Abstract
Ensifer medicae strain WSM1115 forms effective nitrogen fixing symbioses with a range of annual Medicago species and is used in commercial inoculants in Australia. WSM1115 is an aerobic, motile, Gram-negative, non-spore-forming rod. It was isolated from a nodule recovered from the root of burr medic (Medicago polymorpha) collected on the Greek Island of Samothraki. WSM1115 has a broad host range for nodulation and N2 fixation capacity within the genus Medicago, although this does not extend to all medic species. WSM1115 is considered saprophytically competent in moderately acid soils (pH(CaCl2) 5.0), but it has failed to persist at field sites where soil salinity exceeded 10 ECe (dS/m). Here we describe the features of E. medicae strain WSM1115, together with genome sequence information and its annotation. The 6,861,065 bp high-quality-draft genome is arranged into 7 scaffolds of 28 contigs, contains 6,789 protein-coding genes and 83 RNA-only encoding genes, and is one of 100 rhizobial genomes sequenced as part of the DOE Joint Genome Institute 2010 Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria (GEBA-RNB) project.
Similar content being viewed by others
Introduction
The genus Medicago comprises 87 species of annual and perennial legumes, including some that were formerly recognized as Trigonella and Melilotus species [1]. A small number of annual Medicago species that have been domesticated are grown extensively in the sheep-wheat zone of southern Australia, particularly where pasture regeneration after a cropping phase is desirable. Annual Medicago species are grown on more than 20 M ha [2] and are particularly valued for their contribution to farming systems, in which Medicago fix around 25 kg of N per tonne of legume dry matter produced [3].
Medicago are nodulated by two species of root nodule bacteria (Ensifer medicae and Ensifer meliloti) that are recognized as being distinct based on their different nodulation and N2 fixation phenotypes in host interaction studies and more detailed analyses of their genetics [4,5].
Ensifer medicae strain WSM1115 is used in Australia to produce commercial peat cultures (referred to as Group AM inoculants) for the inoculation of several species of annual Medicago (predominantly M. truncatula, M. polymorpha, M. scutellata, M. sphaerocarpus, M. murex, M. rugosa and M. orbicularis). WSM1115 has been used commercially since 2002 [6], when it replaced strain WSM688. WSM1115 was isolated from a nodule from the roots of burr medic (Medicago polymorpha) collected by Prof. John Howieson (Murdoch University, Australia) on the island of Samothraki, Greece.
WSM1115 was selected for use in commercial inoculants having demonstrated good N2-fixation capacity with the relevant medic hosts and adequate saprophytic competence in moderately acidic soil (pH(CaCl2) 5).
Saprophytic competence in acidic soils is a requirement of strains used to inoculate Medicago because several species (M. murex, M. sphaerocarpus and M. polymorpha) are recommended and sown into soils below pH(CaCl2) 5.5, a level that is known to limit both survival of medic rhizobia and nodulation processes [7–10]. Useful variation in saprophytic competence occurs between strains of medic rhizobia [9] and valuable insights into the mechanisms that confer acidity tolerance have been provided by studies using strain WSM419 [11], which has been recently sequenced [12]. However, the complex nature of soil adaptation means that in-situ field studies still provide the most reliable means of selecting an inoculant strain and were used to select WSM1115 for commercial use. In a cross row experiment comparing 15 strains on acidic sand (pH(CaCl2) 5.0; Dowerin, West Australia), the nodulation of plants inoculated with WSM1115 was equal to or better than that of the other strains. This translated to better plant shoot weights, which were similar to those of plants inoculated with WSM688 (the incumbent inoculant strain at time of testing) and 48% greater when compared to former inoculant strain CC169 (J. G. Howieson unpublished data).
The nitrogen fixation capacity (effectiveness) of Medicago symbioses is characterized by strong interactions between the strain of rhizobia and species of Medicago [13–16]. Hence, the ability to form effective symbiosis with the species recommended for inoculation is an important consideration in inoculant strain selection. WSM1115 satisfies this requirement. In greenhouse tests it formed effective symbiosis with 16 genotypes of Medicago and overall produced 48% more shoot dry matter compared to plants inoculated with WSM688, the strain that it replaced (R.A. Ballard and N. Charman, unpublished data).
A limitation of strain WSM1115 is its poor persistence in moderately saline soils (e.g. where summer salinity levels exceed 10 ECe (dS/m)). Poor nodulation of regenerating pasture was first noted in 2004 during the field evaluation and domestication of the salt tolerant annual pasture legume messina (Melilotus siculus syn. Melilotus messanensis). Subsequent studies [17] confirmed that although WSM1115 was able to nodulate and form effective symbiosis with messina, it did not persist as well as other strains (e.g. SRDI554) through the summer months when salinity levels increased.
Here we present a preliminary description of the general features of Ensifer medicae strain WSM1115 together with its genome sequence and annotation.
Classification and features
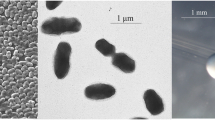
Ensifer medicae strain WSM1115 is a motile, non-sporulating, non-encapsulated, Gram-negative rod in the order Rhizobiales of the class Alphaproteobacteria. The rod-shaped form varies in size with dimensions of approximately 0.5 µm in width and 1.0 µm in length (Figure 1A). It is fast growing, forming colonies within 3–4 days when grown on TY [18] or half strength Lupin Agar (½LA) [19] at 28°C. Colonies on ½LA are opaque, slightly domed and moderately mucoid with smooth margins (Figure 1B).
Minimum Information about the Genome Sequence (MIGS) is provided in Table 1. Figure 2 shows the phylogenetic neighborhood of Ensifer medicae strain WSM1115 in a 16S rRNA gene sequence based tree. This strain has 100% sequence identity (1,366/1,366 bp) at the 16S rRNA sequence level to the fully sequenced Ensifer medicae strain WSM419 [12] and 99% 16S rRNA sequence (1362/1366 bp) identity to the fully sequenced E. meliloti Sm1021 [36].
Phylogenetic tree showing the relationship of Ensifer medicae WSM1115 (shown in bold print) to other Ensifer spp. in the order Rhizobiales based on aligned sequences of the 16S rRNA gene (1,290 bp internal region). All sites were informative and there were no gap-containing sites. Phylogenetic analyses were performed using MEGA, version 5 [33]. The tree was built using the Maximum-Likelihood method with the General Time Reversible model [34]. Bootstrap analysis [35] with 500 replicates was performed to assess the support of the clusters. Type strains are indicated with a superscript T. Brackets after the strain name contain a DNA database accession number and/or a GOLD ID (beginning with the prefix G) for a sequencing project registered in GOLD [32]. Published genomes are indicated with an asterisk.
Symbiotaxonomy
Ensifer medicae strain WSM1115 forms nodules (Nod+) and fixes N2 (Fix+) with a range of annual and perennial Medicago species and Melilotus species (Table 2). Levels of N2 fixation in combination with Medicago littoralis is suboptimal, that species generally forming more effective associations with strains of Ensifer meliloti including strain RRI128 [38]. The level of N2 fixation with Melilotus albus is also noted as positive, but has been observed to vary markedly with different plant accessions.
Genome sequencing and annotation information
Genome project history
This organism was selected for sequencing on the basis of its environmental and agricultural relevance to issues in global carbon cycling, alternative energy production, and biogeochemical importance, and is part of the Community Sequencing Program at the U.S. Department of Energy, Joint Genome Institute (JGI) for projects of relevance to agency missions. The genome project is deposited in the Genomes OnLine Database [32] and a high-quality-draft genome sequence in IMG/GEBA. Sequencing, finishing and annotation were performed by the JGI. A summary of the project information is shown in Table 3.
Growth conditions and DNA isolation
Ensifer medicae strain WSM1115 was cultured to mid logarithmic phase in 60 ml of TY rich medium on a gyratory shaker at 28°C [39]. DNA was isolated from the cells using a CTAB (Cetyl trimethyl ammonium bromide) bacterial genomic DNA isolation method [40].
Genome sequencing and assembly
The genome of Ensifer medicae strain WSM1115 was sequenced at the Joint Genome Institute (JGI) using Illumina [41] data. An Illumina standard paired-end library with a minimum insert size of 270 bp was used to generate 23,080,558 reads totaling 3,462 Mbp and an Illumina CLIP paired-end library with an average insert size of 9,584 + 2,493 bp was used to generate 2,163,668 reads totaling 324 Mbp of Illumina data (unpublished, Feng Chen).
All general aspects of library construction and sequencing performed at the JGI can be found at the JGI user home [40]. The initial draft assembly contained 57 contigs in 11 scaffolds. The initial draft data was assembled with Allpaths, version 38445, and the consensus was computationally shredded into 10 Kbp overlapping fake reads (shreds). The Illumina draft data was also assembled with Velvet, version 1.1.05 [42], and the consensus sequences were computationally shredded into 1.5 Kbp overlapping fake reads (shreds). The Illumina draft data was assembled again with Velvet using the shreds from the first Velvet assembly to guide the next assembly. The consensus from the second VELVET assembly was shredded into 1.5 Kbp overlapping fake reads. The fake reads from the Allpaths assembly and both Velvet assemblies and a subset of the Illumina CLIP paired-end reads were assembled using parallel phrap, version 4.24 (High Performance Software, LLC). Possible mis-assemblies were corrected with manual editing in Consed [43–45]. Gap closure was accomplished using repeat resolution software (Wei Gu, unpublished), and sequencing of bridging PCR fragments. The estimated total size of the genome is 6.9 Mbp and the final assembly is based on 3,654 Mbp of Illumina draft data, which provides an average 530× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [46] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [47]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. These data sources were combined to assert a product description for each predicted protein. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [48], RNAMMer [49], Rfam [50], TMHMM [51], and SignalP [52]. Additional gene prediction analyses and functional annotation were performed within the Integrated Microbial Genomes (IMG-ER) platform [53].
Genome properties
The genome is 6,861,065 nucleotides with 61.16% GC content (Table 4) and comprised of 7 scaffolds (Figures 3a,3b,3c,3d,3e,3f and Figure 3g) From a total of 6,872 genes, 6,789 were protein encoding and 83 RNA only encoding genes. The majority of genes (76.25%) were assigned a putative function whilst the remaining genes were annotated as hypothetical. The distribution of genes into COGs functional categories is presented in Table 5.
Graphical maps of SinmedDRAFT_Scaffold1.2 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical maps of SinmedDRAFT_Scaffold2.1 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical maps of SinmedDRAFT_Scaffold5.3 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical maps of SinmedDRAFT_Scaffold3.7 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical maps of SinmedDRAFT_Scaffold6.5 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical maps of SinmedDRAFT_Scaffold4.6 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
Graphical maps of SinmedDRAFT_Scaffold7.4 of the Ensifer medicae strain WSM1115 genome sequence. From bottom to the top of each scaffold: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew.
References
Small E. Alfalfa and Relatives: Evolution and Classification of Medicago. Ottawa, Canada: NRC Reserach Press; 2011.
Hill MJ, Donald GE. Australian Temperate Pastures Database National Pastures Improvement Coordinating Committee; 1998.
Peoples MB, Baldock JA. Nitrogen dynamics of pastures: nitrogen fixation inputs, the impact of legumes on soil nitrogen fertility, and the contributions of fixed nitrogen to Australian farming systems. Aust J Exp Agric 2001; 41:327–346. http://dx.doi.org/10.1071/EA99139
Rome S, Brunel B, Normand P, Fernandez M, Cleyet-Marel JC. Evidence that two genomic species of Rhizobium are associated with Medicago truncatula. Arch Microbiol 1996; 165:285–288. PubMed http://dx.doi.org/10.1007/s002030050328
Rome S, Fernandez MP, Brunel B, Normand P, Cleyet-Marel JC. Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int J Syst Bacteriol 1996; 46:972–980. PubMed http://dx.doi.org/10.1099/00207713-46-4-972
Bullard GK, Roughley RJ, Pulsford DJ. The legume inoculant industry and inoculant quality control in Australia: 1953–2003. Aust J Exp Agric 2005; 45:127–140. http://dx.doi.org/10.1071/EA03159
Brockwell J, Pilka A, Holliday RA. Soil pH is a major determinant of the numbers of naturally occurring Rhizobium meliloti in non-cultivated soils in central New South Wales. Aust J Exp Agric 1991; 31:211–219. http://dx.doi.org/10.1071/EA9910211
Denton MD, Hill CR, Bellotti WD, Coventry DR. Nodulation of Medicago truncatula and Medicago polymorpha in two pastures of contrasting soil pH and rhizobial populations. Appl Soil Ecol 2007; 35:441–448. http://dx.doi.org/10.1016/j.apsoil.2006.08.001
Howieson JG, Ewing MA. Acid tolerance in the Rhizobium meliloti-Medicago symbiosis. Aust J Agric Res 1986; 37:55–64. http://dx.doi.org/10.1071/AR9860055
Howieson JG, Robson AD, Abbott LK. Acid-tolerant species of Medicago produce root exudates at low pH which induce the expression of nodulation genes in Rhizobium meliloti. Aust J Plant Physiol 1992; 19:287–296. http://dx.doi.org/10.1071/PP9920287
Reeve WG, Brau L, Castelli J, Garau G, Sohlenkamp C, Geiger O, Dilworth MJ, Glenn AR, Howieson JG, Tiwari RP. The Sinorhizobium medicae WSM419 IpiA gene is transcriptionally activated by FsrR and required to enhance survival in lethal acid conditions. Microbiology 2006; 152:3049–3059. PubMed http://dx.doi.org/10.1099/mic0.28764-0
Reeve W, Chain P, O’Hara G, Ardley J, Nandesena K, Brau L, Tiwari R, Malfatti S, Kiss H, Lapidus A, et al. Complete genome sequence of the Medicago microsymbiont Ensifer (Sinorhizobium) medicae strain WSM419. Stand Genomic Sci 2010; 2:77–86. PubMed http://dx.doi.org/10.4056/sigs.43526
Brockwell J, Hely FW. Symbiotic characteristics of Rhizobium meliloti: an appraisal of the systematic treatment of nodulation and nitrogen fixation interactions between hosts and rhizobia of diverse origins. Australian Journal of Agricultural Economics 1966; 17:885–889.
Howieson JG, Nutt B, Evans P. Estimation of hoststrain compatibility for symbiotic N-fixation between Rhizobium meliloti, several annual species of Medicago and Medicago sativa. Plant Soil 2000; 219:49–55. http://dx.doi.org/10.1023/A:1004795617375
Interrante SM, Singh R, Islam MA, Stein JD, Young CA, Butler TJ. Effectiveness of Sinorhizobium inoculants on annual medics. Crop Sci 2011; 51:2249–2255. http://dx.doi.org/10.2135/cropsci2011.02.0076
Terpolilli JJ, O’Hara GW, Tiwari RP, Dilworth MJ, Howieson JG. The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytol 2008; 179:62–66. PubMed http://dx.doi.org/10.1111/j.1469-8137.2008.02464.x
Bonython AL, Ballard RA, Charman N, Nichols PGH, Craig AD. New strains of rhizobia that nodulate regenerating messina (Melilotus siculus) plants in saline soils. Crop Pasture Sci 2011; 62:427–436. http://dx.doi.org/10.1071/CP10402
Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 1974; 84:188–198. PubMed http://dx.doi.org/10.1099/00221287-84-1-188
Howieson JG, Ewing MA, D’antuono MF. Selection for acid tolerance in Rhizobium meliloti. Plant Soil 1988; 105:179–188. http://dx.doi.org/10.1007/BF02376781
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen M, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1.
Garrity GM, Bell JA, Lilburn TG. Class I. Alphaproteobacteria In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second ed. Volume 2: New York: Springer-Verlag; 2005, p. 1.
Validation List No. 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1–6. PubMed http://dx.doi.org/10.1099/ijs.0.64188-0
Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Kreig NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second ed: New York: Springer-Verlag; 2005. p 324.
Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225–420. http://dx.doi.org/10.1099/00207713-30-1-225
Conn HJ. Taxonomic relationships of certain non-sporeforming rods in soil. J Bacteriol 1938; 36:320–321.
Judicial Commission of the International Committee on Systematics of P. The genus name Sinorhizobium Chen et al. 1988 is a later synonym of Ensifer Casida 1982 and is not conserved over the latter genus name, and the species name ‘Sinorhizobium adhaerens’ is not validly published. Opinion 84. International Journal of Systematic and Evolutionary Microbiology 2008;58(Pt 8):1973.
Young JM. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination “Sinorhizobium adhaerens” (Casida 1982) Willems et al. 2003 legitimate? Request for an Opinion. Int J Syst Evol Microbiol 2003; 53:2107–2110. PubMed http://dx.doi.org/10.1099/ijs.0.02665-0
Casida LE. Ensifer adhaerens gen. nov., sp. nov.: a bacterial predator of bacteria in soil. Int J Syst Bacteriol 1982; 32:339–345. http://dx.doi.org/10.1099/00207713-32-3-339
Agents B. Technical rules for biological agents. TRBA (http://www.baua.de):466.
Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475–D479. PubMed http://dx.doi.org/10.1093/nar/gkm884
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731–2739. PubMed http://dx.doi.org/10.1093/molbev/msr121
Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39:783–791. http://dx.doi.org/10.2307/2408678
Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001; 293:668–672. PubMed http://dx.doi.org/10.1126/science.1060966
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Ballard RA, Slattery JF, Charman N. Host range and saprophytic competence of Sinorhizobium meliloti — a comparison of strains for the inoculation of lucerne, strand and disc medics. Aust J Exp Agric 2005; 45:209–216. http://dx.doi.org/10.1071/EA03126
Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, Howieson JG. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology 1999; 145:1307–1316. PubMed http://dx.doi.org/10.1099/13500872-145-6-1307
DOE Joint Genome Institute. http://my.jgi.doe.gov/general/index.html
Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433–438. PubMed http://dx.doi.org/10.1517/14622416.5.4.433
Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Current Protocols in Bioinformatics 2010;Chapter 11:Unit 11 5.
Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175–185. PubMed http://dx.doi.org/10.1101/gr.8.3.175
Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195–202. PubMed http://dx.doi.org/10.1101/gr.8.3.195
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed http://dx.doi.org/10.1186/1471-2105-11-119
Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455–457. PubMed http://dx.doi.org/10.1038/nmeth.1457
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964. PubMed
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100–3108. PubMed http://dx.doi.org/10.1093/nar/gkm160
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439–441. PubMed http://dx.doi.org/10.1093/nar/gkg006
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567–580. PubMed http://dx.doi.org/10.1006/jmbi.2000.4315
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783–795. PubMed http://dx.doi.org/10.1016/j.jmb.2004.05.028
Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271–2278. PubMed http://dx.doi.org/10.1093/bioinformatics/btp393
Acknowledgements
This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. We gratefully acknowledge the funding received from the Murdoch University Strategic Research Fund through the Crop and Plant Research Institute (CaPRI) and the Centre for Rhizobium Studies (CRS) at Murdoch University and the GRDC National Rhizobium Program (Project UMU63).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Reeve, W., Ballard, R., Howieson, J. et al. Genome sequence of Ensifer medicae strain WSM1115; an acid-tolerant Medicago-nodulating microsymbiont from Samothraki, Greece. Stand in Genomic Sci 9, 514–526 (2014). https://doi.org/10.4056/sigs.4938652
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.4938652