Abstract
Transforming growth factor (TGF)-β1 is a cytokine that participates in a broad range of cellular regulatory processes and is associated with various diseases including aortic aneurysm. Increased TGF-β1 levels are linked to Marfan syndrome (MFS) caused by fibrillin1 (FBN1) mutations and subsequent defects in signaling system. FBN1 single nucleotide polymorphisms (SNPs) rs2118181 and rs1059177 do not cause MFS but are associated with dilative pathology of aortic aneurysms (DPAAs). TGF-β1 and FBN1 SNPs rs2118181 and rs1059177 are potential biomarkers for early diagnosis of DPAA. We investigated the relationship between TGF-β1 levels in human blood plasma and FBN1 rs2118181 and rs1059177 in 269 individuals. The results showed a quantitative dependence of SNP genotype and TGF-β1 concentration. Presence of a single rs2118181 minor allele (G) increased the amount of TGF-β1 by roughly 1 ng/mL. Two copies of FBN1 rs1059177 minor allele (G) were required to have an additive effect on TGF-β1 levels. We found higher TGF-β1 concentrations in men compared with women (p = 0.001). A strong correlation between TGF-β1 levels and FBN1 SNPs suggests that a single nucleotide substitution in FBN1 sequence might reduce bioavailability or binding properties of fibrillin-1 and have an effect on TGF-β1 activation and cytokine concentration in blood plasma. By establishing the relationship between TGF-β1 and FBN1 SNPs rs2118181 and rs1059177, we provide evidence that their combination might be used as molecular biomarkers to identify patients at risk for sporadic ascending aortic aneurysm and aortic dissection.
Similar content being viewed by others
Introduction
FBN1 mutations are associated with the development of dilative pathology of ascending aorta (DPAA) (1). It has been shown that fibrillin-1 regulates the bioavailability of transforming growth factor (TGF)-β1 (2). TGF-β1 is a member of the TGF-β family of solubile proteins, cytokines, and, together with other TGF-β isoforms, works through the same signaling systems encompassing various cellular processes such as angiogenesis, proliferation, differentiation, apoptosis, wound healing and modification of extracellular matrix (3). Cellular and molecular mechanisms of aortic dilation have been investigated in the Marfan syndrome (MFS), where aortic aneurysm is one of the main features of a multisystem disorder. Increased TGF-β1 signaling has been associated with defective fibrillin-1 in the pathology of MFS (2). Genome-wide association study identified FBN1 SNPs rs2118181 and rs10519177 to be associated with sporadic DPAA (4). The data were in part replicated by two independent studies (5,6), therefore strengthening the idea that these SNPs contribute to the molecular pathways leading to the sporadic thoracic disease. To uncover the mechanism of sporadic DPAA and to identify potential biomarkers, we investigated the association between TGF-β1 concentration in blood plasma and FBN1 SNPs rs2118181 and rs10519177.
Materials and Methods
Study Subjects
A study group was recruited from three cluster samples of Kaunas population (n = 275). The first cluster consisted of student and staff volunteers from Lithuanian University of Health Sciences (LUHS) (n = 70, age range 18–55 years, mean 48, median 49); the second cluster was composed of volunteers (n = 151, age 55 years and older, mean 64, median 65) appointed to their physician in Dainavos outpatient’s clinic (Kaunas). FBN1 SNPs rs2118181 and rs10519177 genotype frequencies did not differ significantly between the two clusters. Because of the low number of homozygous minor allele carriers of FBN1 SNPs rs2118181 (n = 3) and rs10519177 (n = 14), a third cluster (n = 54, age range 46–72 years, mean 62, median 64) was added that consisted of homozygous minor allele carriers (n = 10, rs2118181, and n = 44, rs10519177) (Table 1). It was selected from a reference sample of the population (n = 840, age range 45–73 years, mean and median 60) screened within the international HAPIEE (Health, Alcohol and Psychosocial factors In Eastern Europe) study (7). None of the study subject reported having aortic dilation.
Ethical approval was obtained from the Ethics Committee of the Lithuanian Health Science University (number BE-2-12), and all subjects provided written informed consent for participation in the study.
Determination of Genotypes
Genomic DNA was isolated from potassium EDTA blood as described (8). Genotyping of FBN1 SNPs rs2118181 and rs10519177 was performed according to the manufacturer’s instructions by using commercially available kits from Applied Biosystems: C_16234705_10 (rs2118181), and C_8932690_10 (rs10519177) and ABI 7900HT real-time PCR Thermocycler (Applied Biosystems). All testing procedures were performed at the laboratory of Molecular Cardiology of the Institute of Cardiology of the Lithuanian University of Health Sciences.
TGF-β1 Quantitative Detection
Blood samples were obtained by using 3-mL Venosafe™ blood collection tubes containing 5.9 mg K2EDTA (Terumo Europe, Belgium). Plasma was obtained by centrifuging blood samples for 15 min at 3,506g within 2 h after phlebotomy. Plasma samples were aliquoted and stored at −20°C. All samples were tested in duplicate with an eBioscience Platinum human TGF-β1 ELISA commercially available kit (Bender Med Systems) based on standard sandwich enzyme-linked immunosorbent assay technology according to the manufacturers’ instructions. Absorbance was measured by using a Stat-fax 4200 microplate reader (Awareness Technology) at 450 nm OD (optical density). TGF-β1 concentration was automatically calculated by using a log-log linear regression. According to manufacturers’ instructions, cross-reactivity with TGF-β2 and TGF-β3 was <0.01%.
Statistical Analysis
A nonparametric Kruskal-Wallis test was used for statistical analysis. The relationship between age and cytokine levels was calculated by using a two-tailed Spearman rank correlation coefficient (R). Multiple linear regression was used to estimate the effect of gender (male), age (years) and genotype on the logarithmically transformed TGF-β1 values.
Results
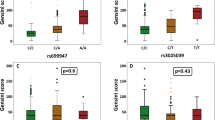
Our study population consisted of 141 male and 128 female subjects. Six samples were rejected from further TGF-β1 testing because of the preanalytical errors. TGF-β1 concentrations differed significantly (p = 0.001) between men (median 8.32 ng/mL, range 1.00–33.10) and women (median 5.81 ng/mL, range 1.10–27.30) and showed dependency on age (R = 0.236, p < 0.001). Genotype distribution and corresponding TGF-β1 values are presented in Table 2. Median TGF-β1 concentrations for each genotype showed quantitative association. The presence of one and two rs2118181 G alleles led to a 1.24 and 3.21 ng/mL increase in median TGF-β1 concentrations, respectively. A single copy of rs10519177 G allele did not seem to cause an increase in TGF-β1, but the presence of two minor allele copies coincided with a 4.08 ng/mL increase in TGF-β1 levels.
A multiple linear regression analysis was adopted to investigate an association between gender, age, FBN1 genotypes and TGF-β1 values (Table 3). Gender, age and rs2118181 AG + GG genotypes or a single-allele G presence were all significant variables to predict log-transformed TGF-β1 values (p < 0.05). An additive effect of G allele correlated to the observed TGF-β1 values in Table 2. We did the same testing for rs10519177 and found that the presence of GG genotype in addition to the gender and age was a significant variable for TGF-β1 values as well (p < 0.05).
Discussion
This is the first report on the association between previously defined FBN1 SNPs (rs2118181 and rs10519177) linked with sporadic thoracic aorta aneurysm and dissection (6) and TGF-β1 concentration in blood plasma of study subjects who were not aware of aortic dilation. Our data demonstrated that presence of rs2118181 minor allele significantly increased median TGF-β1 values, whereas such effect in rs10519177 was achieved only by homozygous carriers of two minor alleles. Moreover, these regularities correspond to the earlier reported associations between DPAA phenotypes and the genotyped FBN1 SNPs (6). According to these data, an additive minor allele model fitted best in describing the association between rs2118181 and aneurysm (odd ratio [OR] 1.70, 95% confidence interval [CI] 1.17–2.46) or Stanford A dissection (OR 2.64, 95% CI 1.66–4.19), whereas a recessive model described the association between rs10519177 and Stanford A dissection (OR 4.31, 95% CI 2.06–9.01).
Chaudhry et al. (2) demonstrated that specific FBN1 sequence encoded by exons 44–49 directly regulates endogenous TGF-β1 functionality without affecting cells or any other intermediate mechanisms, whereas abnormal fibrillin-1 with deficient functionality causes an excessive amount of active TGF-β1 to be released from the extracellular matrix (2). Mutations in FBN1 domains increase proteolytic sensitivity to inflammatory enzymes (9) causing a degradation of microfibrills and subsequent changes in TGF-β1 bioavailability. It is yet unknown how FBN1 SNPs (rs2118181, rs10519177) might affect the fibrillin-1 and TGF-β1 interaction. Both SNPs are in intronic regions of the FBN1 (10). Thus, it might be speculated that they alter FBN1 pre-mRNA thermodynamic and kinetic properties and affect splicing events. FBN1 splicing errors have been shown to cause protein alteration: (a) elimination of an entire exon resulting in a loss of an entire protein domain (11) and (b) activation of alternative splice sites causing an addition of 11 amino acids to the protein (12). It is obvious that the relationship between TGF-β1 and FBN1 SNPs is quantitative. Unaltered fibrillin-1 activates TGF-β1 through interactions between latent TGF-β binding proteins (LTBPs 1–4) and the N terminus of fibrillin-1 (13). If this interaction is somehow disturbed because of changes in fibrillin-1 availability, the amount of LTBPs would increase. In our experiments, we used acid activation to release TGF-β1 from the LTBP complex to measure total TGF-β1 level in blood plasma. Therefore, we do not know whether TGF-β1 increase is due to higher levels of LTBP or increased levels of active TGF-β1. More LTBPs would indicate defective fibrillin-1 interaction with LTBPs and reduced activation of TGF-β1. Higher levels of active TGF-β1 would indicate an increase in the activation process through LTBPs and fibrillin-1 interactions.
The pathogenesis of DPAA remains unclear. It is unknown if a fibrillin-LTBPs junction is needed to protect the large latent complex from proteolytic activation or whether FBN1 functions more directly in controlling assembly or stability of latent TGF-β1 complexes (14). Second, the balance between the need for TGF-β1 in normal development and suppression of its activity in the treatment of disease should be considered (15).
Conclusion
Our study for the first time demonstrates the association between TGF-β1 levels in blood plasma and FBN1 SNPs rs2118181 and rs10519177. We suggest that TGF-β1 levels in blood plasma in combination with FBN1 SNPs might serve as a biomarker to identify patients at risk for sporadic ascending aortic aneurysm and aortic dissection.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Milewicz DM, et al. (1996) Fibrillin-1 (FBN1) mutations in patients with thoracic aortic aneurysms. Circulation. 94:2708–11.
Chaudhry SS, et al. (2007) Fibrillin-1 regulates the bioavailability of TGFβ1. J. Cell Biol. 176:355–367.
Pardali E, Goumans MJ, ten Dijke P. (2010) Signaling by members of the TGF-β family in vascular morphogenesis and disease. Trends Cell Biol. 20:556–67.
LeMaire SA, et al. (2011) Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21. 1. Nat. Genet. 43:996–1000.
Iakoubova OA, et al. (2014) Genetic variants in FBN-1 and risk for thoracic aortic aneurysm and dissection. PLoS One. 17;9:e91437.
Lesauskaite V, et al. (2015) FBN1 polymorphisms in patients with the dilatative pathology of the ascending thoracic aorta. Eur. J. Cardiothorac. Surg. 47:e124–30.
Peasey A, et al. (2006) Determinants of cardiovascular and other non-communicable diseases in Central and Eastern Europe: rationale and design of the HAPIEE study. BMC Public Health. 6:255.
Lesauskaite V, et al. (2007) Matrix metalloproteinase-3 gene polymorphism and dilatative pathology of ascending thoracic aorta. Medicina (Kaunas). 44:386–91.
Ashworth JL, Kelly V, Wilson R, Shuttleworth CA, Kielty CM. (1999) Fibrillin assembly: dimer formation mediated by amino-terminal sequences. J. Cell Sci. 112:3549–3558.
dbSNP: Database for Short Genetic Variations [Internet]. (1998 —). Bethesda (MD): National Center for Biotechnology Information. [updated 2015 Nov 24; cited 2016 Jan 8]. Available from: https://doi.org/www.ncbi.nlm.nih.gov/SNP/
Liu W, Qian C, Comeau K, Brenn T, Furthmayr H, Francke U. (1996) Mutant fibrillm-1 monomers lacking EGF-like domains disrupt microfibril assembley and cause severe Marfan syndrome. Hum. Mol. Genet. 5:1581–1587.
Hutchinson S, Wordsworth BP, Handford PA. (2001) Marfan syndrome caused by a mutation in FBN1 that gives rise to cryptic splicing and a 33 nucleotide insertion in the coding sequence. Hum. Genet. 109:416–20.
Doyle JJ, Gerber EE, Dietaz HC. (2012) Matrix-dependent perturbation of TGFβ signalling in disease. FEBS Lett. 586:2003–15.
Pannu H, et al. (2005) Mutations in transforming growth factor-β receptor type II cause familial thoracic aortic aneurysms and dissections. Circulation. 112:513–20.
Zilberberg L, et al. (2012) Specificity of latent TGF-β binding protein (LTBP) incorporation into matrix: Role of fibrillins and fibronectin. J. Cell Physiol. 12:3828–36.
Acknowledgments
Financial support for the study was provided by the Research Council of Lithuania for National Research Programme “Chronic Noncommunicable Diseases” (Agreement No LIG 05/2012).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this license, visit (https://doi.org/creativecommons.org/licenses/by-nc-nd/4.0/)
About this article
Cite this article
Sepetiene, R., Patamsyte, V., Zukovas, G. et al. Association between Fibrillin1 Polymorphisms (rs2118181, rs10519177) and Transforming Growth Factor β1 Concentration in Human Plasma. Mol Med 21, 735–738 (2015). https://doi.org/10.2119/molmed.2015.00102
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2015.00102