Abstract
Complement activation resulting in significant increases of C4a split product may be a marker of postexertional malaise in individuals with chronic fatigue syndrome (CFS). This study focused on identification of the transcriptional control that may contribute to the increased C4a in CFS subjects after exercise. We used quantitative reverse-transcription polymerase chain reaction to evaluate differential expression of genes in the classical and lectin pathways in peripheral blood mononuclear cells (PBMCs). Calibrated expression values were normalized to the internal reference gene peptidylpropyl isomerase B (PPIB), the external reference gene ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL), or the geometric mean (GM) of the genes ribosomal protein, large, P0 (RPLP0) and phosphoglycerate kinase 1 (PGK1). All nine genes tested, except mannose-binding lectin 2 (MBL2), were expressed in PBMCs. At 1 hour postexercise, C4, mannan-binding lectin serine protease 2 (MASP2) and ficolin 1 (FCN1) transcripts were detected at higher levels (≥2-fold) in at least 50% (4 of 8) of CFS subjects and were detected in 88% (7 of 8) CFS subjects when subjects with overexpression of either C4 or MASP2were combined. Only an increase in the MASP2 transcript was statistically significant (PPIB, P = 0.001; GM, P = 0.047; rbcL, P = 0.045). This result may be due to the significant but transient downregulation of MASP2 in control subjects (PPIB, P = 0.023; rbcL, P = 0.027). By 6 hours postexercise, MASP2 expression was similar in both groups. In conclusion, lectin pathway responded to exercise differentially in CFS than in control subjects. MASP2 down-regulation may act as an antiinflammatory acute-phase response in healthy subjects, whereas its elevated level may account for increased C4a and inflammation-mediated postexertional malaise in CFS subjects.
Similar content being viewed by others
Introduction
Chronic fatigue syndrome (CFS) is a medically-unexplained illness identified by self-reported symptoms and exclusionary conditions. Identification of diagnostic markers for CFS remains challenging. Discovery of biomarkers may have been impeded by the fact that the unique biologic changes responsible for production of the original illness may no longer be present in most prevalent cases of CFS identified by various clinical case definitions (1,2). A consistently observed case-defining symptom in CFS subjects, however, is the exacerbation of symptoms following exercise (postexertional malaise), as opposed to the relief from symptoms following exercise in patients with other fatigue-associated conditions such as depression, rheumatoid arthritis, systemic lupus erthymatosus, and multiple sclerosis (3–6). In addition, postexertional malaise was one of the key symptoms in the Centers for Disease Control (CDC) symptom inventory list that differentiated subjects with CFS from those without the syndrome (7). In a principle component analysis to delineate heterogeneity in medically-unexplained fatiguing illness, postexertional malaise was the highest loading factor among a data set of 38 independent clinical and laboratory measurements (8). Thus, available evidence indicates postexertional malaise to be a unique and a major case-defining symptom, necessitating focused studies on its clinical and molecular characterization. Identification of specific biologic changes associated with postexertional malaise offers a promising approach for the discovery of biomarkers of CFS.
In patients with sickness behavior, clinical evaluation to identify infectious or inflammatory diseases indicated complement activation in subjects with CFS. Therefore we previously used an exercise paradigm to determine alterations in complement split products and found that exercise induced a significant increase of C4a, a putative anaphylatoxin, at 6 hours after exercise only in CFS subjects (9). Mean symptom scores and mean scores for reduced activity and mental fatigue categories of the Multidimensional Fatigue Inventory were also significantly increased in these CFS subjects following exercise. Two complement split products universally accepted as anaphylatoxins, C3a and C5a, were not elevated in the CFS subjects. A microarray study that included probes for 3800 genes and used total RNA from peripheral blood mononuclear cells (PBMCs) of these subjects also identified differences in complement activation between CFS and control subjects following exercise (10). Together, these results demonstrate that complement activation may be a marker of CFS-associated postexertional malaise, and that exercise-induced complement activation, particularly that which leads to the increased level of C4a split product, may be regulated at the transcriptional level.
C4a is generated from the cleavage of the native complement protein C4 via the classical and lectin pathways. In the classical pathway, C4 is cleaved by C1s activated by C1q, whereas in the lectin pathway, C4 is cleaved by mannan-binding lectin serine protease 2 (MASP2), activated by mannose-binding lectins (MBL) or ficolins (FCN). Some proinflammatory cytokines are known to modulate the synthesis of the above proteins both at the mRNA and protein levels. C1q mRNA and proteins are reported to be stimulated by interferon-γ (IFN-γ), IFN-α, IFN-β, and interleukin-6 (IL-6) (11–13). The secretion of the serine protease C1s can be enhanced by either IFN-α or IFN-γ (14). C4 expression both at the protein and mRNA levels can be regulated by IFN-γ, IFN-α, and IL-6 (15–19). On another level, increased C4 may be available for cleavage if C1-inhibitor (SERPING1), which removes the active enzymes C1s and MASP-2 from the respective complexes they form with C1q and FCN, becomes transcriptionally repressed. Increased levels of C4a can thus be hypothesized to result from any one or more transcriptional changes associated with increased amounts of the initiating proteins of the classical and/or lectin pathway, increased amounts of proinflammatory cytokines that may stimulate the production of the initiating proteins of the complement pathway, or repression in the inhibitory regulation of the classical or lectin pathways. On the basis of the above hypothesis, we examined changes in the expression of several genes in the complement pathway by real-time reverse-transcription polymerase chain reaction (real-time RT-PCR) as the first step in testing the molecular basis of altered complement metabolism following exercise.
Materials and Methods
Subjects and Blood Collection
The Centers for Disease Control Institutional Review Board, as required by the Department of Health and Human Services regulations, approved the study. This study used a subset of subjects (eight CFS patients and seven controls) from a previous case-control study that identified a significant increase of C4a split product at 6 h postexercise in CFS subjects (9). All subjects were recruited from the National Jewish Medical and Research Center (NJMRC), Denver, CO, USA. CFS subjects (mean age 37.5 ± 6.61 years; 87.5% females) were volunteers from the outpatient consultation clinic of NJMRC, whereas control subjects (mean age 28.3 ± 3.80 years; 43% females) were derived from the diagnostic complement laboratory of NJMRC. Except for one Asian subject in the control group, all study subjects were white. Although age and sex of the CFS and control subjects were matched in the previous C4a split product study (9), this matching was not retained in the present study. Furthermore, no attempt was made to stratify the data on the basis of age and sex because of the small sample size and a previous report of no difference in the activation of complement pathways associated with age or sex in healthy individuals (20). All subjects performed 20 min of a submaximal steady-state exercise on a stationary bicycle ergometer. Blood was collected from all subjects by venipuncture into tubes containing heparin immediately before exercise (T0), 1 h postexercise (T1), and 6 h postexercise (T2). PBMCs were separated from blood by use of Ficoll gradient and stored as pellets at −70°C until used for RNA extraction.
RNA Isolation and Quantification
Total RNA was isolated from PBMCs using RNAqueous kit (Ambion, Austin, TX, USA) according to the manufacturer’s recommendations, and stored at −70 °C until use. RNA quality and quantity were determined by using the RNA 6000 Nano assay with the Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). RNA quality was also assessed by denaturing agarose gel electrophoresis. All RNA samples were of good quality in terms of intactness of ribosomal RNA bands as determined by both the Bioanalyzer and agarose gel electrophoresis.
LightCycler-Based Real-Time RT-PCR
LightCycler-based real-time RT-PCR with SYBR Green I dye detection and melting-curve analysis for product specificity were used to determine the transcript levels.
cDNA synthesis. A 20-µL RT reaction was set up corresponding to each sample by using 0.5 µg of total RNA spiked with 1 pg (300 copies/cell) of plant mRNA, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) (Stratagene, LaJolla, CA, USA) to determine the efficiency of RT-PCR reactions in general, and as an external reference gene for normalization of gene expression. A singletube reaction method was used to treat total RNA with DNase I to remove traces of contaminating DNA (21). Total RNA containing plant mRNA spike was digested with 4 units of DNase I (Message-Clean Kit, GenHunter, Nashville, TN, USA) in 12.8 µL reaction that contained 1.8 µL 5× first-strand RT buffer (Clontech, Palo Alto, CA, USA). DNase I digestion was performed for 30 min at 37°C, and terminated at 72°C for 2 min followed by incubating on ice immediately. We removed 1 µL of DNase I—treated RNA, diluted 1:10 with water, and set it aside as a no-RT control template for PCR to determine the effectiveness of DNase I treatment. The remainder of the DNase I— treated RNA was supplied with 300 ng of random hexamers (Invitrogen, Carlsbad, CA, USA) and incubated at 72°C for 10 min and then placed on ice. The reaction was finally brought up to 20 µL with 2.2 µL of 5× first-strand buffer (Clontech), 2 µL of 100 mmol/L DTT, 2 µL of 10 mmol/L dNTP (Invitrogen), and 2 µL of PowerScript RT enzyme (Clontech). Each RT reaction was incubated at 42°C for 60 min and then terminated at 72°C for 15 min. To avoid degradation of RT product from repeated freezing and thawing, working stocks (1:5 dilutions of original RT product) were prepared in water, and stored at −20°C until used.
Determining LightCyler PCR conditions and setting up reactions. The optimal annealing and signal-acquisition temperatures for all gene-specific primers (Table 1) and product specificity were determined as described earlier (22) with a calibrator cDNA synthesized as above by using Universal Human Reference RNA (Stratagene) mixed with total RNA from PBMCs of healthy volunteers. These optimal conditions enabled the signal acquisition to be set 1°C–2°C below the melting temperatures of the specific product, minimizing signal interference from nonspecific products. PCR efficiency of each primer set was determined by using the optimal conditions and two-fold serial dilutions (range 0.25–16 ng/PCR) of calibrator cDNA (Table 1).
The LightCycler 2.0 Software version 4.0 (Roche, Indianapolis, IN, USA) was used to set up the reactions. First, 2 µL of DNA Master SYBR Green I mixture (containing Taq DNA polymerase, dNTP, MgCl2, and SYBR Green I dye; Roche) was incubated with 0.16 µL of TaqStart Antibody (Clontech) for 5 min at room temperature prior to the addition of primers and cDNA. Each final 20 µL reaction contained 2 µL of 1:2.5 dilution of cDNA working stock, 0.4 µmol/L of each primer, and 4 µmol/L MgCl2. The thermal cycling conditions were as described previously (22), but incorporated the optimized annealing and signal-acquisition temperatures for each gene-specific primer (Table 1). Effectiveness of DNase I treatment was tested for all cDNA synthesis with primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and no-RT control template.
Experimental design and statistical analysis. Determination of the fold differences in gene expression involved running 4 separate experiments: experiment 1 to generate standard curves for the target genes (run as absolute quantification) by use of calibrator cDNA, experiment 2 to generate standard curves for reference genes (run in absolute quantification) by use of calibrator cDNA, experiment 3 for target gene amplification in unknown samples along with calibrator cDNA (run in relative quantification, monocolor mode), experiment 4 for reference gene amplification in unknown samples along with calibrator cDNA (run in relative quantification, monocolor mode). Data from all four runs were imported to the relative quantification application (monocolor) of the LightCycler software version 4 to determine efficiency-corrected crossing point (Cp) and normalized ratio as represented by the formula below:
Normalized ratio = (Target gene concentration/reference gene concentration in unknown sample)/(Target gene concentration/reference gene concentration in calibrator).
To minimize LightCycler run-to-run variation, we derived a normalized ratio to a calibrator sample (Universal Human Reference RNA, Stratagene) as recommended to assure uniformity from multiple runs and intra- or interlaboratory comparisons (23). Fold-differences in gene expression between cases and controls were determined from their normalized ratios, as described above. We calculated fold differences with respect to normalized ratios derived from individual endogenous reference (peptidylpropyl isomerase B [PPIB]) or external reference (rbcL) genes or with the geometric mean (GM) of the normalized ratios of the reference genes ribosomal protein, large, P0 (RPLP0) and phosphoglycerate kinase 1 (PGK1) determined by using the g-Norm software. The rationales for these different normalization approaches, using PPIB (24), external spike (25,26), or GM (27) for real-time RT-PCR have been published elsewhere.
For the purpose of this study, we defined a gene as over- or underexpressed if 50% or more of CFS subjects showed a ≥two-fold change in expression compared with the median of the control group as determined by at least 2 of the 3 normalization methods. Furthermore, normalized ratios were log10 transformed prior to statistical analyses in StatView (Acton, MA, USA) to test significant differences (P values < 0.05) between CFS and control groups (by using an unpaired t test) or between different time points within each group (by using a paired t test).
Results
Reproducibility and Normalization of Real-Time RT-PCR
Use of optimized annealing and signal-acquisition temperatures for each gene-specific primer pair resulted in high PCR efficiency ranging from 1.95 to 2.00 (Table 1). No product-specific melting curve was observed with GAPDH primers and the no-RT control template, indicating completeness of DNase I digestion. We tested 14 gene-specific primers (9 target genes and 5 reference genes) with cDNA corresponding to 45 samples (44 unknown samples and 1 calibrator cDNA) in duplicate for a total of 1260 reactions (excluding water controls, and loss of 1 cDNA sample corresponding to 6-h postexercise from a CFS subject). Four runs of LightCycler reactions (24 reactions/run) were needed to complete one gene-specific assay with all samples. Mean Cp values with coefficient of variation (CV) for all reference genes (eukaryotic elongation factor 1 [EEF1A1], RPLP0, PGK1, PPIB, rbcL) tested in this study are given in Table 2. Mean Cp values (n = 88; mean 25.33 ± 0.66) of plant spike mRNA (rbcL) over all samples and PCR runs showed good reproducibility (CV, 2.61%) reflecting reproducibility in terms of efficiencies of both cDNA synthesis and PCR. The highest level of variation was observed with the reference gene EF1A1 (mean Cp value 17.79 ± 1.49; CV, 8.37%), whereas the reference gene PPIB showed moderate level of reproducibility (mean Cp value 23.15 ± 1.15; CV, 4.95%), which is in agreement with an earlier study with RNA from PBMCs (24). However, analysis of the reference gene data by g-Norm software selected RPLP0 and PGK1 as the least variable for the purpose of gene expression normalization. Because no single gene or approach is sufficient, we chose three normalization approaches to analyze gene expression data: (a) plant rbcL mRNA spike as external reference, (b) PPIB as the most stable internal reference gene, and (c) the GM of RPLPO and PGK1 by the g-Norm software.
Exercise-Responsive Complement Genes in CFS Subjects
MBL2 transcripts were undetectable by the LightCycler, thus this gene was excluded from further analysis. Of the remaining eight genes analyzed, only MASP2 and IFN-γ were overexpressed at the baseline (T0) according to results of two normalization methods (MASP2 by PPIB and rbcL, and IFN-γ by PPIB and GM), and with the use of fold-change in expression only three genes (C4, MASP2, and FCN1) were exercise responsive in CFS subjects at T1 by all normalization methods (Table 3). In all three normalization methods, C4 showed the highest percentage of CFS subjects (63%–75%; 5–6 of 8 subjects) with overexpression compared with MASP2 or FCN1 (50%–63%; 4–5 of 8 subjects). Furthermore, 88% (7 of 8) of CFS subjects showed ≥two-fold overexpression of either C4 or MASP2 at T1 by at least 2 of the 3 normalization methods.
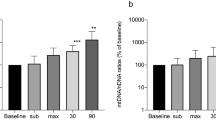
On further analysis, only differential expression in MASP2 was statistically significant between CFS and control groups at T1 for all three normalization methods (PPIB, P = 0.001; GM, P = 0.047; rbcL, P = 0.045) (Figure 1A–C). Differential expression of C4 at T1 was significant (P = 0.032) only with the data normalized by the PPIB (Figure 1D). These analysis results indicate (Figure 1A–D) that MASP2 and C4 in control healthy subjects were downregulated for at least 1 h postexercise, followed by increases in their transcript levels to the baseline by 6 h postexercise. This downregulation at T1 compared with T0 was significant only with MASP2 transcript levels normalized by PPIB (P = 0.023) and rbcL methods (P = 0.027), with borderline significance by GM normalization (P = 0.056). MASP2 transcript level at T2 did not differ between CFS and control subjects or compared with T0 and T1 levels in both CFS and control subjects, except that the increase from T1 and T2 in control subjects was significant when transcript levels were normalized by the rbCL method (P = 0.0143).
Changes in the expression of MASP2 and C4 in CFS and control (CON) healthy subjects at baseline (T0) and at 1 h (T1) and 6 h (T2) following exercise. (A) MASP2 expression level normalized by use of the PPIB method. (B) MASP2 expression normalized by geometric mean (GM) with g-Norm software. (C) MASP2 expression normalized by external plant spike, rbcL transcript. (D) C4 expression normalized by PPIB. P values in bold indicate significant expression differences in MASP2 and C4 between CFS and control subjects at T1. P values in italics indicate significant difference in MASP2 expression between different time points in response to exercise in control subjects.
Discussion
This study reports the first systematic evaluation of complement gene expression in PBMCs. Of nine complement genes tested, transcripts for all genes except MBL2 were detected in PBMCs, indicating that complement gene activity is not restricted to specialized cells such as hepatocytes. Lack of MBL2 expression in PBMCs agrees with previous reports, but MBL2 can be induced in vitro in adherent monocyte and monocyte-derived dendritic cells (28). Although expression for some genes (C1Q, C4, and FCN1) was detected in PBMCs as in earlier reports (29–31), MASP2 expression was not detected previously in PBMCs (28). Initiation of the classical pathway is antibody-dependent, but the lectin pathway is antibody-independent and begins with the recognition and binding of sugars or N-acetyl groups on pathogenic cells (pathogen-associated molecular patterns) by MBL or FCN. Based on the pattern of transcripts detected, genes belonging to both classical (C1QA, C1S) and lectin (ficolins and MASP2) pathways are expressed in PBMCs, with the lectin pathway activated by local synthesis of FCN rather than MBL2.
The major focus of this study was to determine if gene expression differences in the complement system would correlate with the previous observation of increased C4a split product in CFS subjects 6 hours postexercise. Initial analysis identified transcripts of C4, MASP2, and FCN1 at higher levels (≥two-fold) in at least 50% (4 of 8) of CFS subjects at 1 hour postexercise, and the percentage of CFS subjects responding to complement gene expression increased to 88% (7 of 8) when subjects with overexpression of either C4 or MASP2 were combined. Further analysis by t tests identified significant differences in the levels of MASP2 mRNA between CFS and control subjects at 1 hour postexercise. This difference in MASP2 mRNA appears to be due to a significant but transient downregulation in control subjects. By 6 hours postexercise, MASP2 expression was almost similar in both groups. In spite of the small sample size used in this study, this significant change in MASP2 expression at 1 hour postexercise was identified independently of the method used for normalization, indicating that the observed role of the MASP2-mediated lectin pathway in response to exercise is likely to be reproducible. It may be noted, however, that the reference genes (RPLPO and PGK1) used in the GM normalization method may be targets to some extent, as indicated by the apparent inversion of MASP2 expression in CFS compared with control subjects.
The current data do not directly address the activation of complement system during or following exercise. Whatever the trigger, the observed results may involve activation of monocytes, because they may be a common source of C4, MASP2, and FCN. Binding of FCN in conjunction with MASP2 to C4 initiates cleavage of C4, leading to the release of C4a (along with other components of the classical pathway). Both FCN1 (M-ficolin) and FCN2 (L-ficolin), located in chromosome 9q34 and separated by 22 kb, were detected in PBMCs, with FCN1 expressed at higher level in PBMCs than FCN2 and also differentially expressed between CFS and control subjects. Although FCN1 has been (until recently) solely associated with monocytes, neutrophils, and spleen cells and hypothesized to be a secretory protein (32), it has recently been found in serum in low concentrations (33). If released by monocytes, FCN1 could bind to a variety of its ligands, acetylated sugars, or proteins, which are abundant during or following exercise (34,35).
Previous reports of serum or plasma levels of complement activation products suggest that C activation takes place immediately with exhaustive exercise (36), but more slowly in the subjects in this study undergoing submaximal exercise (9). The trend of downregulation of expression with return to baseline levels in control subjects that precedes by several hours the activated proteins is in keeping with built-in regulatory processes inherent in the complement system. By contrast, expression of several complement genes remained at higher levels in CFS subjects before and after exercise, indicating a lack of acute-phase transcriptional response by these genes, which may lead to localized and uncontrollable inflammation-mediated tissue damage (37). The lectin pathway may also be activated by translocation of microbial products from the gut, a process that is postulated to occur during exercise (38) or other events such as infection, injury, or vaccine and trigger local inflammation and a variety of autoimmune diseases (39). Either situation may lead to increased C4a in CFS subjects compared with controls, and this C4a may have a regulatory role in inflammation by inhibiting monocyte chemotaxis (40) or through functions similar to C3a “activator and inhibitor” sequences as proposed by Erdei et al. (2004) (41). An antiinflammatory role would coincide with the downregulation of MASP2 expression in control subjects that occurred 1 hour postexercise.
Mechanisms regulating the expression of MASP2 are currently unknown, except for a report on a weak stimulatory effect by IL-1B that is abolished by IL-6 (42), and a putative role for the transcription factor Stat 3 in MASP2 expression (43). Our bioinformatics analysis of MASP2 promoter by MatInpsector revealed several transcription-factor binding sites, including two potential glucocorticoid-responsive elements (GRE) located within nucleotide positions −154 to −136 and between positions −989 to −971 (positions numbered with respect to transcription start site). A central role for the hypothalamic-pituitary-adrenal (HPA) axis in modulating postexertional malaise in CFS subjects in the context of hypocortisolism has been discussed in the literature (44–46) without mechanistic explanations. Steroid treatment studies report a reduction or a general downregulation of complement activation by glucocorticoids (47–49). A hypothesis based on the presence of GRE in MASP2 promoter and the antiinflammatory response of glucocorticoids in general is that in control subjects exercise induces cortisol secretion that inhibits MASP2 transcription through GRE, whereas at least in a subgroup of CFS subjects, exercise-induced cortisol secretion is below a threshold (hypocortisolism) required to inhibit MASP2 expression. Although cortisol was not measured in this study, other studies have reported significant increases in cortisol and its correlation with postexercise performance in healthy subjects (50–53). Accumulating evidence suggests that physical exercise, depending on the kind of exercise and subject characteristics, can act as a powerful modulator of the HPA axis and influence central nervous system functions in general (54,55). The complement system, either alone or interacting with proinflammatory cytokines, may be a link in the bidirectional communication between the HPA axis and immune functions that are likely to be altered in subjects with CFS (56).
Our findings should be generalized only in light of limitations of the study design. The previous study that reported the significant increase in C4a split product (9) and the current study examining the transcriptional changes in complement genes used subjects from the same recruitment. Age and sex of the CFS and control subjects were matched in the C4a split-product study, but this matching was not retained in the present study. We examined gene expression changes in PBMCs, a noninvasive source of sample for biomarker discovery and validation. PBMCs, however, are mixtures of lymphocytes and monocytes, and as such the findings do not reflect cell-type-specific responses. Furthermore, whether the observed transcriptional changes in complement genes are reflected in their plasma protein levels remains to be investigated.
In conclusion, this study detected expression of both classical and lectin pathways in PBMCs of normal healthy and CFS subjects, but transcripts for components of the lectin pathway (C4 and MASP2) were observed at higher levels in CFS subjects 1 hour postexercise. Higher expression of C4 and MASP2 may contribute to the increased C4a split product in CFS subjects 6 hours postexercise. MASP2 expression was significantly downregulated in control subjects 1 hour postexercise, and this downregulation may be mediated by the antiinflammatory effect of cortisol in response to exercise. Further studies are needed to replicate the differential expression of complement genes and its potential link with inflammation and cortisol secretion in response to exercise.
Disclosure
We declare that the authors have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
References
Fukuda K, et al. (1994) The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann. Intern. Med. 121:953–59.
Reeves WC, et al. (2005) Chronic fatigue syndrome— a clinically empirical approach to its definition and study. BMC. Med. 3:19.
Dunn AL, Trivedi MH, Kampert JB, Clark CG, Chambliss HO. (2005) Exercise treatment for depression: efficacy and dose response. Am. J. Prev. Med. 28:1–8.
Bearne LM, Scott DL, Hurley MV. (2002) Exercise can reverse quadriceps sensorimotor dysfunction that is associated with rheumatoid arthritis without exacerbating disease activity. Rheumatology (Oxford) 41:157–66.
Robb-Nicholson LC, et al. (1989) Effects of aerobic conditioning in lupus fatigue: a pilot study. Br. J. Rheumatol. 28:500–5.
Mostert S, Kesselring J. (2002) Effects of a short-term exercise training program on aerobic fitness, fatigue, health perception and activity level of subjects with multiple sclerosis. Mult. Scler. 8:161–8.
Wagner D, et al. (2005) Psychometric properties of the CDC Symptom Inventory for assessment of chronic fatigue syndrome. Popul. Health Metr. 3:8.
Vollmer-Conna U, Aslakson E, White PD. (2006) An empirical delineation of the heterogeneity of chronic unexplained fatigue in women. Pharmacogenomics 7:355–64.
Sorensen B, et al. (2003) Complement activation in a model of chronic fatigue syndrome. J. Allergy Clin. Immunol. 112:397–403.
Whistler T, Jones JF, Unger ER, Vernon SD. (2005) Exercise responsive genes measured in peripheral blood of women with chronic fatigue syndrome and matched control subjects. BMC. Physiol. 5:5.
Gulati P, Lemercier C, Guc D, Lappin D, Whaley K. (1993) Regulation of the synthesis of C1 subcomponents and C1-inhibitor. Behring Inst. Mitt. 196–203.
Kolosov M, Kolosova I, Zhou A, Leu RW. (1996) Autocrine induction of macrophage synthesis of complement subcomponent C1q by endogenous interferon-alpha/beta. J. Interferon Cytokine Res. 16:209–15.
Walker DG. (1998) Expression and regulation of complement C1q by human THP-1-derived macrophages. Mol. Chem. Neuropathol. 34:197–218.
Drouet C, Reboul A. (1989) Biosynthesis of C1r and C1s subcomponents. Behring Inst. Mitt. 84:80–8.
Collins T, Winkelstein JA, Sullivan KE. (1996) Regulation of early complement components C3 and C4 in the synovium. Clin. Diagn. Lab. Immunol. 3:5–9.
Kulics J, Circolo A, Strunk RC, Colten HR. (1994) Regulation of synthesis of complement protein C4 in human fibroblasts: cell- and gene-specific effects of cytokines and lipopolysaccharide. Immunology 82:509–15.
Sacks S, Zhou W, Campbell RD, Martin J. (1993) C3 and C4 gene expression and interferongamma-mediated regulation in human glomerular mesangial cells. Clin. Exp. Immunol. 93:411–7.
Seelen MA, Brooimans RA, van der Woude FJ, van Es LA, Daha MR. (1993) IFN-gamma mediates stimulation of complement C4 biosynthesis in human proximal tubular epithelial cells. Kidney Int. 44:50–7.
Vincent F, et al. (1993) Synthesis and regulation of complement components by human monocytes/macrophages and by acute monocytic leukemia. DNA Cell Biol. 12:415–23.
Zimmermann-Nielsen E, Svehag SE, Thorlacius-Ussing O, Baatrup G. (2001) ELISA for evaluating the incorporation of plasma derived complement split-products C3b/iC3b into solid-phase immune complexes. J. Immunol. Methods 249:43–51
Huang Z, Fasco MJ, Kaminsky LS. (1996) Optimization of Dnase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. Biotechniques 20:1012–20.
Rajeevan MS, Ranamukhaarachchi DG, Vernon SD, Unger ER. (2001) Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25:443–51.
Skrzypski M. (2008) Quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR) in translational oncology: lung cancer perspective. Lung Cancer 59:147–54.
Pachot A, Blond JL, Mougin B, Miossec P. (2004) Peptidylpropyl isomerase B (PPIB): a suitable reference gene for mRNA quantification in peripheral whole blood. J. Biotechnol. 114:121–4.
Bower NI, Moser RJ, Hill JR, Lehnert SA. (2007) Universal reference method for real-time PCR gene expression analysis of preimplantation embryos. Biotechniques 42:199–206.
Gilsbach R, Kouta M, Bonisch H, Bruss M. (2006) Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. Biotechniques 40:173–7.
Vandesompele J, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1–11.
Seyfarth J, Garred P, Madsen HO. (2006) Extrahepatic transcription of the human mannose-binding lectin gene (mbl2) and the MBL-associated serine protease 1–3 genes. Mol. Immunol. 43:962–71.
Fan Q, Weill B, Delpech M. (1994) Study of C4A mRNA in mononuclear blood cells from a patient with SLE and C4A homozygous deficiency without C4A gene deletion. Clin. Exp. Rheumatol. 12:657–60.
Lu J, Le Y, Kon OL, Chan J, Lee SH. (1996) Biosynthesis of human ficolin, an Escherichia coli-binding protein, by monocytes: comparison with the synthesis of two macrophage-specific proteins, C1q and the mannose receptor. Immunology 89:289–94.
Moosig F, et al. (2006) Reduced expression of C1q-mRNA in monocytes from patients with systemic lupus erythematosus. Clin. Exp. Immunol. 146:409–16.
Endo Y, Matsushita M, Fujita T. (2007) Role of ficolin in innate immunity and its molecular basis. Immunobiology 212:371–9.
Honore C, et al. (2008) The innate pattern recognition molecule Ficolin-1 is secreted by monocytes/macrophages and is circulating in human plasma. Mol. Immunol. 45:2782–9.
Watt MJ, Heigenhauser GJ, Dyck DJ, Spriet LL. (2002) Intramuscular triacylglycerol, glycogen and acetyl group metabolism during 4 h of moderate exercise in man. J. Physiol. 541:969–78.
White LJ, et al. (2006) Accumulation of acetyl groups following cycling: a 1H-MR spectroscopy study. Int. J. Sports Med. 27:100–4.
Dufaux B, Order U. (1989) Complement activation after prolonged exercise. Clin. Chim. Acta. 179:45–9.
Markiewski MM, Lambris JD. (2007) The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am. J. Pathol. 171:715–27.
Shek PN, Shephard RJ. (1998) Physical exercise as a human model of limited inflammatory response. Can. J. Physiol. Pharmacol. 76:589–97.
Nancy AL, Shoenfeld Y. (2008) Chronic fatigue syndrome with autoantibodies: the result of an augmented adjuvant effect of hepatitis-B vaccine and silicone implant. Autoimmun. Rev. 8:52–5.
Tsuruta T, et al. (1993) Novel function of C4a anaphylatoxin. Release from monocytes of protein which inhibits monocyte chemotaxis. Am. J. Pathol. 142:1848–57.
Erdei A, Andrasfalvy M, Peterfy H, Toth G, Pecht I. (2004) Regulation of mast cell activation by complement-derived peptides. Immunol. Lett. 92:39–42.
Endo Y, et al. (2002) Functional characterization of human mannose-binding lectin-associated serine protease (MASP)-1/3 and MASP-2 promoters, and comparison with the C1s promoter. Int. Immunol. 14:1193–201.
Unterberger C, et al. (2007) Stat3 is involved in control of MASP2 gene expression. Biochem. Biophys. Res. Commun. 364:1022–5.
Baschetti R. (2005) Chronic fatigue syndrome, exercise, cortisol and lymphadenopathy. J. Intern. Med. 258:291–2.
Glass JM, et al. (2004) The effect of brief exercise cessation on pain, fatigue, and mood symptom development in healthy, fit individuals. J. Psychosom. Res. 57:391–8.
Van Den EF, Moorkens G, Van HB, Cosyns P, Claes SJ. (2007) Hypothalamic-pituitary-adrenal axis function in chronic fatigue syndrome. Neuropsychobiology 55:112–20.
Ernst E, Espersen GT, Andersen MV. (1990) Reduction of complement activation in rheumatoid arthritis by steroid treatment. Int. J. Tissue React. 12:77–9.
Packard BD, Weiler JM. (1983) Steroids inhibit activation of the alternative-amplification pathway of complement. Infect. Immun. 40:1011–4.
Zimmermann-Nielsen E, Gronbaek H, Dahlerup JF, Baatrup G, Thorlacius-Ussing O. (2005) Complement activation capacity in plasma before and during high-dose prednisolone treatment and tapering in exacerbations of Crohn’s disease and ulcerative colitis. BMC. Gastroenterol. 5:31.
Li TL, Cheng PY. (2007) Alterations of immunoendocrine responses during the recovery period after acute prolonged cycling. Eur. J. Appl. Physiol. 101:539–46.
Minetto MA, et al. (2007) Corticotropin axis sensitivity after exercise: comparison between elite athletes and sedentary subjects. J. Endocrinol. Invest. 30:215–23.
Rojas VS, et al. (2006) Acute BDNF and cortisol response to low intensity exercise and following ramp incremental exercise to exhaustion in humans. Brain Res. 1121:59–65.
Minetto MA, et al. (2008) Changes in awakening cortisol response and midnight salivary cortisol are sensitive markers of strenuous training-induced fatigue. J. Endocrinol. Invest. 31:16–24.
Leal-Cerro A, et al. (2003) Mechanisms underlying the neuroendocrine response to physical exercise. J. Endocrinol. Invest. 26:879–85.
Dishman RK, et al. (2006) Neurobiology of exercise. Obesity (Silver Spring) 14:345–56.
Lorusso L, et al. (2008) Immunological aspects of chronic fatigue syndrome. Autoimmun. Rev. 2008, Sep 15. [Epub ahead of print].
Acknowledgments
Support for B Sorensen was provided by the research participation program at the Centers for Disease Control and Prevention (CDC), National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Division of Viral and Rickettsial Diseases, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the CDC. The authors wish to acknowledge the laboratory support of I Dimulescu, bioinformatics support of V Falkenberg, and the valuable comments and critical reading of the manuscript by ER Unger.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sorensen, B., Jones, J.F., Vernon, S.D. et al. Transcriptional Control of Complement Activation in an Exercise Model of Chronic Fatigue Syndrome. Mol Med 15, 34–42 (2009). https://doi.org/10.2119/molmed.2008.00098
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2008.00098