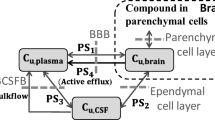
Abstract
Machine learning (ML) approaches have been applied to predicting drug pharmacokinetic properties. Previously, we predicted rat unbound brain-to-plasma ratio (Kpuu,brain) by ML models. In this study, we aimed to predict human Kpuu,brain through animal ML models. First, we re-evaluated ML models for rat Kpuu,brain prediction by using trendy open-source packages. We then developed ML models for monkey Kpuu,brain prediction. Leave-one-out cross validation was utilized to rationally build models using a relatively small dataset. After establishing the monkey and rat ML models, human Kpuu,brain prediction was achieved by implementing the animal models considering appropriate scaling methods. Mechanistic NeuroPK models for the identical monkey and human dataset were treated as the criteria for comparison. Results showed that rat Kpuu,brain predictivity was successfully replicated. The optimal ML model for monkey Kpuu,brain prediction was superior to the NeuroPK model, where accuracy within 2-fold error was 78% (R2 = 0.76). For human Kpuu,brain prediction, rat model using relative expression factor (REF), scaled transporter efflux ratios (ERs), and monkey model using in vitro ERs can provide comparable predictivity to the NeuroPK model, where accuracy within 2-fold error was 71% and 64% (R2 = 0.30 and 0.52), respectively. We demonstrated that ML models can deliver promising Kpuu,brain prediction with several advantages: (1) predict reasonable animal Kpuu,brain; (2) prospectively predict human Kpuu,brain from animal models; and (3) can skip expensive monkey studies for human prediction by using the rat model. As a result, ML models can be a powerful tool for drug Kpuu,brain prediction in the discovery stage.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
During the last two decades, machine learning approaches have been gradually applied to drug discovery, including the prediction on drug pharmacokinetic (PK) properties [1,2,3,4,5,6,7]. Amongst various PK parameters, unbound brain-to-plasma ratio (Kpuu,brain) is a critical one for drugs targeting central nervous system (CNS) [8]. It represents the ability of a drug to cross blood-brain barrier (BBB) after systemic administration and consequently to trigger pharmacological effects. Accurate prediction on Kpuu,brain of the drug candidates can drastically reduce the drug discovery cycles by facilitating the candidate screening process. However, the traditional determination of Kpuu,brain through in vivo studies is expensive and time-consuming. With the development of in silico methodology, it turns out to be an alternative for this purpose [8,9,10].
As part of artificial intelligence, the origin of machine learning can be dated back to 1950s [11]. The evolution of machine learning framework has never stopped since then. Recent years, low-code automated machine learning modules such as PyCaret and scikit-learn lead the trends and were widely used in various fields [12]. We previously have reported machine learning models that can successfully predict Kpuu,brain in rats by using StarDrop (Optibrium, UK), a commercially available machine learning software [13]. In this study, we re-evaluated rat Kpuu,brain prediction through current trendy open-source automated machine learning library, PyCaret, in order to assess the flexibility of machine learning approaches under different conditions [14]. In parallel, we tested rat Kpuu,brain prediction using graph convolutional network (GCN) approach, which has been reported for chemical structure representation with deep learning [15,16,17,18]. Additionally, limited information regarding in silico prediction on monkey and human Kpuu,brain involving machine learning approaches could be found at present. Therefore, we expanded the machine learning methodology to develop and to evaluate machine learning models for monkey Kpuu,brain prediction. Likewise, we generated Kpuu,brain dataset along with in vitro permeability data of membrane transporters, multiple drug resistance 1 (MDR1), and breast cancer resistance protein (BCRP), as additional features. The reason was MDR1 and BCRP had been recognized as key factors that impacting drug disposition through BBB [19,20,21,22]. After setting up the animal machine learning models, human Kpuu,brain prediction was achieved by using the animal models with appropriate scaling methods. Moreover, we previously reported physiologically based NeuroPK models which relied on MDR1 and BCRP’s properties for predicting Kpuu,brain among different species [23]. In this study, we used NeuroPK models as the criteria to further evaluate the performance of machine learning models on monkey and human Kpuu,brain prediction.
Materials and Methods
Chemicals
Commercially available compounds (reagent grade and above) and internal compounds from Takeda Pharmaceutical Company Limited (Fujisawa, Kanagawa, Japan) were used in this study.
Animal Studies
Animal studies were conducted under the approvals and guidance of Institutional Animal Care and Use Committee of the Shonan Health Innovation Park and Takeda Pharmaceutical Company Limited. Animal studies were conducted in animal research facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
In Vitro Permeability Assays
The method has been described previously [13, 23]. In brief, 1 μM of the testing compound in transport buffer (Hanks’ balanced salt solution with 10 mM HEPES, pH 7.4) was added to the apical or basolateral side of the transwell chamber, which was cultured with confluent cell monolayers of MDCK-MDR1 cells (NIH, USA) or MDCK-BCRP cells (Solvo Biotechnology, Szeged, Hungary). After incubating for 1 h at 37°C with 5% CO2, solution from both donor and receiver side was collected. The concentrations of the testing compounds were quantified by liquid chromatography (LC-20 or Nexera, Shimadzu, Kyoto, Japan)–mass spectrometry (API 4000 or API 5000, AB Sciex LLC, Toronto, Canada) (LC/MS/MS). Permeability of the testing compounds from apical to basolateral (A to B) direction or B to A direction and the efflux ratio (ER) were then determined. The apparent permeability coefficient Papp (cm/s) was calculated by Eq. 1:
where dCr/dt is the cumulative concentration of the compound in the receiver chamber (μM/s), Vr is the solution volume in the receiver chamber, A is the surface area for transport; and C0 is the initial concentration in the donor chamber (μM).
Efflux ratio (ER) was then determined by Eq. 2:
In Vivo Administration and Sampling
In vivo study condition of part of the monkey data (Table S1) was described previously [23]. For the newly added, the compounds (mainly internal compounds), cassette dosing solution of up to 10 testing compounds with a final dose of 0.1 mg/kg each was administered intravenously as bolus to the monkeys. Newly added compounds exhibited fast brain equilibrium in rodent in general (Figure S1 and Table S2) and overall efflux capability, the driving factor for brain disposition, is stronger in rodents than in monkeys [23,24,25,26]. As a result, the risk to have delayed brain steady state in monkeys for these compounds would be low. Together with the consideration of animal welfare, monkeys were euthanized after 1 h post dose in this study. Blood and brain samples were collected. The blood samples were transferred to polypropylene tubes immediately, cooled on ice, and centrifuged (4°C, 6000 × g for 2 min). Approximately 120 μL of plasma was obtained for each sample. As for the brain samples, 20% brain homogenate in deionized water (w/v) were prepared under 4°C condition. Samples were stored at −80°C until use. The concentrations of the testing compounds in plasma and brain samples were measured by LC-MS/MS.
Unbound Fraction in Brain and Plasma and Determination of Kpuu,brain
The unbound fraction in plasma and brain for each compound was evaluated using the equilibrium dialysis method. Details have been reported previously [13, 23, 27]. Briefly, testing compounds solubilized in dimethyl sulfoxide (DMSO) were diluted to 1 μM by monkey plasma or 20% (w/v) monkey brain homogenate in 100 mM sodium phosphate buffer (pH 7.4). Dialysis was conducted with an equal volume of sodium phosphate buffer at 37°C for 16–20 h. The buffer obtained from the apparatus was mixed with an equal volume of either control plasma or control brain homogenate. The samples were mixed with three volumes of acetonitrile. Supernatant was stored at −80°C until LC-MS/MS analysis.
Plasma unbound fraction was defined by Eq. 3:
Unbound fraction in brain (fu,brain) was calculated by Eq. 4:
where D and fu,brain′ represent the dilution factor for the brain homogenate and unbound fraction determined in the 20% (w/v) brain homogenate, respectively.
Kpuu,brain was then defined by Eq. 5:
where Cbrain and Cplasma represent testing compound’s concentration in brain and plasma, respectively.
Re-evaluation of Rat Kpuu,brain Model Using Current Machine Learning Modules
Re-evaluation of the rat dataset used PyCaret machine learning library in Python 3.8 (Python Software Foundation, Delaware, USA) with 2D descriptors generated by alvaDesc software (Alvascience Srl, Lecco, Italy) [14, 28]. Six hundred forty compounds were split into 512 training data and 128 test data by cluster characteristics, which were described before [13]. 5-fold cross validation was performed for model optimization. Final model was selected by prediction accuracy within 2-fold error and coefficient of determination (R2) of the test data set. R2 was defined by Eq. 6:
where N is the sample size, i is the sample number, y is the prediction value, ӯ is the mean value; and ŷ is the true value.
Rat Kpuu,brain Prediction Using GCN Approach
The same rat Kpuu,brain training and test datasets were used. Compounds’ chemical structures were represented by graphs using DGL-LifeSci in Python 3.8 (Python Software Foundation, Delaware, United States) [29]. Rat Kpuu,brain prediction was fulfilled through deep learning with GCN models using Deep Graph Library with PyTorch backend [30, 31]. The training set was used for model establishment, which was further randomly split into model training set and model validation set. Model hyperparameter optimization was tuned with Optuna [32]. Under this deep learning framework, model was selected by the prediction accuracy of the validation set. Final model performance was evaluated by the prediction accuracy within 2-fold error and coefficient of determination (R2) of the test dataset.
Monkey Kpuu,brain Model Establishment Using Machine Learning Approach
Due to the limitation of PyCaret for leave-one-out cross validation (LOOCV) settings, monkey machine learning models were built in Dataiku (New York City, USA), a commercial machine learning platform using a 2-step approach. Monkey dataset of 51 compounds was used (Table S1). Compound-dependent 2D descriptors were generated by AlvaDesc software according to chemical structures [28]. In vitro MDR1 ER and BCRP ER were also included as additional features. Three thousand eight hundred sixty-four qualified features were finally used for step 1 model establishment. A total of 14 regressors were utilized (Table S3). After the first round model training, top 20 features with the highest importance or correlation were selected from the 7 models with mean absolute error (MAE) less than 0.5 (Table S3). It is noted no feature importance was generated for LightGBM, K nearest neighbors model, and support vector machines models. Eighty features were finally used for step 2 model establishment. MAE was defined by Eq. 7:
where N is the sample size, i is the sample number, y is the prediction value, and ŷ is the true value.
Consider the small size of the dataset, LOOCV was implemented during model training and hyperparameter tuning. Corresponding feature importance was calculated for the tunned models. The final model was selected by prediction accuracy within 2-fold error and R2 of the whole dataset. To evaluate ERs’ role, we further checked step 2 model performance with ERs, without BCRP ER, without MDR1 ER, and without both ERs as features.
Human Kpuu,brain Prediction by Extrapolation from Animal Machine Learning Models
The developed rat model and monkey model were applied to human Kpuu,brain prediction of 14 compounds [8, 33, 34]. Information of compounds was summarized in Table S4. MDR1 and BCRP ERs scaling from human to rat or monkey using the ratio of relative expression factor (REF) and relative activity factor (RAF) between species were examined in addition to the original in vitro ER values (Eqs. 8 and 9). RAF and REF parameters were referred to our previous work [23].
Kpuu,brain Prediction by the NeuroPK Model
The same monkey and human dataset were also fitted and assessed by NeuroPK models using MATLAB (MathWork Inc., MA, USA) curve fitting toolbox [23]. Kpuu,brain in NeuroPK model was calculated by Eq. 10:
where α represents a ratio to extrapolate the in vitro efflux activity of MDR1 in MDCK cells into that of MDR1 in vivo and β represents a ratio to extrapolate the in vitro efflux activity of BCRP in MDCK cells into that of BCRP in vivo [27]. Specially for the NeuroPK model, in order to avoid negative values in the equation, those ER values lower than 1 were adjusted to 1.
Results
Re-evaluation of the Rat Model
LightGBM model was the best for rat Kpuu,brain prediction by using the PyCaret machine learning library. Prediction accuracy within 2-fold error reached 79% and R2 was 0.53 for the test set. This result was no inferior to the previous published data (Table I and Figure S2) [13]. Furthermore, rat model can select MDR1 and BCRP ERs as the most important features in this study as well (Fig. 1).
Rat Kpuu,brain Prediction Through GCN Approach
The GCN model resulted in a 65% prediction accuracy within 2-fold error and R2 of 0.36 on rat Kpuu,brain prediction for the test set. This result was inferior to that from the machine learning models (Table I and Figure S2).
Monkey Dataset Analysis
Monkey dataset was summarized in Table S1. The dataset included both inhouse data and references information, where part of the dataset has been described previously [23, 35,36,37]. Fast brain equilibrium was confirmed for most newly tested compounds in rodents (Figure S1). Statistical distribution of monkey Kpuu,brain data of 51 compounds’ basic chemical properties were evaluated (Table II and Figure S3). The distribution of Kpuu,brain, in silico LogP calculated by ACD/Lab (Toronto, Canada), MW, and ERs covered a wide range. Both substrates (ER > = 2) and non-substrates of MDR1 and BCRP were included.
Monkey Kpuu,brain Prediction
Total 80 features were finally extracted from step 1 models and subsequently were used for Step 2 model establishment. Top 3 models with the best accuracy after step 2 training were summarized in Table S5. Among them, Ridge L2 regressor exhibited the best performance on monkey Kpuu,brain prediction, with a 2-fold error accuracy of 78% and R2 = 0.76 for the whole dataset (Table III and Figure S4). Top 10 highly correlated features were shown in Fig. 2a.
Unlike the rat machine learning model, MDR1 ER and BCRP ER were not selected as top features by the Ridge L2 model. Because of this, we further tested the model establishment without one or both ERs as features. After excluding ERs, Ridge L2 model delivered inferior predictability to the model with ERs, where accuracy within 2-fold error was 65% and R2 = 0.57 after excluding both ERs. When only excluding one of the ERs, accuracy within 2-fold error was 69% and 65% together with R2 equaling to 0.69 and 0.56 for models without MDR1 ER or BCRP ER as features, respectively. Highly correlated features for model without ERs as features were shown in Fig. 2b. Top 3 features were the same as the model with ERs features, which were VE2sign_Dz(p) (average coefficient of the last eigenvector from Barysz matrix weighted by polarizability), JGI2 (mean topological charge index of order 2), and VE2_D/Dt (average coefficient of the last eigenvector (absolute values) from distance/detour matrix). These features describe compounds’ chemical structure properties. For feature comparison, we also summarized the top 10 features of the best 3 monkey machine learning models in Table S6.
NeuroPK Model Performance on Monkey and Human Kpuu,brain Data
Identical monkey dataset of 51 compounds were also used for NeuroPK model. The results demonstrated that the best within 2-fold error accuracy was 61% and R2 was 0.41 (Table III). As for the human dataset, the best fitted model showed a 2-fold error accuracy of 71% and R2 of 0.35 (Table IV and Figure S5).
Human Kpuu,brain Prediction
Human Kpuu,brain prediction was achieved by using the established rat Kpuu,brain LightGBM model and monkey Kpuu,brain Ridge L2 model with the consideration of scaled MDR1 and BCRP ERs in addition to the original in vitro ER values (Table IV and Figure S6). For the rat model, REF scaling gave the highest accuracy for human Kpuu,brain prediction with a 2-fold error accuracy of 71% and R2 of 0.30 (Table IV and Table S7). On the other hand, best human Kpuu,brain prediction by the monkey model was achieved without scaling ERs. Within 2-fold error accuracy was 64% and R2 equaled to 0.52.
Discussion
Machine learning has been treated as one effective strategy to predict drug pharmacokinetic properties. We reported its use for small molecule Kpuu,brain prediction in rats before [13]. In this study, we expanded the application of machine learning models to monkey and human Kpuu,brain prediction. Monkey Kpuu,brain prediction was achieved by establishing machine learning model using actual monkey data. On the other hand, good human Kpuu,brain prediction was achieved through the projection from the animal machine learning models with appropriate scaling method. For comparison analysis, monkey and human Kpuu,brain described by the physiologically based NeuroPK models were used as the standard criteria for evaluating machine learning model performance.
Re-evaluated rat model under the trendy open-source machine learning module provide good prediction accuracy (Table I). Moreover, it can select MDR1 and BCRP ER as key features like previously reported models, which indicated the machine learning approach could recognize physiological parameters with a relatively abundant dataset (Fig. 1). It also revealed the performance consistency among different machine learning modules in terms of rat Kpuu,brain prediction (Table I). With the development of machine learning frameworks, refinement and optimization of existing models using new modules is possible to deliver consistent predictability, which is important for long term application in practice of such machine learning models.
In parallel, GCN approach was examined. Chemical structure information of the compounds was described by graphs with subsequent Kpuu,brain prediction utilizing deep learning neural network model. Although this approach can provide acceptable prediction accuracy, it was inferior to the machine learning models (Table I). One potential reason is that MDR1 ER and BCRP ER cannot be included in this approach. They are the additional in vitro experimental information not directly deriving from chemical structures; therefore, they cannot be translated and involved as part of the generated graphs to be used for model establishment. Besides, deep learning typically requires thousands of data entries in order to establish a promising model, much more than that required by the machine learning models. Consider these facts, we decided to continue with the machine learning approaches for monkey Kpuu,brain prediction at this stage.
Contrary to the rat dataset, one hurdle for the monkey Kpuu,brain prediction lied in the small size of available data input. Monkey dataset (Table S1) contained 51 compounds from both literatures and inhouse studies [23, 35,36,37]. One potential concern on the brain steady state of those compounds with 1-h collection time point was first confirmed through rodent data (Figure S1 and Table S2). As described, most compounds can reach brain equilibrium quickly within 1 h in rodents, where the differences to a later time point were within 2-fold error. It is known that efflux transporters are one major force for brain penetration and Kpuu,brain [19,20,21,22, 38]. Various studies have demonstrated that rodent’s overall brain efflux transporter expression and activity are stronger than monkey, which provided the rationale to have early brain steady state in monkeys for those compounds as well [23,24,25,26].
To provide more confidence on our monkey dataset for machine learning use, we conducted principal component analysis of all the generated features for monkey dataset versus rat dataset, where principal component 1 (PC1)–principal component 2 (PC2) were shown in Fig. 3. PC1 and PC2 had explained variance of 0.34 and 0.09, respectively. Cumulative explained variance was shown in Figure S7. In spite of a smaller data size, monkey dataset possessed wider distribution than the rat dataset, revealing the coverage of chemical space in the monkey dataset is not limited. LOOCV was adopted and turned out to be a feasible strategy to offer reasonable prediction on Kpuu,brain in monkey. So far, the machine learning model showed better performance than the mechanistic NeuroPK model according to 2-fold error prediction accuracy and R2 (Table III). Nevertheless, we previously reported the predictive performance on the external compounds, which suggested mechanistic NeuroPK model exhibited slightly better prediction in terms of R2 than the optimal machine learning model [13]. Due to lack of additional data for monkey or human Kpuu,brain, it is hard to validate this point in this study. Yet, similar to the observation in the rat models, it is possible that chemotypes not well covered by current chemical space and descriptors could be better explained by the NeuroPK model.
When it comes to important features of the final monkey Kpuu,brain models in this study, a number of overlapped features were observed in the best 3 models (Table S6). For example, SIC (structural information content index for neighborhood symmetry), B09_N-S (presence/absence of N-S at topological distance 9), and SpMin1_Bh(i) (smallest eigenvalue n. 1 of Burden matrix weighted by ionization potential) might highly contribute to monkey brain penetration. But these descriptors are tiny pieces of 2-dimensional (2D) chemical structure information, which cannot be well linked to specific chemotypes or functional groups. Further evaluation is required to confirm the importance of these structure pieces with different dataset. Moreover, we will try different feature generation methods such as using fingerprint approach to better describe spatial structure properties in the future.
Correlation matrix of top 10 features from the Ridge L2 model and ERs can indicate whether certain features could represent ERs’ roles in the monkey Kpuu,brain model (Figure S8). Results suggested that BCRP ER statistically correlated to the feature B05_O-Cl_ (presence/absence of O–Cl at topological distance 5). We have confirmed that excluding the ERs led to worse prediction accuracy (Table III). Excluding BCRP ER or both ERs showed more impacts on prediction accuracy than excluding MDR1 ER. It is noted that BCRP ER was selected as top 10 features by the other monkey model (Lasso L1 model) (Table S6). Based on this information, BCRP ER may play an important role in monkey Kpuu,brain prediction. As for MDR1 ER, there was limited correlation to the top features selected by the Ridge L2 Kpuu,brain model. Removing MDR1 ER impaired the model predictivity but not as much as excluding BCRP ER. Similarly, MDR1 ER was not selected as crucial features by the other 2 models as well. One potential explanation was the expression level of MDR1 in monkey is much lower than that in rodents so that its importance was not affirmed by the machine learning models [25, 39]. Overall, BCRP ER exhibited more influences than MDR1 ER on monkey Kpuu,brain prediction.
We further analyzed the monkey Ridge L2 model outliers (outside 2-fold error) in relation to unbound fraction fu values. Outliers had fu values generally ranged from very small values to medium values (< 0.5) while large fu compounds actually can be predicted within 2-fold error under this model (Figure S9). It should be pointed out that limited data were available for large fu (> 0.5) range. In spite of this, the monkey machine learning model can cover compounds with a wide range of fu.
By using MDR1 and BCRP REF as scaling factor among different species, rat machine learning model well predicted human Kpuu,brain (Table IV). Monkey machine learning model also showed the ability to predict human Kpuu,brain with comparable performance. These findings suggested that machine learning model of preclinical species could be properly translated to human, which will be a powerful tool during CNS drug discovery and development. Merits of machine learning models for human Kpuu,brain prediction include high-throughput and low demands on in vivo studies. In this study, we found REF as scaling factor worked better than RAF in the rat model (Table S7). It implied that transporter’s expression level could be a more appropriate factor than its activity when dealing with species differences between rodent and human. A potential reason is that RAF used here was from our previous study that fitting the Kpuu,brain and Kpuu,CSF values with a set of compounds while REF was calculated by calibration of proteomic expression level from scaler compounds [23, 40]. It was also claimed that RAF ought to be calculated through transporter-selective substrate probe [41]. It was likely that RAF value extracted from other resources might not fit the current datasets well so that the cross-species translation was worse than REF.
When trying to utilize the monkey model to predict human Kpuu,brain, prediction accuracy was comparable to that of rat machine learning model or NeuroPK model. In practice, a compound’s Kpuu,brain for human often tends to be translated from that of monkey or other non-rodent animals [42]. Closer physiological similarity between monkey and human compared with rodent is one underlying reason for more promising translation [43]. It was claimed that monkey would be a more appropriate animal model for CNS than rodent to understand human neurobiology not only from anatomical and physiological aspects but also due to cognitive and behavioral differences [44]. Thus, it was expected that monkey model can provide reasonable prediction on human Kpuu,brain. Probably because of the similar expression patterns and activities of BCRP and MDR1 between monkey and human, no benefits were observed by using REF or RAF scaled ERs [25, 39, 45].
Kpuu,brain were fitted by the NeuroPK models through mathematical equation using MDR1 and BCRP ERs in a retrospective manner. In both monkey and human cases, machine learning models can provide comparable to better predictivity than the NeuroPK model (Table III and IV). For the monkey models, even the one excluding ERs can provided slightly better accuracy than the NeuroPK model. One advantage is that machine learning models involve a wide range of physicochemical features of a compound. The combination of physicochemical and physiological properties may generate more comprehensive models rather than just relying on one aspect. This may explain why monkey machine learning model with ERs can provide significantly better predictivity than the NeuroPK model.
For the human Kpuu,brain prediction, both rat and monkey machine learning models can deliver comparable predictivity to that of the mechanistic NeuroPK models. One advantage of utilizing machine learning models lies in indirect prediction could be achieved instead of using actual human dataset to generate a reliable model. It should be noted that using the small human dataset is technically hard to generate a stable NeuroPK model from data fitting process. Therefore, it is encouraging that machine learning models totally have the potentials to be used as prospective tools for human Kpuu,brain prediction. In other words, unlike the NeuroPK model, machine learning models can directly predict human Kpuu,brain based on the established rat or monkey model instead of fitting the limited human data. Especially, using the rat model will greatly diminish the demands on monkey studies so as to cost-effectively accelerate the drug discovery timeline. Consider above advantages, it is rational to continuously refine and optimize machine learning models for human translation.
We further evaluated the Kpuu,brain of the same compounds among the three species. Both monkey and rat Kpuu,brain showed more or less distinctions from human Kpuu,brain without better correlation from either species (Figure S10). In empirical, monkey data are typically chosen for human PK prediction owing to better PK translation between primates [42]. Other than this fact, rat Kpuu,brain with appropriate scaling method can also be a good option for human projection just like what our machine learning model suggested. To some extent, rat model for Kpuu,brain prediction will be more attractive means because monkey studies have higher barriers than rodent studies. Translating from rat model to human can re-allocate the resources during the drug discovery stages.
Conclusion
To summarize, in silico models using machine learning approaches were able to deliver good monkey and human Kpuu,brain prediction with reasonable accuracy. Compared to the mechanistic NeuroPK model, machine learning models have several attractive advantages: (1) can provide reasonable prediction on monkey Kpuu,brain even without MDR1 and BCRP ERs; (2) can act prospectively for human Kpuu,brain prediction from preclinical species models; and (3) can skip expensive monkey studies for human translation by using the rat machine learning model. In conclusion, machine learning models can be a useful tool for CNS compound selection in the drug discovery stage.
References
Tao L, Zhang P, Qin C, Chen SY, Zhang C, Chen Z, et al. Recent progresses in the exploration of machine learning methods as in-silico ADME prediction tools. Adv Drug Deliv Rev. 2015;86:83–100.
Goller AH, Kuhnke L, Montanari F, Bonin A, Schneckener S, Ter Laak A, et al. Bayer’s in silico ADMET platform: a journey of machine learning over the past two decades. Drug Discov Today. 2020;25(9):1702–9.
Jia L, Gao H. Machine learning for in silico ADMET prediction. Methods Mol Biol. 2022;2390:447–60.
Patel L, Shukla T, Huang X, Ussery DW, Wang S. Machine learning methods in drug discovery. Molecules. 2020;25(22).
Gupta R, Srivastava D, Sahu M, Tiwari S, Ambasta RK, Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers. 2021;25(3):1315–60.
Kosugi Y, Hosea N. Prediction of oral pharmacokinetics using a combination of in silico descriptors and in vitro ADME properties. Mol Pharm. 2021;18(3):1071–9.
Kosugi Y, Hosea N. Direct comparison of total clearance prediction: computational machine learning model versus bottom-up approach using in vitro assay. Mol Pharm. 2020;17(7):2299–309.
Liu H, Dong K, Zhang W, Summerfield SG, Terstappen GC. Prediction of brain:blood unbound concentration ratios in CNS drug discovery employing in silico and in vitro model systems. Drug Discov Today. 2018;23(7):1357–72.
Watanabe R, Esaki T, Ohashi R, Kuroda M, Kawashima H, Komura H, et al. Development of an in silico prediction model for P-glycoprotein efflux potential in brain capillary endothelial cells toward the prediction of brain penetration. J Med Chem. 2021;64(5):2725–38.
Zhu L, Zhao J, Zhang Y, Zhou W, Yin L, Wang Y, et al. ADME properties evaluation in drug discovery: in silico prediction of blood-brain partitioning. Mol Divers. 2018;22(4):979–90.
Fradkov AL. Early history of machine learning. IFAC-PapersOnLine. 2020;53(2):1385–90.
Gain U, Hotti V, editors. Low-code autoML-augmented data pipeline–a review and experiments. Journal of Physics: Conference Series; 2021: IOP Publishing.
Kosugi Y, Mizuno K, Santos C, Sato S, Hosea N, Zientek M. Direct comparison of the prediction of the unbound brain-to-plasma partitioning utilizing machine learning approach and mechanistic neuropharmacokinetic model. AAPS J. 2021;23(4):72.
Ali M. PyCaret: an open source, low-code machine learning library in Python. 2020.
Kearnes S, McCloskey K, Berndl M, Pande V, Riley P. Molecular graph convolutions: moving beyond fingerprints. J Comput Aided Mol Des. 2016;30(8):595–608.
Duvenaud DK, Maclaurin D, Iparraguirre J, Bombarell R, Hirzel T, Aspuru-Guzik A, et al. Convolutional networks on graphs for learning molecular fingerprints. Advances in neural information processing systems. 2015;28:2224–32.
Kojima R, Ishida S, Ohta M, Iwata H, Honma T, Okuno Y. kGCN: a graph-based deep learning framework for chemical structures. J Cheminform. 2020;12(1):32.
Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:160902907. 2016.
Katagiri Y, Kawaguchi H, Umemura K, Tadano J, Miyawaki I, Takano M. Investigation of the role and quantitative impact of breast cancer resistance protein on drug distribution into brain and CSF in rats. Drug Metab Pharmacokinet. 2022;42:100430.
Enokizono J, Kusuhara H, Ose A, Schinkel AH, Sugiyama Y. Quantitative investigation of the role of breast cancer resistance protein (Bcrp/Abcg2) in limiting brain and testis penetration of xenobiotic compounds. Drug Metab Dispos. 2008;36(6):995–1002.
Schinkel AH, Smit JJ, van Tellingen O, Beijnen JH, Wagenaar E, van Deemter L, et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77(4):491–502.
Sakata A, Tamai I, Kawazu K, Deguchi Y, Ohnishi T, Saheki A, et al. In vivo evidence for ATP-dependent and P-glycoprotein-mediated transport of cyclosporin A at the blood-brain barrier. Biochem Pharmacol. 1994;48(10):1989–92.
Sato S, Matsumiya K, Tohyama K, Kosugi Y. Translational CNS steady-state drug disposition model in rats, monkeys, and humans for quantitative prediction of brain-to-plasma and cerebrospinal fluid-to-plasma unbound concentration ratios. AAPS J. 2021;23(4):81.
Uchida Y. Quantitative proteomics-based blood-brain barrier study. Biol Pharm Bull. 2021;44(4):465–73.
Chu X, Bleasby K, Evers R. Species differences in drug transporters and implications for translating preclinical findings to humans. Expert Opin Drug Metab Toxicol. 2013;9(3):237–52.
Kodaira H, Kusuhara H, Fuse E, Ushiki J, Sugiyama Y. Quantitative investigation of the brain-to-cerebrospinal fluid unbound drug concentration ratio under steady-state conditions in rats using a pharmacokinetic model and scaling factors for active efflux transporters. Drug Metab Dispos. 2014;42(6):983–9.
Sato S, Tohyama K, Kosugi Y. Investigation of MDR1-overexpressing cell lines to derive a quantitative prediction approach for brain disposition using in vitro efflux activities. Eur J Pharm Sci. 2020;142:105119.
Mauri A. alvaDesc: a tool to calculate and analyze molecular descriptors and fingerprints. Ecotoxicological QSARs: Springer; 2020. p. 801–20.
Li M, Zhou J, Hu J, Fan W, Zhang Y, Gu Y, et al. DGL-LifeSci: an open-source toolkit for deep learning on graphs in life science. ACS Omega. 2021;6(41):27233–8.
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Pytorch: an imperative style, high-performance deep learning library. Advances in neural information processing systems. 2019;32:8026–37.
Wang M, Zheng D, Ye Z, Gan Q, Li M, Song X, et al. Deep graph library: a graph-centric, highly-performant package for graph neural networks. arXiv preprint arXiv:190901315. 2019.
Akiba T, Sano S, Yanase T, Ohta T, Koyama M, editors. Optuna: a next-generation hyperparameter optimization framework. Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery & data mining; 2019.
Summerfield SG, Lucas AJ, Porter RA, Jeffrey P, Gunn RN, Read KR, et al. Toward an improved prediction of human in vivo brain penetration. Xenobiotica. 2008;38(12):1518–35.
Bauer M, Karch R, Wulkersdorfer B, Philippe C, Nics L, Klebermass EM, et al. A proof-of-concept study to inhibit ABCG2- and ABCB1-mediated efflux transport at the human blood-brain barrier. J Nucl Med. 2019;60(4):486–91.
Nagaya Y, Nozaki Y, Kobayashi K, Takenaka O, Nakatani Y, Kusano K, et al. Utility of cerebrospinal fluid drug concentration as a surrogate for unbound brain concentration in nonhuman primates. Drug Metab Pharmacokinet. 2014;29(5):419–26.
Uchida Y, Wakayama K, Ohtsuki S, Chiba M, Ohe T, Ishii Y, et al. Blood-brain barrier pharmacoproteomics-based reconstruction of the in vivo brain distribution of P-glycoprotein substrates in cynomolgus monkeys. J Pharmacol Exp Ther. 2014;350(3):578–88.
Kido Y, Nanchi I, Fusamae Y, Matsuzaki T, Akazawa T, Sawada H, et al. Species difference in brain penetration of P-gp and BCRP substrates among monkey, dog and mouse. Drug Metab Pharmacokinet. 2022;42:100426.
Kikuchi R, de Morais SM, Kalvass JC. In vitro P-glycoprotein efflux ratio can predict the in vivo brain penetration regardless of biopharmaceutics drug disposition classification system class. Drug Metab Dispos. 2013;41(12):2012–7.
Ito K, Uchida Y, Ohtsuki S, Aizawa S, Kawakami H, Katsukura Y, et al. Quantitative membrane protein expression at the blood-brain barrier of adult and younger cynomolgus monkeys. J Pharm Sci. 2011;100(9):3939–50.
Trapa PE, Troutman MD, Lau TY, Wager TT, Maurer TS, Patel NC, et al. In vitro-in vivo extrapolation of key transporter activity at the blood-brain barrier. Drug Metab Dispos. 2019;47(4):405–11.
Kumar AR, Prasad B, Bhatt DK, Mathialagan S, Varma MVS, Unadkat JD. In vivo-to-in vitro extrapolation of transporter-mediated renal clearance: relative expression factor versus relative activity factor approach. Drug Metab Dispos. 2020;49(6):470–8.
Loryan I, Reichel A, Feng B, Bundgaard C, Shaffer C, Kalvass C, et al. Unbound brain-to-plasma partition coefficient, Kp, uu, brain—a game changing parameter for CNS drug discovery and development. Pharmaceutical research. 2022;39(7):1321–41.
Passingham R. How good is the macaque monkey model of the human brain? Curr Opin Neurobiol. 2009;19(1):6–11.
Izpisua Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, et al. Brains, genes, and primates. Neuron. 2015;86(3):617–31.
Billington S, Salphati L, Hop C, Chu X, Evers R, Burdette D, et al. Interindividual and regional variability in drug transporter abundance at the human blood-brain barrier measured by quantitative targeted proteomics. Clin Pharmacol Ther. 2019;106(1):228–37.
Acknowledgements
The authors would like to acknowledge our DMPK colleagues, especially Sho Sato, Masanori Nakakariya, our intern Asahi Adachi, and our former temporary staff Misato Mori at Takeda Pharmaceutical Company Limited for their comments and supports on this study.
Funding
The study used internal budget of Takeda Pharmaceutical Company Limited.
Author information
Authors and Affiliations
Contributions
Siyu Liu: substantial contributions to the conception and design of the work; conducting the study and interpretation of data for the work; drafting the work manuscript; and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Yohei Kosugi: substantial contributions to the conception and design of the work; review the manuscript; and final approval of the version to be published.
Corresponding author
Ethics declarations
Conflict of Interest
The authors are employees of Takeda Pharmaceutical Company Limited.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 1401 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, S., Kosugi, Y. Human Brain Penetration Prediction Using Scaling Approach from Animal Machine Learning Models. AAPS J 25, 86 (2023). https://doi.org/10.1208/s12248-023-00850-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-023-00850-1