method has shown improvement over a UNet trained on wholly annotated datasets and over six UNets trained for each organ individually, improving statistically significantly over the overall Dice score. The proposed CUNet improves the segmentation with respect to a traditional UNet when keeping the rest of parameters constant.
We have tested training with partially labeled datasets in the context of body composition measurements from axial images in CT scans. However, the proposed method is generalizable to any other context where multiple labels in different cases are present and could be used to train multi-organ segmentation method by leveraging single-class labeled data. In the current experiments, we are assuming that each image has only been labeled with a single organ. However, Eq. 1 could enable a variable number of classes to be present in each image. We have focused on 2D images. However, extensions to 3D are straightforward, for instance using a v-net instead of a u-net [14, 16].
The proposed method segments pectoralis and subcutaneous fat with high average dice coefficients, enabling its use for large cohort research. However, when presented with images with poor quality, cases with thin pectoralis or with dense breasts, the segmentation can be mislead, as shown in Fig. 5. Further analysis of such outliers, and an importance sampling strategy that over-represents such fringe cases, could be used to improve the performance of the algorithm.
We have chosen as cost function the average of the per-structure dice coefficient, which is independent of the size of the structure being segmented. This might pose problems with structures that are small or too difficult to segment. An extension of the proposed method would be to modulate the cost function with weights that take into account such structural properties. Such analysis is left for future work. We have trained with a balanced dataset, in the sense that each structure had the same number of annotated images in the partial database. Modifications of Eq. 1 and data augmentation strategies can be made to compensate for unbalanced datasets.
Deep learning segmentation methods have conquered most of single organ segmentation problems. The next challenge in medical image segmentation would be to segment complex images, such as CT scans entirely. With this work, we have demonstrated that we can create multi-organ segmentation algorithms from partially labeled datasets that are equivalent or better than algorithms trained with wholly labeled datasets. This could be extrapolated to the creation of multi-organ segmentation networks from the already existing per-organ segmentation databases.n.
References
Kayalibay, B., Jensen, G., van der Smagt, P.: CNN-based segmentation of medical imaging data. arXiv preprint arXiv:1701.03056 (2017)
Cai, J., Lu, L., Xie, Y., Xing, F., Yang, L.: Improving deep pancreas segmentation in CT and MRI images via recurrent neural contextual learning and direct loss function. arXiv preprint arXiv:1707.04912 (2017)
Fidon, L., et al.: Scalable multimodal convolutional networks for brain tumour segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 285–293. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_33
Drozdzal, M., Chartrand, G., Vorontsov, E.: Learning normalized inputs for iterative estimation in medical image segmentation. Med. Image Anal. 44, 1–13 (2018)
Roth, H.R., et al.: Hierarchical 3D fully convolutional networks for multi-organ segmentation. arXiv preprint arXiv:1704.06382 (2017)
Fidon, L., et al.: Generalised wasserstein dice score for imbalanced multi-class segmentation using holistic convolutional networks. arXiv preprint arXiv:1707.00478 (2017)
Sudre, C.H., Li, W., Vercauteren, T., Ourselin, S., Jorge Cardoso, M.: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Cardoso, M.J., et al. (eds.) DLMIA/ML-CDS -2017. LNCS, vol. 10553, pp. 240–248. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67558-9_28
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Drozdzal, M., Vorontsov, E., Chartrand, G., Kadoury, S., Pal, C.: The importance of skip connections in biomedical image segmentation. In: Carneiro, G., et al. (eds.) LABELS/DLMIA -2016. LNCS, vol. 10008, pp. 179–187. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46976-8_19
McDonald, M.L.N., et al.: Quantitative computed tomography measures of pectoralis muscle area and disease severity in chronic obstructive pulmonary disease. A cross-sectional study. Ann. Am. Thorac. Soc. 11(3), 326–334 (2014)
Kinsey, C.M., San Josée Estéepar, R., Van der Velden, J., Cole, B.F., Christiani, D.C., Washko, G.R.: Lower pectoralis muscle area is associated with a worse overall survival in non- small cell lung cancer. Cancer Epidemiol., Biomark. Prev.: Publ. Am. Assoc. Cancer Res., Cosponsored Am. Soc. Prev. Oncol. 26(1), 38–43 (2017)
Harmouche, R., Ross, J.C., Washko, G.R., San José Estépar, R.: Pectoralis muscle segmentation on CT images based on bayesian graph cuts with a subject-tailored atlas. In: Menze, B., et al. (eds.) MCV 2014. LNCS, vol. 8848, pp. 34–44. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-13972-2_4
Moreta-Martinez, R., Onieva-Onieva, J., Pascau, J., San Jose Estépar, R.: Pectoralis muscle and subcutaneous adipose tissue segmentation on CT images based on convolutional networks. In: Computer Assisted Radiology and Surgery. Springer (2017)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D u-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Regan, E.A., et al.: Genetic epidemiology of copd (copdgene) study design. COPD 7(1), 32–43 (2010)
Milletari, F., Navab, N., Ahmadi, S.A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
González, G., Washko, G.R., San José Estépar, R. (2018). Multi-structure Segmentation from Partially Labeled Datasets. Application to Body Composition Measurements on CT Scans. In: Stoyanov, D., et al. Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO BIA TIA 2018 2018 2018. Lecture Notes in Computer Science(), vol 11040. Springer, Cham. https://doi.org/10.1007/978-3-030-00946-5_22
Download citation
DOI: https://doi.org/10.1007/978-3-030-00946-5_22
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00945-8
Online ISBN: 978-3-030-00946-5
eBook Packages: Computer ScienceComputer Science (R0)