Abstract
Background
An intraductal papillary mucinous neoplasm (IPMN) is a pancreatic tumor with malignant potential. Although we anticipate a sensitive method to diagnose the malignant conversion of IPMN, an effective strategy has not yet been established. The combination of probe electrospray ionization-mass spectrometry (PESI-MS) and machine learning provides a promising solution for this purpose.
Methods
We prospectively analyzed 42 serum samples obtained from IPMN patients who underwent pancreatic resection between 2020 and 2021. Based on the postoperative pathological diagnosis, patients were classified into two groups: IPMN-low grade dysplasia (n = 17) and advanced-IPMN (n = 25). Serum samples were analyzed by PESI-MS, and the obtained mass spectral data were converted into continuous variables. These variables were used to discriminate advanced-IPMN from IPMN-low grade dysplasia by partial least square regression or support vector machine analysis. The areas under receiver operating characteristics curves were obtained to visualize the difference between the two groups.
Results
Partial least square regression successfully discriminated the two disease classes. From another standpoint, we selected 130 parameters from the entire dataset by PESI-MS, which were fed into the support vector machine. The diagnostic accuracy was 88.1%, and the area under the receiver operating characteristics curve was 0.924 by this method. Approximately 10 min were required to perform each method.
Conclusion
PESI-MS combined with machine learning is an easy-to-use tool with the advantage of rapid on-site analysis. Here, we show the great potential of our system to diagnose the malignant conversion of IPMN, which would be a promising diagnostic tool in clinical settings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
An intraductal papillary mucinous neoplasm (IPMN) of the pancreas is characterized by the production of mucin with a dilated pancreatic duct.1,2,3 This category of pancreatic tumor was first conceptualized in the 2nd section of the World Health Organization classification in 1996, which made clinicians aware of IPMN as a differential diagnosis. Since then, the number of IPMN cases has increased because of the enhanced accuracy of imaging technology and recognition of the disease.4,5
An intraductal papillary mucinous neoplasm (IPMN) of the pancreas is characterized by the production of mucin with a dilated pancreatic duct.1,2,3 This category of pancreatic tumor was first conceptualized in the 2nd section of the World Health Organization classification in 1996, which made clinicians aware of IPMN as a differential diagnosis. Since then, the number of IPMN cases has increased because of the enhanced accuracy of imaging technology and recognition of the disease.4,5
IPMN is pathologically divided into three classes: low-grade dysplasia (IPMN-LGD), high-grade dysplasia (IPMN-HGD), and invasive carcinoma (IPMN-IC).6,7,8 IPMN-HGD is considered potentially malignant and is therefore treated as an indication for surgery, similar to invasive carcinoma.9 The international Fukuoka criteria coined words, such as “high-risk stigmata (HRS)” and “worrisome feature (WF),” to discern IPMN-HGD from IPMN-LGD.10 However, there are no definitive methods to predict their properties more accurately, and the latest method achieves a diagnostic accuracy of no more than 63–76%.11,12,13
Recently, liquid biopsy has received great attention worldwide because of its relatively higher sensitivity in detecting the early phase of malignant conversion. Serological analysis by conventional mass spectrometry still contributes to cancer screening, but because of its inherent drawbacks in procedures, such as low throughput analysis, complicated sample preparation, and chromatographic separation, these techniques play a supporting role. Probe electrospray ionization-mass spectrometry (PESI-MS) is a derivative of ESI that uses a unique needle ion emitter without a troublesome capillary.14 We took advantage of PESI-MS to develop a medical device that is useful for cancer diagnosis.14,15 Based on this perspective, we applied this to various malignancies, such as head and neck, stomach, liver, and breast cancer.16,17,18,19
The aim of this study was to verify the diagnostic power of this system to accurately predict IPMN subtypes by detecting tumor-specific serological changes.
Materials and Methods
Patient Selection and Data Collection
This study was conducted in accordance with the ethical standards of the Declaration of Helsinki, and the protocol was approved by the ethics committees of the University of Tokyo and the University of Yamanashi [approval numbers: 2019370NI-(2) and 2086]. Each patient provided written informed consent before participating in this study, and all clinicopathological data were anonymized.
We examined 42 IPMN patients who did not have histologically proven cancer before resection, and who underwent pancreatic resection between February 2020 and November 2021 at the University of Tokyo Hospital and the University of Yamanashi Hospital. Preoperative tumor evaluation was basically performed by endoscopic ultrasonography. Endoscopic retrograde cholangiopancreatography was added as needed. We regularly assessed the diameter of the cyst and the presence of an enhanced mural nodule by endoscopic ultrasonography, but we did not always perform aspiration for the cyst. If endoscopic retrograde cholangiopancreatography was performed, pancreatic juice cytology was simultaneously performed. Surgical indications were as follows: main-duct type IPMN, combined-duct type IPMN, branched-duct type IPMN with high-risk stigmata, and malignant suspicious branched-duct type IPMN with worrisome features.10 Clinicopathological data from patients were acquired from a prospectively maintained database. Pathological findings were evaluated based on IPMN international guidelines.10 We defined IPMN-HGD and IPMN-IC as advanced IPMN (Ad-IPMN) and compared IPMN-LGD and Ad-IPMN.
Sample Collection and Preparation
The blood draw was performed just prior to the surgery. Venous blood samples from 42 IPMN patients were preoperatively infused into thrombin-containing tubes by experienced phlebotomists. After the tube remained upright for 15–60 min at room temperature, it was centrifuged at 3000 rpm for 5 min. Then, the supernatant was collected and frozen at −80 °C until PESI-MS analysis.
Ten microliters of dissolved serum samples were added to 390 µl 50% ethanol and stirred using a vortex mixer (Scientific Industries, Inc.) at 10 °C for 5 min. After being placed on ice for 5 min, the sample was centrifuged at 15,000 rpm for 10 min at 4 °C. Nine microliters of supernatant were added to a sample plate (Shimadzu, Corp.)
PESI-MS Analysis and Data Processing
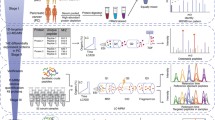
A Direct Probe Ionization-Mass Spectrometer-8060 (DPiMS, Shimadzu, Corp.) was used for the analysis. The detailed method of operation of this instrument has been described in previous reports.18,19,20,21 The ion intensity was obtained in positive ion mode using this mass spectrometer and analyzed by LabSolutions software (Shimadzu, Corp.). The mass spectrum was visualized by m/z (the mass to charge ratio) in the x-axis and ion intensity (A.U.) in the y-axis. To obtain the peak value defined as the “peak intensity,” the ion-intensity value was added to the maximum ion intensity within one m/z and the values before and after it. The corresponding m/z was rounded down to an integer bin. An example of data processing is shown in Supplementary Fig. 1. Bins with a peak intensity less than 500,000 A.U. were excluded from the analysis. This value almost corresponded to the quartile of IPMN data and corresponded to the maximum data of the solvent-only sample (Supplementary Fig. 2). With the above processing, 328 out of 1191 peak intensities were excluded, and the remaining 863 were used for the database for the support vector machine (SVM) analysis. This flowchart is described in Fig. 1.
The patient flowchart for this study. A total of 1191 peak intensities were obtained from each patient. Among them, 328 peak intensities with values influenced by electronic noise were excluded. The remaining 863 peak intensities were analyzed. IPMN, intraductal papillary mucinous neoplasm; LGD, low-grade dysplasia; Ad, Advanced
Statistical Analysis
Continuous clinicopathological variables were expressed as the median and range. The data were analyzed using the Mann–Whitney U test and Fisher exact test or Pearson’s chi-square test, depending on data characteristics.
Diagnostic accuracy was analyzed using machine learning as follows. First, partial least square (PLS) regression was performed for all 863 peak intensities to visually understand the difference in distribution between IPMN-LGD and Ad-IPMN. PLS regression is a statistical method used to project high-dimensional data into a series of linear subspaces of the explanatory variables. In this study, each serum sample had 863 explanatory variables (i.e., 863 peak intensities after data processing), and new one-dimensional variables that discriminate IPMN-LGD from Ad-IPMN are made by combining the coefficients for all 863 variables. The most discriminating new variable is defined as component 1, and the second most discriminating one is defined as component 2.22 Second, a discrimination test was performed using SVM. The variables used for the analysis by SVM were determined as follows. The explanatory variables (represented as m/z) that differed between IPMN-LGD and Ad-IPMC groups for the objective variables were identified by Student’s t-test and sorted by P-value. The SVM model was optimized by sequentially adding the corresponding explanatory variables in order from the one with the lowest P-value. The optimized SVM model determined the possibility score of each serum sample as a continuous value between 0 and 1, corresponding to IPMN-LGD and Ad-IPMN, respectively. The possibility score is shown as a box plot graph in Supplementary Fig. 4. To calculate the diagnostic accuracy, the threshold value used for the judgment was set to 0.5. If the value was closer to 1 than 0.5, the sample was diagnosed as IPMN-LGD, and if it was closer to 0 than 0.5, it was diagnosed as Ad-IPMN. This possibility score was evaluated using a random sub-sampling method, a type of cross validation. Finally, the receiver operating characteristic curve (ROC) according to the possibility value was described, and the area under the curve (AUC) was calculated.
Furthermore, the discriminant accuracy of the diagnostic algorithm was validated using 7 independent serum samples, which were obtained from patients who underwent pancreatectomy in 2022. In the validation set, we applied the same 130 variables obtained from training set analysis for SVM. Sensitivity, specificity, and accuracy were analyzed in the same way for seven samples.
Statistical analyses were performed using SPSS Statistics, version 25.0 (IBM Corp., Armonk, NY, USA) and EZR (The R Foundation for Statistical Computing, Vienna, Austria), and hierarchical analysis was conducted using MetaboAnalyst 5.0 (Xia Lab) [Nature protocols (2011), 6, 743-760, Jianguo Xia and David S Wishart]. P < 0.05 was considered statistically significant.
Results
Patient Backgrounds
Samples were collected from the University of Tokyo (n = 26) and the University of Yamanashi (n = 16). Seventeen patients were diagnosed with IPMN-LGD, while 25 patients were diagnosed with Ad-IPMN. Preoperative chemotherapy was not administered for any participants. In the endoscopic ultrasonography exam, an enhanced mural nodule was identified in 64.7% of IPMN-LGD, and 76.0% of Ad-IPMN. Of the entire cohort, cytology was performed in 27 patients. Atypical cells (more than class 3) were found in 70.0% of IPMN-LGD (7/10), and 94.1% of Ad-IPMN (16/17). Of the patient who underwent cytology (10 patients in IPMN-LGD and 17 patients in Ad-IPMN), three patients in LGD-IPMN and five patients in Ad-IPMN were evaluated for KRAS mutation, and two patients in LGD-IPMN and four patients in Ad-IPMN presented a mutation of KRAS status. The clinicopathological backgrounds of all patients are described in Table 1. No variables were significantly different between IPMN-LGD and Ad-IPMN patients, except for the carcinoembryonic antigen value (median IPMN-LGD 1.7 and Ad-IPMN 2.6, P = 0.003).
Data Acquired by PESI-MS and Processing
Using PESI-MS, 1191 peak intensities from each serum sample were acquired. Among them, 328 peak intensities were excluded because these average intensities were less than 500,000 A.U. The remaining 863 peak intensities were integrated for further analysis. The mass spectrum comprising all average peak intensities is presented in Fig. 2. The red line in this figure shows the 500,000 A.U. level. The m/z values with peak intensities lower than this red line were excluded from the analysis.
PLS Regression Analysis
A two-dimension plot generated by PLS regression is shown in Fig. 3. Although the plotted regions of IPMN-LGD and Ad-IPMN partially overlapped, they were well separated by component 1 and 2 macroscopically.
Partial least squares regression. The pink plot reflects IPMN-LGD, and the green plot indicates Ad-IPMN. Component 1 and component 2 were weighted 12.1% and 9.8% of all components. Pink and green areas were the 95% confidence intervals of each plot. IPMN, intraductal papillary mucinous neoplasm; LGD, low-grade dysplasia; Ad, Advanced
SVM and Cross Validation
The ROC curve generated by SVM is presented in Fig. 4. The selected number of variables for SVM was 130. The m/z of these 130 variables and the corresponding P-values of the Student’s t-test are shown in Supplementary Table 1. The AUC generated by the SVM model using the database of the 130 variables was 0.924 (95% confidence interval: 0.831–0.981). The specificity, sensitivity, and diagnostic accuracy calculated according to the possibility scores of cross validations using the repeat random sub-sampling method were 88.2% (15/17), 88.0% (22/25), and 88.1% (37/42), respectively. We show these scores for each group with box plot graphs in Supplementary Fig. 4A.
Validation Analysis
In the validation set, one patient was classified into IPMN-LGD, and 6 patients were classified into Ad-IPMN by the pathological diagnosis. In total, 5 out of 7 serum samples were correctly diagnosed by our method, the sensitivity was 66.7% (4/6), the specificity was 100.0% (1/1), and the accuracy rate was 71.4% (5/7). The box plot of possibility score is shown in Supplementary Fig. 4B. The possibility score of IPMN-LGD was quite low, 0.00003.
Discussion
This study demonstrated that the combination of PESI-MS and machine learning had the potential to detect IPMN canceration by analyzing a small amount of patient serum. The two-dimensional figure provided by PLS regression enabled the visualization of the two diseases separately. When 130 parameters out of the entire dataset acquired by PESI-MS were applied to the database for SVM, the system showed that the discrimination rate of Ad-IPMN from IPMN-LGD was 88.1%, and the AUC was 0.924. In the validation set, sensitivity, specificity, and accuracy were 66.7, 100.0, and 71.4%, respectively. Additionally, this diagnostic tool required ~10 min without special preparation other than drawing patient blood. Conversely, HRS and WF proposed in the International Consensus Guideline in 2018 require invasive examination and have a diagnostic accuracy of 63–75%. This diagnostic tool has a high ability to diagnose IPMN canceration, providing a promising diagnostic modality in clinical practice.
Although histopathology remains the gold standard for diagnosing cancer, integrated molecular analysis using mass spectrometry has gained attention in recent years for detecting malignant tumors because of its high throughput ability, sensitivity and specificity.23,24 Unlike other mass spectrometry methods, PESI-MS provides a spectrum by ionizing molecules with a distinct probe motion, which requires minimal sample preparation, thereby addressing the time-consuming processing of mass spectrometry.15 Other recent methods of mass spectrometry, including matrix assisted laser desorption/ionization-mass spectrometry, desorption electrospray ionization-mass spectrometry or MasSpec Pen, have produced precise results and promising biomarkers; however, they require complex pretreatment and are not suitable for routine medical care.25,26 In this study, PESI-MS analysis, data processing and machine learning required ~10 min in total, which is sufficient for clinical situations, such as outpatient examination or rapid intraoperative diagnosis. Preoperative pathological diagnosis of IPMN canceration is difficult oncologically. PESI-MS analysis is considered meaningful from that point of view.
This study is also significant in that only serum was used for the sample rather than malignant tissue. The collection of tumor tissue is not minimally invasive. Additionally, the concept of intratumoral heterogeneity has recently become widely accepted.27 Thus, examinations using tumor tissue, including needle biopsy, can lead to false-negative results. In contrast, serum samples can be obtained with no invasiveness, and their components do not change regardless of where the serum sample is collected. Accordingly, this diagnostic modality is easy to repeat and may have high reproducibility.
The calculation method with maximization of separating margins is named SVM, a type of machine learning. The classification feature of SVM is expanding its use mainly in cancer genomics.28 Multi-omics data obtained from clinical specimens contain a lot of information, and it is possible to extract latent features that lead to various clinical questions and the elucidation of pathological conditions by performing integrated analysis in combination with medical information. However, the omics data obtained from clinical samples have large individual differences and variations in distribution between samples, and it is difficult to ensure the robustness and semantic interpretability of results. Therefore, it requires optimal variable selection to maximize differences between two targeted factors. In this study, PESI-MS analysis first provided 11,910 ion intensity values, which were reduced to one value for one integer, resulting in 1191 peak intensities by data processing as described above. Among them, variables with the largest difference were selected in the comparative test, and the number of variables that maximized the difference was determined. Finally, 130 variables were selected. Previous studies selected SVM variables using the same approach and demonstrated a good discriminative ability with reproducibility.29,30 An additional important point to note regarding the interpretation is that the greater the number of variables, the higher the probability of overfitting. In this study, cross validation was performed to decrease overfitting.31 Further, analysis of the validation set gave an accuracy rate of 71.4%. Although it was lower than the training set analysis, perhaps due to small samples, a probability score of 0.00003 for IPMN-LGD was a notable value. Therefore, this diagnostic modality may provide a good specificity for IPMN-LGD, which could avoid over-surgery for benign IPMN patients.
In this study, the largest difference between IPMN-LGD and Ad-IPMN was identified at an m/z of 1035 (see Supplementary Table 1). According to the Human Metabolome Database version 5.0, this m/z is composed of phosphatidylinositol phosphates, asparagoside F, uttronin A, and triglycerides.32 Considering that phosphatidylinositol phosphates act intracellularly, and asparagoside F and uttronin A are present in plants, triglycerides were a representative molecule at this m/z. High serum triglyceride concentrations have been considered to be associated with metabolic syndrome. A few cohort studies revealed that they were related to a risk of colon, breast, or cervical cancer.33,34,35 As shown in Fig. 2, although the peak intensity of m/z 1035 was not large among all plots, a slight difference in these molecules was detected by PESI-MS analysis. The ROC curve drawn by only m/z 1035 is shown in Supplementary Fig. 3. The AUC was 0.775, which was equivalent to the value reported by previous clinical research. 11,12,13 However, by adding 129 peak intensities that were almost statistically different between IPMN-LGD and Ad-IPMN, the AUC increased to 0.924, considerably higher than that seen in previous reports. It should be noted that m/z 1035 did not reflect actual serum triglycerides (ref. Table 1). For example, both m/z 1035 and m/z 1024 (13th lower P-value in Supplementary Table 1) consist of triglycerides. That is, PESI-MS discriminates finer part differences in triglycerides.
There are several limitations to this work. First, this study included a small sample of patients. In general, small size prediction research should be accepted only to show the potential of new biological insights. Although this research applied cross validation to reduce overfitting, more data and validation by other datasets are needed for future clinical application. Second, the Ad-IPMN group in this study included both carcinoma in situ and invasive adenocarcinoma. Invasive adenocarcinoma can change serum components more significantly. Third, when multiple comparisons are performed, the false discovery rate should be controlled. Supplementary Table 1 also shows a “q value” calculated according to the Benjamini-Hochberg procedure. If the significant criterion is set as 0.05, all q values were more than significant criteria. Thus, this study cannot conclude the difference between IPMN-LGD and Ad-IPMN as for the molecular identification.
Conclusions
The new diagnostic system consisting of PESI-MS and SVM discriminated Ad-IPMN from IPMN-LGD with a high accuracy using the top 130 variables with a potential biomarker. Variable selection using more data will increase the robustness of this diagnostic tool and facilitate its clinical application.
Data Availability
The data that support the findings of this study are available on reasonable request from the corresponding author.
References
Werner J, Fritz S, Büchler MW. Intraductal papillary mucinous neoplasms of the pancreas—a surgical disease. Nat Rev Gastroenterol Hepatol. 2012;9(5):253–9.
Tanaka M. Clinical management and surgical decision-making of IPMN of the pancreas. Methods Mol Biol. 2019;1882:9–22.
Adsay V, Mino-Kenudson M, Furukawa T, et al. Pathologic evaluation and reporting of intraductal papillary mucinous neoplasms of the pancreas and other tumoral intraepithelial neoplasms of pancreatobiliary tract: recommendations of verona consensus meeting. Ann Surg. 2016;263(1):162–77.
Klöppel G, Solcia E, Longnecker DS. Histological typing of tumours of the exocrine pancreas. Cham: Springer; 1996.
Marchegiani G, Andrianello S, Pollini T, et al. “Trivial” cysts redefine the risk of cancer in presumed branch-duct intraductal papillary mucinous neoplasms of the pancreas: a potential target for follow-up discontinuation? Am J Gastroenterol. 2019;114(10):1678–84.
Facciorusso A, Crinò SF, Ramai D, et al. Association between pancreatic intraductal papillary mucinous neoplasms and extrapancreatic malignancies: a systematic review with meta-analysis. Eur J Surg Oncol J Eur Soc Surg Oncol Br Assoc Surg Oncol. 2022;48(3):632–9.
Baiocchi GL, Molfino S, Frittoli B, et al. Increased risk of second malignancy in pancreatic intraductal papillary mucinous tumors: review of the literature. World J Gastroenterol. 2015;21(23):7313–9.
Pugliese L, Keskin M, Maisonneuve P, et al. Increased incidence of extrapancreatic neoplasms in patients with IPMN: fact or fiction? A critical systematic review. Pancreatol Off J Int Assoc Pancreatol (IAP). 2015;15(3):209–16.
Basturk O, Esposito I, Fukushim N. Pancreatic intraductal papillary mucinous neoplasm. In: WHO classification of tumors. WHO classification of tumors. Digestive system tumors. Lyon: IARC Press; 2019. p. 310–8.
Tanaka M, Fernández-Del Castillo C, Kamisawa T, et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatol Off J Int Assoc Pancreatol (IAP). 2017;17(5):738–53.
Shimizu Y, Hijioka S, Hirono S, et al. New model for predicting malignancy in patients with intraductal papillary mucinous neoplasm. Ann Surg. 2020;272(1):155–62.
Marchegiani G, Andrianello S, Borin A, et al. Systematic review, meta-analysis, and a high-volume center experience supporting the new role of mural nodules proposed by the updated 2017 international guidelines on IPMN of the pancreas. Surgery. 2018;163(6):1272–9.
Crippa S, Fogliati A, Valente R, et al. A tug-of-war in intraductal papillary mucinous neoplasms management: comparison between 2017 International and 2018 European guidelines. Dig Liver Dis. 2021;53(8):998–1003.
Hiraoka K, Nishidate K, Mori K, Asakawa D, Suzuki S. Development of probe electrospray using a solid needle. Rapid Commun Mass Spectrom. 2007;21(18):3139–44.
Yoshimura K, Chen LC, Asakawa D, Hiraoka K, Takeda S. Physical properties of the probe electrospray ionization (PESI) needle applied to the biological samples. J Mass Spectrom. 2009;44(6):978–85.
Iwano T, Yoshimura K, Inoue S, et al. Breast cancer diagnosis based on lipid profiling by probe electrospray ionization mass spectrometry. Br J Surg. 2020;107(6):632–5.
Saito R, Yoshimura K, Shoda K, et al. Diagnostic significance of plasma lipid markers and machine learning-based algorithm for gastric cancer. Oncol Lett. 2021;21(5):405.
Kiritani S, Yoshimura K, Arita J, et al. A new rapid diagnostic system with ambient mass spectrometry and machine learning for colorectal liver metastasis. BMC Cancer. 2021;21(1):262.
Ashizawa K, Yoshimura K, Johno H, et al. Construction of mass spectra database and diagnosis algorithm for head and neck squamous cell carcinoma. Oral Oncol. 2017;75:111–9.
Iwano T, Yoshimura K, Watanabe G, et al. High-performance collective biomarker from liquid biopsy for diagnosis of pancreatic cancer based on mass spectrometry and machine learning. J Cancer. 2021;12(24):7477–87.
Takeda S, Yoshimura K, Tanihata H. Sample preparation for probe electrospray ionization mass spectrometry. J Vis Exp. 2019. https://doi.org/10.3791/59942.
Ghosh T, Zhang W, Ghosh D, Kechris K. Predictive modeling for metabolomics data. Methods Mol Biol. 2020;2104:313–36.
Macklin A, Khan S, Kislinger T. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research. Clin Proteomics. 2020;17:17.
Banerjee S. Empowering clinical diagnostics with mass spectrometry. ACS Omega. 2020;5(5):2041–8.
de Figueiredo Junior AG, Serafim PVP, de Melo AA, et al. Analysis of the lipid profile in patients with colorectal cancer in advanced stages. Asian Pac J Cancer Prev. 2018;19(5):1287–93.
Brown HM, Pirro V, Cooks RG. From DESI to the MasSpec Pen: ambient ionization mass spectrometry for tissue analysis and intrasurgical cancer diagnosis. Clin Chem. 2018;64(4):628–30.
Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16(1):41.
Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics. 2018;15(1):41–51.
Moler EJ, Chow ML, Mian IS. Analysis of molecular profile data using generative and discriminative methods. Physiol Genomics. 2000;4(2):109–26.
Segal NH, Pavlidis P, Noble WS, et al. Classification of clear-cell sarcoma as a subtype of melanoma by genomic profiling. J Clin Oncol Off J Am Soc Clin Oncol. 2003;21(9):1775–81.
Jia Z. Controlling the overfitting of heritability in genomic selection through cross validation. Sci Rep. 2017;7(1):13678.
Wishart DS, Guo A, Oler E, et al. HMDB 5.0: the human metabolome database for 2022. Nucleic Acids Res. 2022;50(D1):D622–31.
Tande AJ, Platz EA, Folsom AR. The metabolic syndrome is associated with reduced risk of prostate cancer. Am J Epidemiol. 2006;164(11):1094–102.
Furberg AS, Veierød MB, Wilsgaard T, Bernstein L, Thune I. Serum high-density lipoprotein cholesterol, metabolic profile, and breast cancer risk. J Nat Cancer Inst. 2004;96(15):1152–60.
Cust AE, Kaaks R, Friedenreich C, et al. Metabolic syndrome, plasma lipid, lipoprotein and glucose levels, and endometrial cancer risk in the European prospective investigation into cancer and nutrition (EPIC). Endocr Relat Cancer. 2007;14(3):755–67.
Acknowledgment
We thank Ayumi Manita and Setsuko Fukuda (Department of Anatomy and Cell Biology, Interdisciplinary Graduate School of Medicine and Engineering, University of Yamanashi) for assisting with this project. We thank Melissa Crawford, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Funding
Open access funding provided by University of Tokyo. This work was not supported by the JSPS KAKENHI (Grant Number 22K16529).
Author information
Authors and Affiliations
Contributions
SK, TI, KY, and HH analyzed and interpreted the mass spectra obtained by PESI-MS. RS, DY, and GW collected samples and clinical data. SK was the major contributor in writing the manuscript. KY and JA were major revisers of the manuscript. NA, JK, and DI read and revised the manuscript, and equally contributed to this manuscript. ST supervised the whole project scientifically. KH had final responsibility for the decision to submit for publication. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
DISCLOSURE
This research was financially supported by Shimadzu Corporation to S.T. and K.H.
Ethics Approval and Consent to Participate
This clinical research was approved by the University of Tokyo Institutional Review Board; the approval number was 11262. Written informed consent was obtained from all participants. This study was performed in accordance with the Declaration of Helsinki.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kiritani, S., Iwano, T., Yoshimura, K. et al. New Diagnostic Modality Combining Mass Spectrometry and Machine Learning for the Discrimination of Malignant Intraductal Papillary Mucinous Neoplasms. Ann Surg Oncol 30, 3150–3157 (2023). https://doi.org/10.1245/s10434-022-13012-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-13012-y