Abstract
Quantitative evaluation of potential pharmacodynamic (PD) interactions is important in tuberculosis drug development in order to optimize Phase 2b drug selection and ultimately to define clinical combination regimens. In this work, we used simulations to (1) evaluate different analysis methods for detecting PD interactions between two hypothetical anti-tubercular drugs in in vitro time-kill experiments, and (2) provide design recommendations for evaluation of PD interactions. The model used for all simulations was the Multistate Tuberculosis Pharmacometric (MTP) model linked to the General Pharmacodynamic Interaction (GPDI) model. Simulated data were re-estimated using the MTP–GPDI model implemented in Bliss Independence or Loewe Additivity, or using a conventional model such as an Empirical Bliss Independence-based model or the Greco model based on Loewe Additivity. The GPDI model correctly characterized different PD interactions (antagonism, synergism, or asymmetric interaction), regardless of the underlying additivity criterion. The commonly used conventional models were not able to characterize asymmetric PD interactions, i.e., concentration-dependent synergism and antagonism. An optimized experimental design was developed that correctly identified interactions in ≥ 94% of the evaluated scenarios using the MTP–GPDI model approach. The MTP–GPDI model approach was proved to provide advantages to other conventional models for assessing PD interactions of anti-tubercular drugs and provides key information for selection of drug combinations for Phase 2b evaluation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Combination therapies are standard in treating tuberculosis (TB). Current drug development paradigms focus substantially on pharmacokinetic (PK) interactions between drugs in combinations, but focus less on pharmacodynamic (PD) interactions, although these may also contribute to therapeutic failure or success and are thus at least as important. Three classes of PD drug–drug interactions are currently defined as additive, synergistic, and antagonistic interaction, where the latter two are defined as having an increased or decreased effect compared to expected additivity (1). However, the definition of additivity is not trivial, and several competing criteria to define additivity have been used: pure effect summation (2), Bliss Independence (3), and Loewe Additivity (4). In order to explore PD interactions, a commonly used experiment is the conventional microdilution checkerboard design in in vitro drug screening to evaluate PD interactions. However, a full eight-by-eight or even ten-by-ten checkerboard design that explores the entire combined concentration space is quite labor-intensive. Hence, screening studies frequently use only three-by-three designs, which are less labor-intensive, but may also provide significantly less information on the (joint) exposure–response relationships of the drugs.
The Multistate Tuberculosis Pharmacometric (MTP) model, which predicts the change in bacterial number for fast- (F), slow- (S), and non-multiplying (N) bacteria, with and without drug effects, is a semi-mechanistic PK-PD model for studying exposure–response relationships for anti-tubercular drugs and was first developed using in vitro data (5). The MTP model has successfully been implemented by Chen et al. to estimate drug efficacy in a murine model (6,7), to quantify human early bacterial activity with clinical trial simulations (8) and for predicting early bacterial activity in humans using only in vitro information (9).
The General Pharmacodynamic Interaction (GPDI) model is a model-based evaluation of PD interactions by Wicha et al. (10) that provides model-based estimates of PD interactions given as multi-directional shifts of drug potency and/or efficacy. A key advantage is its compatibility with different additivity criteria, making the GPDI model an appealing approach for studying and comparing various underlying additivity criteria. Moreover, the GPDI model allows for characterization and quantification of a novel class of PD interactions called asymmetric interactions (10). Asymmetric synergistic or antagonistic interactions are concentration-dependent, i.e., the interaction changes in magnitude depending on the concentration of the perpetrator and victim drug. Theoretically, two drugs with an asymmetric interaction could display a situation in which the two drugs change from perpetrator to victim and/or the interaction changes from synergistic to antagonistic, depending on the drug concentrations. The GPDI model has been successfully applied to data from high-throughput screening experiments (10), linked to the MTP model for estimating PD interactions in TB using in vitro data by Clewe et al. (11) and using murine data by Chen et al. (6).
The objectives of this work were (1) to evaluate different analysis methods, including the GPDI model, an Empirical Bliss Independence-based model, and the Greco model based on Loewe Additivity, for detecting PD interactions between two hypothetical anti-tubercular drugs in time-kill experiments of colony-forming unit (CFU) versus time data, and (2) to identify an optimized experimental study design in order to more efficiently assess PD interactions, including asymmetric interactions.
MATERIALS AND METHODS
The MTP–GPDI Simulation Model
The MTP model (5), displayed in Fig. 1, was used as the underlying model for all simulations. It consists of three bacterial states, i.e., fast- (F), slow- (S), and non-multiplying (N) bacteria, and was used for predicting CFU data from each hypothetical drug. The differential equation systems for F (Eq. 1), S (Eq. 2), and N (Eq. 3) bacterial states were as follows:
where kG is the growth rate of F; kFS, kSF, kFN, kSN, kNS are transfer rates between each bacterial state; and F, S, and N represent fast-, slow- and non-multiplying bacteria. The parameters in the MTP model used for simulation are shown in Table I.
The multistate tuberculosis pharmacometric (MTP) model. F, fast-multiplying state; S, slow-multiplying state; N, non-multiplying state; kG, growth rate of the fast-multiplying state bacteria; kFS, time-dependent linear rate parameter describing transfer from fast- to slow-multiplying state; kSF, first-order transfer rate between slow- and fast-multiplying states; kFN, first-order transfer rate between fast- and non-multiplying states; kSN, first-order transfer rate between slow- and non-multiplying states; kNS, first-order transfer rate between non-multiplying and slow-multiplying states
The MTP model (5) was linked to the GPDI model (10) to describe joint drug effects and drug interactions in the hypothetical two-drug combination. Killing of the bacterial sub-state F was assumed (E; Eq. 4). No drug effect was assumed on other bacterial states (killing of S or N bacterial sub-state or inhibition of growth of F bacterial sub-state) in the MTP model. The GPDI model was implemented on both Bliss Independence (Eqs. 5–7) and Loewe Additivity (Eq. 8) to allow for shifts of each EC50 in presence of the combination partner.
with
and
or
where EmaxA and EmaxB are the maximal achievable drug effects for drug A and drug B, respectively. For Loewe Additivity (Eq. 8), solely a mutual Emax can be modeled. EC50A and EC50B are the drug concentrations at 50% of EmaxA and EmaxB. HA and HB are sigmoidicity parameters for each drug. INTAB and INTBA characterized the maximum fractional change of the respective PD parameters of single drug EC50A and EC50B. A value of zero of INTAB or INTBA defined no interaction; a positive value of one INT parameter defined mono-directional antagonism; positive values of both INT parameters bidirectional antagonism, while a value of one INT parameter between − 1 and 0 defined mono-directional synergy; values between − 1 and 0 for both INT parameters defined bidirectional synergy. INT values of opposite polarity indicate asymmetric type interactions with concentration-dependent antagonism and/or synergy. EC50INT,AB and EC50INT,BA represent the interaction potencies and HINT,AB and HINT,BA the interaction sigmoidicities. Note that Loewe Additivity cannot be solved explicitly for E when EC50A ≠ EC50B and/or HA ≠ HB, but implicitly by root finding methods. The technical aspects of solving the Loewe Additivity models are provided in the supplementary material.
Conventional Rich and Reduced Time-Kill Curve Designs for PD Interaction Studies
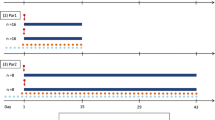
Two different designs based on the conventional microdilution checkerboard technique of two hypothetical anti-tubercular drugs, drug A and drug B, were used. A conventional rich study design with a ten-by-ten checkerboard, including 1 scenario of natural growth (no treatment), 9 scenarios of monotherapy with each single drug, and 81 scenarios of combinations, was employed based on 2-fold increasing static in vitro concentrations, ranging from 0.25 to 64 mg/L for drug A and drug B with a daily sample to 14 days after first administration (Fig. 2). A conventional reduced study design with a four-by-four checkerboard, including one scenario of natural growth (no treatment), three scenarios of monotherapy with each single drug and nine scenarios of combinations was employed with 8-fold increasing concentration, including 0, 8, 16, and 64 mg/L for drug A and drug B and with daily sampling to 14 days. All treatments started 4 days after infection and lasted for 14 days with daily treatment.
Illustration of the combinations of concentrations of drugs A and B in the a conventional rich design, b conventional reduced design, and c newly proposed optimized design. The sampling time points for each design are shown in (d). The conventional rich and reduced designs were evaluated for the analysis of pharmacodynamic interaction between drugs A and B using conventional models linked to the Multistate Tuberculosis Pharmacodynamic (MTP) model and the General Pharmacodynamic interaction (GPDI) model
Optimized Time-Kill Curve Design for PD Interaction Studies
An optimized design for the evaluation of PD interactions of drug combinations based on exposure levels at 0, EC20, EC50, and EC80 of each single drug is proposed in this study. The rationale behind this optimized design was to better capture changes in the potencies (EC50) of each drug, using information from the exposure–response relationships of each drug in monotherapy. The optimized design included one scenario of natural growth (no treatment), three scenarios of monotherapy with each single drug, and nine scenarios of combinations. Sampling time points for the optimized design are the same as the conventional rich and reduced study design (Fig. 2). All treatments started 4 days after infection and lasted for 14 days with daily treatment.
Explorative Studies with Conventional Models for Quantifying PD Interactions
In order to evaluate different analysis methods, including the GPDI model, an Empirical Bliss Independence-based model and the Greco model based on Loewe Additivity, the estimated interaction on a parameter level (EC50) obtained from estimating PD interactions using these models linked to the MTP model was evaluated. Only the conventional rich and reduced designs were used. Four different scenarios were simulated using the MTP-GPDI model with the same exposure–response relationship for hypothetical drug A (EmaxA = 1 day−1 and EC50A = 10 mg/L) in all evaluated scenarios, but a varied exposure–response relationship was used for hypothetical drug B, compared with drug A, including both different EmaxB and EC50B (EmaxB = 3 day−1 and EC50B = 16 mg/L), different EmaxB (EmaxB = 3 day−1) and same EC50B, same EmaxB and different EC50B (EC50B = 16 mg/L), and both same EmaxB and EC50B. Sigmoidicity constants HA and HB were set to 1 in each scenario. Details of each scenario are shown in Table II. For Loewe Additivity, only scenarios 3 and 4 were included, as Loewe Additivity is not defined for drugs exhibiting different maximum effects.
To explore the behavior of conventional models to assess different types of PD interactions (antagonism, synergism, or asymmetric interaction) or additivity within each scenario, the two interaction parameters INTAB and INTBA in the GPDI model were varied in the simulations and set to − 0.9, − 0.75, − 0.5, − 0.25, 0, 1.5, 4, 6.5, or 9 equivalent to − 90%, − 75%, − 50%, − 25%, 0%, 250%, 500%, 750%, or 1000% shift in EC50 of each single drug, respectively (Table II and Fig. 3). The simulated data were re-estimated using the MTP model linked only to conventional models such as an Empiric Bliss Independence-based model (Eq. 9) and the Greco model (Eq. 10) based upon Loewe Additivity:
where α and β are the PD interaction parameters to be estimated in Eqs. 9 and 10 for the conventional models. In Eq. 9, β = 1 indicated Bliss Independence; β < 1 indicated Bliss antagonism; β > 1 indicated Bliss synergy. EA and EB represent the effect generated from drug A or drug B in monotherapy using sigmoidal maximum effect models, which were then linked to stimulation of death of F bacterial state in the MTP model. In Eq. 10, α = 0 indicated Loewe Additivity, α < 0 indicated Loewe antagonism, or α > 0 indicated Loewe synergy. CA and CB are the drug A and B concentrations. EC50A and EC50B are drug concentrations of drug A and B that trigger the half-maximum effect, and Emax is the maximum effect in the Greco model. HA and HB are sigmoidicity parameters for each drug.
Classification on the parameter level by the Empirical Bliss model based on Bliss Independence using the conventional rich design (left) or the conventional reduced design (right). The color of each dot indicates the classification of the pharmacodynamic interaction by the Empirical Bliss Independence model: Bliss Independence (ADD in blue), antagonism (ANT in red), and synergy (SYN in green). The true classification is given by the four different sectors in each plot: the upper right sector shows ANT, the lower left sector shows SYN, and the upper left and lower right sectors show asymmetric interaction (ANT+SYN) Time-kill experiments of CFU versus time of two hypothetical drugs A and B with four different scenarios of different exposure–response relationships (1–4) were simulated using the conventional rich or reduced design (Fig. 2) and the Multistate Tuberculosis Pharmacometric (MTP) model linked to the General Pharmacodynamic Interaction (GPDI) model. In each scenario (1–4) of different exposure–response parameters of drugs A and B, the type of pharmacodynamic interaction (ANT, SYN, asymmetric interaction (ANT+SYN)) and no interaction were varied, resulting in 81 (9 × 9) different pharmacodynamic interactions where each simulated pharmacodynamic interaction is one dot in each plot. The x-axis shows the value (nine different values) of the interaction parameter INTAB of the GPDI model, presented as percent shift of the EC50 of drug A caused by drug B. The y-axis shows the distribution (nine different values) of the interaction parameter INTBA of the GPDI model, presented as percent shift of the EC50 of drug B caused by drug A.
The MTP model and exposure–response parameters for monotherapy (EmaxA, EC50A, HA, EmaxB, EC50B, and HB) were fixed in the re-estimations. The likelihood ratio test, based on the objective function value, was used to judge whether an interaction was significantly different from additivity. If estimation of the interaction parameters α or β resulted in a decrease of 3.84 (α < 0.05, df = 1, χ2 distribution) compared to a reduced model assuming additivity (β fixed to 1 for the Empiric Bliss Independence model and α fixed to 0 for the Greco model), the interaction was deemed statistically significant.
The stochastic simulation and estimation method (SSE) in NONMEM 7.3 (ICON Development Solution, Ellicott City, MD, USA) (12) and Perl-speaks-NONMEM (PsN, version 4.6.12; Department of Pharmaceutical Biosciences, Uppsala University, Sweden) (13) were used.
As the Loewe Additivity-based models in this study, i.e., the MTP-GPDI model in Loewe Additivity (Eq. 7) and the Greco model based on Loewe Additivity (Eq. 10), do not have an explicit solution, but require numeric techniques to be solved, a root finder function was written in C using the GNU scientific library (14). The C function was called from NONMEM® via NM-TRAN abbreviated functions and a Fortran interface using the Fortran 2003 iso_c_binding intrinsic module. Everything was compiled and linked to the NONMEM executable using version 5.1.1 of gcc and gfortran. The C code as well as the wrapper functions and instructions on how to link C code to NONMEM® are available in the supplementary material.
Evaluation of Conventional and Optimized Designs for PD Interaction Assessment
After the explorative evaluation of different analysis methods, including the GPDI model, an Empirical Bliss Independence-based model, and the Greco model, the performance of the GPDI model and conventional PD interaction models in a broader context with conventional (rich and reduced designs) and optimized designs were evaluated. The simulated data using the Loewe Additivity-based or Bliss Independence-based GPDI model were re-estimated using the MTP model linked to the GPDI model (based on Bliss Independence or Loewe Additivity), the Empiric Bliss Independence-based model, or the Greco model based on Loewe Additivity, in order to compare the estimated PD interactions to the true (simulated) interactions.
The MTP model and exposure–response parameters for monotherapy (EmaxA, EC50A, HA, EmaxB, EC50B, and HB) were fixed in the re-estimation. The PD interaction parameters (INTAB and INTBA) were estimated using the GPDI model. In order to provide more general conclusions for a variety of hypothetical drugs, EmaxA and EmaxB were randomly sampled from 1 to 5, EC50A and EC50B were randomly sampled from 0.1 to 64 mg/L, and HA and HB were randomly sampled from 1 to 2. As before, EmaxA and EmaxB were set to the same value in the case of Loewe Additivity-based models. The PD interaction parameters, EC50A and EC50B, were fixed to the true monotherapy EC50A and EC50B values. The parameters INTAB and INTBA were randomly sampled from − 0.9 to − 0.5 (synergistic interaction), from 0.5 to 25 (antagonistic interaction), and 0 (additivity, no interaction). Details of the parameter values in the simulations are shown in Table III.
In order to handle datasets that are dependent on the model parameters (optimized design), the SSE was performed in R software (version 3.2.4; R Foundation for Statistical Computing, Vienna, Austria). Differential equations were solved using the ‘deSolve’ package (15,16) (version 1.13). To improve performance, differential equations were encoded in C, compiled as a dynamically linked library (.dll) and linked to the ‘deSolve’ interface. Model parameters were estimated with ‘optim’ from ‘stats’ (version 3.2.4) using maximum-likelihood estimation. The GNU scientific library for C (14) was used to provide root finding functionality within the Ordinary Differential Equation system for the Loewe Additivity models. The R code and the C code can be obtained from the authors upon request.
Relative bias (rBias), relative imprecision (rRMSE), and correct classification rate of interaction parameters in the GPDI model using conventional rich design, reduced design, or optimized design were calculated with the true and estimated parameter values. The rBias (Eq. 11), rRMSE (Eq. 12), and correct classification rate (Eq. 13) were derived as:
where estimationi denotes the estimated parameter i value, truei is the true parameter i value used in the initial simulations, and N is the number of simulations for each set of truei (N = 1000).
RESULTS
Explorative Studies with Conventional Models for PD Interaction Studies
Figure 3 displays the results obtained from estimating the Empirical Bliss Independence model in a conventional rich and reduced design for four different scenarios of Emax and EC50 of the two hypothetical drugs using the MTP–GPDI model as the underlying “true” model. The true interactions on a parameter level (EC50) of the simulated combinations are displayed in Fig. 3 as percent shift of EC50A or EC50B. In each plot, four sectors are shown as true antagonistic interactions on a parameter level (i.e., both EC50 values shifted < 0%), true synergistic interaction (i.e., both EC50 values shifted > 0%), true asymmetric interaction (i.e., one EC50 value shifted to < 0% and the other EC50 value shifted to > 0%, leading to concentration-dependent antagonism and/or synergy on the effect level), and additivity (i.e., both EC50 values shift at 0%). The estimated interactions on a parameter level from the Empiric Bliss Independence model are color-coded, i.e., antagonism (ANT in red), synergy (SYN in green), and no interaction (ADD in blue) as quantified by the interaction parameter α. Figure 4 displays the result from estimating the Greco model using the Loewe Additivity-based MTP–GPDI model while the same visualization modalities were applied as in Fig. 3, presenting the result of the estimated interaction parameter β.
Classification by the Greco model based on Loewe Additivity using the conventional rich design (left) or the conventional reduced design (right). The color of each dot indicates the classification of the pharmacodynamic interaction by the Greco model using the GPDI model as underlying true model: Loewe Additivity (ADD in blue), antagonism (ANT in red), and synergy (SYN in green). The true classification is given by the four different sectors in each plot: the upper right sector shows ANT, the lower left sector shows SYN, and the upper left and lower right sectors show asymmetric interaction (ANT+SYN). Numbers 1, 2, and 3 on the right side of dots are arbitrary chosen for Fig. 5 to illustrate the interaction surface for symmetric (1 and 3) and an asymmetric (2) interaction scenario. Time-kill experiments of CFU versus time of two hypothetical drugs A and B with two different scenarios of different exposure–response relationships (3 and 4) were simulated using the conventional rich or reduced design (Fig. 2) and the Multistate Tuberculosis Pharmacometric (MTP) model linked to the General Pharmacodynamic Interaction (GPDI) model. In each scenario (3 and 4) of different exposure–response parameters of drugs A and B, the type of pharmacodynamic interaction (ANT, SYN, asymmetric interaction (ANT+SYN)) and no interaction were varied, resulting in 81 (9 × 9) different pharmacodynamic interactions where each simulated pharmacodynamic interaction is one dot in each plot. The x-axis shows the distribution (nine different values) of the interaction parameter INTAB of the GPDI model, presented as percent shift of the EC50 of drug A caused by drug B. The y-axis shows the distribution (nine different values) of the interaction parameter INTBA of the GPDI model, presented as percent shift of the EC50 of drug B caused by drug A
In the conventional rich design, the Empirical Bliss model (Fig. 3) and the Greco model (Fig. 4) correctly captured antagonism and synergy. However, in situations with asymmetric interactions, the conventional models only estimated symmetric antagonism (ANT in red), synergy (SYN in green), and defined additivity (ADD in blue) and were intrinsically unable to estimate asymmetric interactions. We have arbitrarily selected three points from Fig. 4 representing synergy, asymmetric interaction, and antagonism simulated from the GPDI model (Fig. 5), but estimated as synergy, additivity, and antagonism by the Greco model. It is apparent that the asymmetric case (scenario 2) displays regions of antagonism and synergy, which cannot be captured by the Greco model that wrongly classified this scenario as additive. Moreover, the obtained classification in the asymmetric region of the interaction space was apparently dependent on the maximum effect as well as the potency of the interacting drug A and B, and it was difficult to foresee if an interaction would result in synergy, antagonism, or no interaction in the asymmetric sectors.
Predicted log10 CFU/mL in numbers and log10 CFU/mL deviation from Loewe Additivity (in shaded areas) for hypothetical drug A and drug B in different combinations at 7 days after treatment. Plots 1, 2, and 3 represent corresponding selected point 1—estimated synergy, 2—estimated additivity, and 3—estimated antagonism by the Greco Model in Fig. 4. In each plot, concentrations of drug A and drug B are shown on x-axis and y-axis, respectively. White areas in the figure show Loewe Additivity, whereas red shaded areas show higher log10 CFU/mL (antagonism) than additivity and green shaded areas show lower log10 CFU/mL (synergism) than additivity
Similar patterns of interaction classification were also observed in the conventional reduced design in the scenario with differing Emax between drug A and B. In the scenarios with the same Emax for drug A and B, most of the estimations from conventional models were not significant to estimate and therefore defined additivity as indicated by the blue color coding, regardless which PD interactions were simulated from the MTP–GPDI model.
Evaluation of Conventional and Optimized Designs for PD Interaction Assessment
After the explorative studies, we assessed whether conventional rich and reduced designs were suitable to support estimation of parameters from the GPDI model using SSE for 1000 combinations of the two hypothetical drugs A and B. In addition, the correct interaction classification rate was calculated for the GPDI models, as well as for the Empiric Bliss Independence and the Greco model for comparison.
Regardless of the underling additivity criterion, the rBias and rRMSE of the conventional rich design were lowest overall, as shown in Table IV. The conventional reduced design was the least informative design for estimating GPDI parameters, as seen from the highest rBias up to 7.56% and highest rRMSE up to 78.9%. The optimized design based on EC20, EC50, and EC80 of the respective single drugs resulted in lower rBias and lower rRMSE than the reduced design at the same sample size. The rRMSE of the optimized designs was up to 1.6-fold lower than those in the reduced design.
In Table V, the correct classification rate (%) is presented for the conventional rich, reduced, and optimized design. The rich and optimal designs provided similarly high correct classification rates of > 93.5%, including the asymmetric type of interaction. In contrast, the conventional models could not classify the asymmetric interactions at all, shown as a 0% correct classification rate. However, “pure” antagonism or synergy were also correctly classified by the Empiric Bliss Independence and the Greco model in > 97.5% of the scenarios.
DISCUSSION
Combination therapies represent a major challenge in drug development. In particular, when drugs are added to existing clinical combination regimens, as done in anti-tubercular drug development, clinical evaluation of PD drug interactions is barely possible. Consequently, pre-clinical information has to be exploited and integrated along the drug development process to inform clinical drug development and selection for Phase 2a/b regimens.
The present study provides a systematic investigation of the novel MTP–GPDI model approach, its performance in comparison to conventional models for PD interactions, and experimental design recommendations. In this study, we demonstrated that the GPDI model is an advantageous model-based method for evaluating PD interactions for TB drugs compared to conventional models, as it also allows for quantification of asymmetric interactions, which were not quantifiable by the conventional methods. Although the present study focused on TB as the therapeutic area, the concepts behind the GPDI model are applicable to other therapeutic areas where combination therapies are prevalent, such as oncology, neurology, or anesthesia.
The MTP model allows for quantification of anti-tubercular drug effects on different distinct subpopulations, i.e., fast-, slow-, and non-multiplying bacteria. To be able to directly relate the estimates of the GPDI model to the estimates of the conventional models, we only evaluated a single effect site—stimulation of the death of fast-multiplying bacteria—in the present simulation study. However, in the case of multiple effect sites, conclusions on synergy or antagonism may not directly relate to parameter estimates pointing to synergy or antagonism in the GPDI model as interactions can occur at several different mechanistic levels in the MTP model, i.e., inhibition of the growth of F bacterial state and/or stimulation of the death of F, S, or N bacterial states. Therefore, conclusions on drug interactions should be drawn on the biomarker level in such situations, i.e., predicted change in CFU based on the interaction model compared to a model with only expected additivity of the monotherapy effects of the drugs. Moreover, future studies should evaluate optimized designs for situations with multiple effect sites, which may require the addition of further concentrations to the design if drug potencies at the effect sites differ.
The term “asymmetric interaction” was introduced earlier (10) and summarizes concentration-dependent synergistic and antagonistic behavior of interacting drugs, which can only be quantified with the GPDI model approach. The GPDI concept (10) was developed using 200 drug combination experiments emanating from a high-throughput screening investigation and up to 38% of the observed interactions were asymmetric. Hence, the asymmetric interaction type might represent an important and relevant class of PD interactions that conventional model-based approaches, including the evaluated Empirical Bliss model and the Greco model based on Loewe Additivity, cannot intrinsically evaluate. In the first part of the present work, we explored how conventional models compare to the GPDI model in assessment of anti-tubercular drug PD interactions on a parameter level. Thereafter, different scenarios based on Emax and EC50 for hypothetical drugs A and B were created and mimicked realistic situations. While there was good agreement overall between the conventional models and the GPDI models for “pure” synergy and antagonism, the conventional models were not able to classify the asymmetric interactions correctly and there was no clear pattern of what conventional models would classify in the asymmetric region of the PD interaction space (Figs. 3 and 4). Hence, conventional models may provide erratic classification results in these scenarios and may likely lead to wrong decisions on the nature of a PD interaction, which may harm decision-making in a drug development program. Moreover, the conventional models are tied to a single additivity criterion. For instance, the Empirical Bliss model is based on Bliss Independence and the Greco model is based on Loewe Additivity, whereas the GPDI model is compatible with various additivity criteria, including both of the latter criteria. The advantage of compatibility with different additivity criteria is that no assumption about additivity has to be made and several criteria can be evaluated. In addition, INTAB and INTBA characterized the maximum fractional change of the respective PD parameters from a single drug, here EC50. Hence, the interaction parameters of the GPDI model are interpretable, which is not possible when conventional models are applied.
The second part of this study focused on the performance of the MTP-GPDI model under different experimental designs. In addition to conventionally used checkerboard designs, termed rich and reduced design in the present work, an optimized design was proposed, which was based on EC20, EC50, and EC80 of each single drug defined in the monotherapy. The rich design was based on the ten-by-ten microdilution checkerboard design, resulting in a labor-intensive set of 81 scenarios of two-drug combination experiments. The reduced design was as a small version of the rich design and spanned the same concentration range as the rich design, but with a four-by-four checkerboard at lower granularity than the rich design yielding nine scenarios of two-drug combination experiments. The proposed optimized design, based on 0, EC20, EC50, and EC80 of each drug, also resulted in nine scenarios of two-drug combination experiments. However, our analysis demonstrated that the optimized design was much more informative than the reduced design and enabled estimates of both the GPDI interaction parameters INTAB and INTBA at lower rBias and lower rRMSE compared to the conventional reduced design. Notably, rBias was even 0.25-fold lower in the optimized design than that in the conventional rich design. Thus, our study reflected a very wide range of potential interacting drugs with varied Emax from 1 to 5 day−1, EC50 from 0.1 to 64 mg/L, and H from 1 to 2 for each single drug (17). In addition to the superior rBias and rRMSE estimates for the optimized design, the classification rate was also very close to the rich design and even > 93.5% for the asymmetric interaction type that was not possible to identify with the conventional PD interaction models at all. The optimized design proposed in this study thus provides more information on PD interactions than the conventional reduced design.
CONCLUSION
The GPDI model, which provides model-based estimates of PD interactions given as symmetric and asymmetric percent shift of PD parameters of single drug effects and is compatible with different additivity criteria, was shown to be advantageous to other conventional models, as it correctly quantifies asymmetric interactions. The optimized design, which was based on EC20, EC50, and EC80 of each single drug, provided lower rBias and lower rRMSE than a conventional reduced four-by-four checkerboard study design at the same total number of samples required. The MTP–GPDI model is an advantageous approach to other conventional models for assessing PD interactions of anti-tubercular drugs and provides key information for selection of drug combinations for Phase 2b evaluation.
References
Greco WR, Park HS, Rustum YM. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1990;50:5318–27.
Boucher AN, Tam VH. Mathematical formulation of additivity for antimicrobial agents. Diagn Microbiol Infect Dis. 2006;55:319–25.
Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol [Internet]. 1939;26:585–615. Available from: https://doi.org/10.1111/j.1744-7348.1939.tb06990.x
Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–90.
Clewe O, Aulin L, Hu Y, Coates ARM, Simonsson USH. A multistate tuberculosis pharmacometric model: a framework for studying anti-tubercular drug effects in vitro. J Antimicrob Chemother [Internet]. 2016;71:964–74. Available from: http://www.jac.oxfordjournals.org/lookup/doi/10.1093/jac/dkv416
Chen C, Wicha SG, de Knegt GJ, Ortega F, Alameda L, Sousa V, et al. Assessing pharmacodynamic interactions in mice using the multistate tuberculosis pharmacometric and general pharmacodynamic interaction models. CPT Pharmacometrics Syst Pharmacol [Internet]. 2017;6:787–97. Available from: http://doi.wiley.com/10.1002/psp4.12226
Chen C, Ortega F, Rullas J, Alameda L, Angulo-Barturen I, Ferrer S, et al. The multistate tuberculosis pharmacometric model: a semi-mechanistic pharmacokinetic–pharmacodynamic model for studying drug effects in an acute tuberculosis mouse model. J Pharmacokinet Pharmacodyn [Internet]. 2017;44:133–41. Available from: http://link.springer.com/10.1007/s10928-017-9508-2
Svensson RJ, Simonsson USH. Application of the multistate tuberculosis pharmacometric model in patients with rifampicin-treated pulmonary tuberculosis. CPT Pharmacometrics Syst Pharmacol. 2016;5:264–73.
Wicha SG, Clewe O, Svensson RJ, Gillespie SH, Hu Y, Coates ARM, et al. Forecasting clinical dose-response from pre-clinical studies in tuberculosis research—translational predictions with rifampicin. Clin Pharmacol Ther [Internet]. 2018; Available from: http://www.ncbi.nlm.nih.gov/pubmed/29700814
Wicha SG, Chen C, Clewe O, Simonsson USH. A general pharmacodynamic interaction model identifies perpetrators and victims in drug interactions. Nat Commun. 2017;8:2129.
Clewe O, Wicha SG, de Vogel CP, de Steenwinkel JEM, USH S. A model-informed preclinical approach for prediction of clinical pharmacodynamic interactions of anti-TB drug combinations. J Antimicrob Chemother [Internet]. 2018;73:437–47. Available from: http://academic.oup.com/jac/article/doi/10.1093/jac/dkx380/4609339
Beal SL, Sheiner LB, Boeckmann AJ, Bauer RJ. NONMEM User’s Guides (1989–2009). Ellicott City: Icon Development Solutions; 2009.
Keizer RJ, Karlsson MO, Hooker A. Modeling and simulation workbench for NONMEM: tutorial on Pirana, PsN, and Xpose. CPT Pharmacometrics Syst Pharmacol. 2013;2:e50.
Galassi M. GNU Scientific Library Reference Manual. 3rd ed.
Soetaert K, Petzoldt T, Setzer RW. Solving differential equations in R. R J. 2010;2:5–15.
Soetaert K, Petzoldt T, Setzer RW. R Package deSolve, Writing Code in Compiled Languages. :1–51.
Kirby S, Brain P, Jones B. Fitting E(max) models to clinical trial dose-response data. Pharm Stat. 2011;10:143–9.
Acknowledgments
The work was supported by the Swedish Research Council, the Innovative Medicines Initiative Joint Undertaking (www.imi.europa.eu) under grant agreement no. 115337, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in-kind contribution. The funding parties were not involved in the study design, the analysis, and interpretation of data, preparation, review, or approval of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, C., Wicha, S.G., Nordgren, R. et al. Comparisons of Analysis Methods for Assessment of Pharmacodynamic Interactions Including Design Recommendations. AAPS J 20, 77 (2018). https://doi.org/10.1208/s12248-018-0239-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-018-0239-0