Abstract
Background
Intrinsic pathway of apoptosis is generally mediated by BCL-2 (B cell lymphoma 2) family of proteins; they either induce or inhibit the apoptosis. Overexpression of BCL-2 in cancer cell may lead to delay in apoptosis. BCL-w is the pro-survival member of the BCL-2 family. BCL2L2 gene is present on chromosome number 14 in humans, and it encodes BCL-w protein; BCL-w protein is 193 amino acids residues in length. Interactions among the BCL-2 proteins are very specific. The fate of cell is determined by the ratio of pro-apoptotic proteins to pro-survival proteins. BCL-w promotes cell survival. Studies suggested that overexpression of BCL-w protein is associated with many cancers including DLBCL, BL, colorectal cancers, gastric cancers, and many more. The cause of overexpression is translocations or gene amplification which will subsequently result in cancerous activity.
Process
For in-silico analysis, BCL2L2 gene was retrieved from UniProt (UniProt ID: Q92843). 54 missense variants have been collected in BCL-w proteins from COSMIC database. Different tools were used to detect the deleteriousness of the variants.
Result
In silico mutational study reveals how the non-synonymous mutations directly affect the protein’s native structure and its function. Variant mutational analysis with PolyPhen-2 revealed that out of 55 variants, 28 of the missense mutations was probably damaging with a score ranging from 0.9 to 1, while 24 variants were benign with a score ranging from 0 to 0.4.
Conclusions
This in silico work aims to determine how missense mutations in BCL-w protein affect the activity of the protein, the stability of the protein, and to determine the pathogenicity of the variants. Prediction of pathogenicity of variants will reveal if the missense mutation has a damaging effect on the native structure of protein or not. Prediction of protein stability will reveal whether the mutation has a stabilizing or destabilizing effect on the protein.
Similar content being viewed by others
Background
BCL-2 family of proteins are associated with mitochondrial-mediated cell death. The proteins of BCL-2 family either inhibits or induces cell death. On the basis of BH domain, members are classified into three groups [1]. The pro-survival proteins possess BH1-4 domains e.g. BCL-2, BCL-XL, MCL1 [2,3,4], BCL-w, and A1/BFL-1. Multi-domain pro-apoptotic proteins contains BH1-3 domains, e.g., BAX and BAK [2,3,4,5], and lastly the BH3 only pro-apoptotic proteins which are further classified as activators or sensitizers. BAD, BIK, BMF are sensitizers and BIM, tBID, and PUMA are activators [2, 6]. Here, sensitizers do not bind to BAK and BAX [2, 7, 8] while the BH3 domain of the activators binds to BAK and BAX and induces conformational change that results in the oligomerization of these proteins in the outer membrane of the mitochondria, this oligomerization results in MOMP formation [2, 9]. In cytosol, cytochrome c (released from mitochondria intermembraned space) with Apaf-1, caspase 9, and ATP [10,11,12] forms a complex also known as apoptosome. This complex cleaves off and activates the caspase 3 that results in apoptosis.
BCL-w is the pro-survival protein in the BCL-2 family. BCL2L2 gene present on chromosome number 14 in humans encodes the BCL-w protein and this protein is 193 amino acids residues in length [2, 13]. BCL-w protein is generally found on the outer membrane of the mitochondria [2, 14]. The BCL-w protein consists of nine α helices with flanking amphipathic helices α1 (10−24 residues), α2 (43−56), α3 (62−68), α4 (76−87), α6 (116−132), α7 (134−141), α8 (144−150), α9 (157−173), and central hydrophobic groove formed by helix, α5 (93−111).
BCL-w is found in the testes, colon, brains, and cells with lymphoid and myeloid origin [2, 13, 15]. Studies suggested that BCL-w is involved in spermatogenesis [2, 15] and is majorly expressed in spermatocytes, Leydig cells, Sertoli cells and spermatogonia, BCL-w also promotes their survival [2, 16, 17]. Experimental studies also suggest that overexpression of this protein might results in spermatocytes degeneracy, decline in the number of spermatogonia and vacuolization of sertoli cells [2, 18]. BCL-w also promotes the survival of gut epithelial cells [2, 15], prevents small intestine cells and mid-colon cells from death [2, 19], it also promotes enterocyte survival and B lymphocyte survival [2, 20]. High level of BCL-w also estimated in some areas of brain such as mature brain, sensory neurons, hippocampus and cerebellum [2, 21, 22]. BCL-w has also been involved in the development of dendrite and it controls the morphogenesis of mitochondria. BCL-w has also been involved in disorders of nervous system such as Alzheimer’s disease and Parkinson’s diseases, the cause of these diseases is the increased level of BCL-w. Overexpression of BCL-w is associated with ischemic brain [2, 23]. Overexpression of the BCL2L2 results in the survival of megakaryocytes and increased platelet formation [2, 24].
Genetic alterations in BCL2L2 contributes to many cancers such as copy number variations in small [2, 25] and non-small [2, 26] lung cancer, high level of BCL-w contributes to gastric carcinomas, and low BCL-w expression contributes to colorectal cancer [2, 27]. Patients with breast cancers significantly have high BCL-w mRNA level [2, 28, 29]. BCL-w has significantly involved with the cancer of urinary system [2, 30]. Overexpression of BCL-w is associated with cervical cancer, prostate cancer, hepatocellular carcinoma (HCC) and leiomyosarcomas. Expression of BCL-w is significantly higher in DLBCL, BL, CML [2, 31], and B-CLL [2, 32].
The interaction of pro-survival protein, i.e., BCL-w with pro-apoptotic proteins initiates the process of apoptosis but any dysregulation in these interactions will block the apoptotic pathway. Any chemical or amino acid alterations in the protein will interrupt the interactions between pro-survival proteins and pro-apoptotic proteins. Understanding of these mutations will help us to understand if the mutation is involved in any disease. This in silico study helps us to define the role of missense variants of BCL-w, which may alter proteins native structure and its function. By examining the role of mutation on biological function, we can determine the correlation between the mutation and the disease. The missense variants retrieved from this study were subjected to some in silico prediction tools such as Polyphen-2, SIFT, Provean, FATHMM, mutation assessor and stability prediction namely I-mutant 2.0, iStable, SAAFEC, SDM, DUET, and mCSM (Table 1).
Method
Data collection—selection of the BCL-w variants
For in silico analysis, BCL2L2 gene was retrieved from UniProt (UniProt ID: Q92843). 54 missense variants have been collected in BCL-w proteins from COSMIC database. Among these, neither of the variants were listed in the ClinVar.
Variants pathogenicity prediction
For predicting the deleteriousness of the variants, the in silico pathogenicity prediction tools that were used were PolyPhen-2 [33], SIFT [34], Provean [35,36,37], FATHMM [38], and Mutation Assessor [39].
Protein stability analysis
For predicting the of effect of amino acid change on the native BCL-w protein, I-mutant 2.0 [40], MUpro [41], and iStable [42], SAAFEC [43], SDM [44], DUET [45], and mCSM [46] web servers were used. I-mutant 2.0 is a web server that determines the change in stability due to point mutation or missense mutation. MUpro web server is a program that predicts the protein stability due to alteration in the sequence. Integrated predictor iStable was used for the predicting the stability of the protein, iStable may require both the sequence and the structure as an input. SAAFEC is a web server used to compute the energy changes due to single mutation. SDM (site-directed mutator) is an online server is that is also used for predicting the effect of point mutation on the protein stability. DUET is a web tool for the estimation of consequence of single mutation on proteins stability and its function. mCSM, a web tool used to estimate the impact of point mutation on protein stability, protein-protein-binding, and protein-DNA binding.
Result
Pathogenecity prediction of BCL-w missense variants
Variant mutational analysis with PolyPhen-2 revealed that out of 55 variants 28 of the missense mutations was probably damaging with score ranging from 0.9 to 1, while 24 variants were benign with score ranging from 0 to 0.4. PolyPhen-2 evaluates the damaging effect of point mutation by mapping SNPs to gene transcripts. From SIFT analysis, 28 out of 55 variants were deleterious, i.e., not tolerant with score ranging from 0 to 0.76, remaining 27 variants were tolerant (score range 0.76–1). Provean analysis revealed that 34 of the variants were neutral rest 20 were deleterious (one mutation, i.e., Q133R shows error) (Table 2). FATHMM analysis shows that 49 of the variants were deleterious, i.e., with score ≥ 0.67 rest 6 variants were neutral, i.e., no impact on the proteins native structure and function. Mutation assessor tool predicts the impact of point mutation on protein sequence and has revealed that 29 variants have low value while 15 variants have medium effect and 11 mutations have neutral effect.
Note: PolyPhen-v2 score less than 0.5 is considered to be tolerated and more than 0.5 is considered to be deleterious. SIFT score ranges from 0.0 to 0.05 are considered to be deleterious while score near 1.0 are considered to be tolerated; Provean score equals to or below − 2.5 are considered to be deleterious while score above − 2.5 are considered to be neutral; FATHMM score equals to or above 0.67 are deleterious; mutation assessor score prediction: 0–1 is neutral, 1–2 low, and above 2 medium.
Protein stability analysis
Pathogenic missense mutations cause change in free energy which further leads to alteration in protein stability. Here, BCL-w variants were subjected to various protein stability tools for analyzing change in free energy due to point mutation. I-Mutant 2.0, MUpro, iStable, SAAFEC, SDM, DUET, and mCSM tools were used for determining the protein stability. The tools revealed that the variants decrease the protein stability by showing a destabilizing or decreasing energy as result. I-Mutant2.0, MUpro, mCSM, SDM, DUET, and SAAFEC tools shows the more negative ΔΔG value (ΔΔG > 0) shows the more destabilizing effect of the mutation, while the more positive ΔΔG value (ΔΔG <0) shows stability decrease in case of iStable tool.
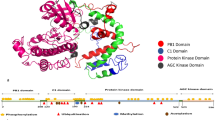
Some of the servers require fasta format while some require PDB structure or PDB ID as an input. I-Mutant 2.0, MUpro, iStable, and SAAFEC use fasta format while SDM, DUET, and mCSM need PDB structure or PDB ID as an input. Some post-translational modifications that takes place during the conversion of peptide sequence to 3D structure may cause deletion of amino acids residue, i.e., some part of the protein may not be included in the crystallographic structure, as small peptide sequence yields a better crystal quality or structure of a protein is extracted from a crystal structure from proteins complex and isolating some proteins from complex of proteins may cause differences in the sequence in fasta format to sequence in PDB structure. Now, the fasta format of BCL-w starts from MATPA, while amino acid sequence in PDB structure starts from ATP, as shown in Fig. 1 for this reason, mutation given in DUET, SDM, and mCSM as A158V instead of A159V, besides this some of the amino acids are not included in the sequence of PDB structure due to these modifications are Q132R, V185A, and A187P as shown in Table 3.
Discussion
Present in silico mutational study reveals how the non-synonymous mutations directly affect the proteins native structure and its function. The activity of the protein complex and its function depends on the complex formed between proteins; the interactions between proteins might be necessary for molecular features like cell signaling and cell regulation. The protein complex formed may be homodimer or heterodimer are formed due to interactions between proteins. The missense mutations at the interface of the protein-protein interaction (PPI) causes disruption in the shape, size, and secondary structure of the complex. For the specific function of the protein complex, there should be presence of stable interaction between proteins. Moreover, mutation of large amino acids into a smaller amino acid causes gaps while mutation of smaller one leads to bumps or inter-molecular clashes. BCL-w, has a pro-survival function, and is also involved in normal as well as diseased cells and disorders of nervous system and cancer. The protein–protein interactions gets disturbed due to non-synonymous mutation which may lead to diseased state. The structure of the protein is directly influenced by its function and its stability. The genetic variations, i.e., amino acid change that represses its property directly influences all other properties. The hydrogen bonds within amino acid residues maintains the protein stability, i.e., reduced H-bonds may cause loss of stability of the protein while higher H-bonds may increase the protein stability. The structural changes caused due to variants corresponds to physicochemical properties of the proteins like size, charge, hydrophobicity, molecular weight, and side chains. These changes further causes alteration in the chemical properties which may be necessary for maintaining secondary, tertiary, and quaternary structure of proteins.
Most pathogenic variants destabilizes the 3D structure, stability, and folding-free energy of the protein, which subsequently results in disruption in proteins function and regulation [47, 48].
Conclusion
Proteins are dynamic in nature as they are flexible in nature due to temperature, pH, and interaction with other molecule may be a ligand. Understanding of proteins native conformation may reveal the role of variants in diseased condition. The activity and function of the protein complex is determined by its interaction with other proteins. However, the stability of a protein complex can disrupt due to mutations in the protein. This in silico study has estimates the efficiency of various pathogenicity prediction tools and stability analysis tools for BCL-w variants and the study may help in characterization of mutations in the protein complex and molecular level. Furthermore, the result indicates that the missense mutation alters the stability of BCL-w.
Availability of data and materials
NA
Abbreviations
- MCL-1:
-
Myeloid cell leukemia-1
- BCL-w:
-
B cell lymphoma-w
- A1/BFL-1:
-
BCL-2-related protein A1/BCL-2 related isolated from fetal liver-11
- BAX:
-
BCL-2-associated X protein
- BAK:
-
BCL-2 antagonist/killer
- BAD:
-
BCL-2-associated agonist of cell death
- BIK:
-
BCL-2 interacting killer
- BMF:
-
BCL-2-modifying factor
- BIM:
-
BCL-2-interacting mediator of cell death
- tBID:
-
Truncated form of BH3-interacting domain death agonist
- PUMA:
-
p53-upregulated modulator of apoptosis
- MOMP:
-
Mitochondrial outer membrane permeabilization
- CML:
-
Chronic myeloid lymphoma
- Apaf-1:
-
Apoptosis protease activating factor 1
- B-CLL:
-
B cell chronic lymphocytic leukemia
- PolyPhen-2:
-
Polymorphism phenotyping-2
- SIFT:
-
Sorting intolerant from tolerant
- Provean:
-
Protein variation effect analyzer
- FATHMM:
-
Functional analysis through hidden Markov models
References
Wong RSY (2011) Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30(1):1–14. https://doi.org/10.1186/1756-9966-30-87
Hartman ML, Czyz M (2020) BCL-w: apoptotic and non-apoptotic role in health and disease. Cell Death Dis 11:4. https://doi.org/10.1038/s41419-020-2417-0
Singh R, Letai A, Sarosiek K (2019) Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20(3):175–193. https://doi.org/10.1038/s41580-018-0089-8
Hartman ML, Czyz M (2012) Pro-apoptotic activity of BH3-only Proteins and BH3 Mimetics : from Theory to Potential Cancer Therapy, pp 966–981
Hartman ML, Czyz M (2013) Anti-apoptotic proteins on guard of melanoma cell survival. Cancer Lett. 331(1):24–34. https://doi.org/10.1016/j.canlet.2013.01.010
Dutta S, Gullá S, Chen TS, Fire E, Grant RA, Keating AE (2010) Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J. Mol. Biol. 398(5):747–762. https://doi.org/10.1016/j.jmb.2010.03.058
Knight T, Luedtke D, Edwards H, Taub JW, Ge Y (2019) A delicate balance – The BCL-2 family and its role in apoptosis, oncogenesis, and cancer therapeutics. Biochem. Pharmacol. 162:250–261. https://doi.org/10.1016/j.bcp.2019.01.015
Shamas-Din A et al (2014) Multiple partners can kiss-and-run: Bax transfers between multiple membranes and permeabilizes those primed by tBid. Cell Death Dis. 5(6):e1277–e1277. https://doi.org/10.1038/cddis.2014.234
Gama V, Deshmukh M (2015) Life after MOMP. Mol. Cell 58(2):199–201. https://doi.org/10.1016/j.molcel.2015.03.035
Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1(7):949–957. https://doi.org/10.1016/S1097-2765(00)80095-7
Singh K, Briggs JM (2016) Mutation research / reviews in mutation research functional implications of the spectrum of BCL2 mutations in lymphoma. Mutat. Res. Mutat. Res. 769:1–18. https://doi.org/10.1016/j.mrrev.2016.06.001
Li P et al (1997) Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell 91(4):479–489. https://doi.org/10.1016/S0092-8674(00)80434-1
Gibson L et al (1996) bcl-w, a novel member of the bcl-2 family, promotes cell survival. Oncogene 13(4):665–675 [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed/8761287
Lapham A, Adams JE, Paterson A, Lee M, Brimmell M, Packham G (2009) The Bcl-w promoter is activated by β -catenin / TCF4 in human colorectal carcinoma cells. Gene 432(1–2):112–117. https://doi.org/10.1016/j.gene.2008.12.002
Reilly LAO et al (2001) Tissue expression and subcellular localization of the pro-survival molecule Bcl-w, pp 486–494
Yan W, Samson M, Je B, Toppari J (2015) Bcl-w forms complexes with Bax and Bak, and elevated ratios of Bax / Bcl-w and Bak / Bcl-w correspond to spermatogonial and spermatocyte apoptosis in the testis, pp 682–699
Russell LD et al (2001) Spermatogenesis in Bclw-deficient mice1. Biol. Reprod. 65(1):318–332. https://doi.org/10.1095/biolreprod65.1.318
Yan WEI et al (2015) Overexpression of Bcl-w in the testis disrupts spermatogenesis : revelation of a role of BCL-W in. Mol Endocrinol 17:1868–1879. https://doi.org/10.1210/me.2002-0389
Pritchard DM, Print C, Reilly LO, Adams JM, Potten CS, Hickman JA (2000) Bcl-w is an important determinant of damage-induced apoptosis in epithelia of small and large intestine
Stern LE et al (2000) Epidermal growth factor alters the bax : bcl-w ratio following massive small bowel resection. J Surg Res 1(42):38–42. https://doi.org/10.1006/jsre.2000.5897
Skoglo Y (1999) Differential expression of bcl-w and bcl-x messenger rna in the developing and adult rat nervous system. Neuroscience 91(2):673–684
Middleton G, Wyatt S, Ninkina N, Davies AM (2001) Reciprocal developmental changes in the roles of Bcl-w and Bcl-x(L) in regulating sensory neuron survival. Development. 128(3):447–57. https://doi.org/10.1242/dev.128.3.447
Zhu X et al (2004) Neuroprotective properties of Bcl-w in Alzheimer disease, pp 1233–1240. https://doi.org/10.1111/j.1471-4159.2004.02416.x
Pease-raissi SE et al (2017) Article paclitaxel reduces axonal Bclw to initiate IP 3 R1-dependent axon degeneration. Neuron 96(2):373–386.e6. https://doi.org/10.1016/j.neuron.2017.09.034
Kim YH et al (2006) Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene 2005:130–138. https://doi.org/10.1038/sj.onc.1208997
Kawasaki T et al (2007) BCL2L2 is a probable target for novel 14q11.2 amplification detected in a non-small cell lung cancer cell line. Cancer Sci 98(7):1070–1077 10.1111/j.1349-7006.2007.00491.x
Zhang B et al (2014) Loss of Smad4 in colorectal cancer induces resistance to 5-fluorouracil through activating Akt pathway. Br. J. Cancer 110(4):946–957. https://doi.org/10.1038/bjc.2013.789
Kim ES, Choi JY, Hwang SJ, Bae IH (2019) Hypermethylation of miR-205-5p by IR governs aggressiveness and metastasis via regulating Bcl-w and Src. Mol. Ther. Nucleic Acid 14:450–464. https://doi.org/10.1016/j.omtn.2018.12.013
Ding W, Ren J, Ren H, Wang D (2017) Long noncoding RNA HOTAIR modulates MiR-206-mediated Bcl-w signaling to facilitate cell proliferation in breast cancer. Sci. Rep.:1–9. https://doi.org/10.1038/s41598-017-17492-x
Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu D (2011) microRNA-203 suppresses bladder cancer development by repressing bcl-w expression. FEBS J 278:786–792. https://doi.org/10.1111/j.1742-4658.2010.07997.x
Raimondo S et al (2015) Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism, pp 1–12. https://doi.org/10.1186/s12964-015-0086-x
Sanz L, Garcia-marco A, Casanova B, Garc M, Garcia-pardo A, Silva A (2004) Bcl-2 family gene modulation during spontaneous apoptosis of B-chronic lymphocytic leukemia cells. Biochem Biophys Res Commun 315:562–567. https://doi.org/10.1016/j.bbrc.2004.01.095
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet 76:1. https://doi.org/10.1002/0471142905.hg0720s76
Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40(W1):W452–W457. https://doi.org/10.1093/nar/gks539
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7(10):e46688. https://doi.org/10.1371/journal.pone.0046688
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31(16):2745–2747. https://doi.org/10.1093/bioinformatics/btv195
Choi Y (2012) A fast computation of pairwise sequence alignment scores between a protein and a set of single-locus variants of another protein, in Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine - BCB. 12:414–417. https://doi.org/10.1145/2382936.2382989
Shihab HA et al (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34(1):57–65. https://doi.org/10.1002/humu.22225
Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39(17):e118–e118. https://doi.org/10.1093/nar/gkr407
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33:W306–W310. https://doi.org/10.1093/nar/gki375
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. 1132(2005):1125–1132. https://doi.org/10.1002/prot.20810
Chen C-W, Lin J, Chu Y-W (2013) iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinformatics 14(S2):S5. https://doi.org/10.1186/1471-2105-14-S2-S5
Li G, Panday SK, Alexov E (2021) SAAFEC-SEQ: a sequence-based method for predicting the effect of single point mutations on protein thermodynamic stability. Int. J. Mol. Sci. 22(2):606. https://doi.org/10.3390/ijms22020606
Worth CL, Preissner R, Blundell TL (2011) SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 39(suppl):W215–W222. https://doi.org/10.1093/nar/gkr363
Pires DEV, Ascher DB, Blundell TL (2014) DUET : a server for predicting effects of mutations on protein stability using an integrated computational approach. 42:314–319. https://doi.org/10.1093/nar/gku411
Pires DEV, Ascher DB, Blundell TL (2014) Structural bioinformatics mCSM : predicting the effects of mutations in proteins using graph-based signatures. 30(3):335–342. https://doi.org/10.1093/bioinformatics/btt691
Ajabnoor MA, Elango R, Banaganapalli B (2019) Ac ce pt e cr t. J. Biomol. Struct. Dyn. 0(0):000. https://doi.org/10.1080/07391102.2019.1671899
Hijikata A, Tsuji T, Shionyu M, Shirai T (2017) Decoding disease-causing mechanisms of missense mutations from supramolecular structures. Sci. Rep. 7(1):8541. https://doi.org/10.1038/s41598-017-08902-1
Acknowledgements
I would like to express my sincere gratitude to Dr. Indrakant K Singh, assistant professor, Deshbandhu College, University of Delhi, for giving us the opportunity to work on this topic. It would never be possible for us to take this research work to this level without his innovative ideas and his relentless support and encouragement.
Funding
No grant has been received.
Author information
Authors and Affiliations
Contributions
Both authors have contributed in research work and writing the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No wet lab experiment is used so not required in the present work.
Consent for publication
Taken from co-author
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumari, P., Rameshwari, R. In silico mutational analysis to identify the role and pathogenicity of BCL-w missense variants. J Genet Eng Biotechnol 20, 120 (2022). https://doi.org/10.1186/s43141-022-00389-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-022-00389-2