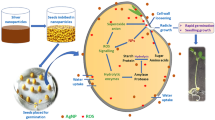
Abstract
Background
Microalgae and nanoparticles are currently considered promising tools for numerous agricultural and biotechnological applications. The green microalga Chlorella sp. MF1 and its biosynthesized silver nanoparticles (AgNPs) were used in this study as biofortification agents to enhance glucosinolate and kaempferol levels in Eruca sativa. UV–visible spectroscopy, XRD, FTIR and TEM were comprehensively used for characterizing Chlorella-based AgNPs.
Results
The biosynthesized AgNPs were found to be spherical in shape, with size ranging from 1.45 to 5.08 nm. According to FTIR measurements, silver ions were reduced to AgNPs by functional groups such as amide, hydroxyl and carboxylate. Different experimental treatments were conducted, including either soaking seeds of E. sativa or foliar spray with various concentrations of Chlorella suspension (1, 2, 3 and 4 g L−1) and AgNPs (5, 10, 20 and 40 mg L−1). Expression levels of five key genes in the biosynthetic pathway of glucosinolates (MAM1, SUR1, MYB34 and MYB51) and kaempferol (CHS) were assessed using qRT-PCR. The results indicated an upregulation in the gene expression levels in all treatments compared to control, recording the highest level at 40 mg L−1 AgNPs and 4 g L−1 Chlorella suspension. In addition, high glucosinolates and kaempferol content was detected in plants whose leaves were sprayed with AgNPs and Chlorella suspension (40 mg L−1 and 4 g L−1) based on HPLC analysis. Sequence analysis of amplified CHS fragments from E. sativa plants treated with AgNPs (40 mg L−1) showed high sequence similarity to A. thaliana CHS gene. However, there were several CHS regions with sequence polymorphism (SNPs and Indels) in foliar sprayed plants.
Conclusions
Results of this study evidenced that the application of AgNPs and Chlorella suspension increased glucosinolates and kaempferol content in E. sativa through upregulation of key genes in their biosynthetic pathway.
Similar content being viewed by others
1 Background
The interest in the bioavailability and biological effects of secondary metabolites in plants has been increased in the past two decades [78, 102]. These are identified by their direct free radical-scavenging and antioxidant activities. They can also induce expression of various genes encoding metabolic enzymes, which are thought to decrease the risk of various diseases and disorders [7, 57, 66].
Eruca sativa Mill. (arugula or rocket) is an edible annual plant from the Brassicaceae family. Because of its fresh peppery taste, it is utilized as a leaf vegetable. It is native to the Mediterranean area, but it is found all over the world [32]. Rocket leaves are a source of various bioactive compounds such as carotenoids, fibers, flavonoids, glucosinolates, polyphenols and vitamin C [25, 97]. Glucosinolates are nitrogen- and sulfur-containing plant secondary metabolites present in plants of the order Brassicales [40]. Glucosinolates and numerous decomposition products possess several biological activities [55]. They are frequently studied as part of the plant defense system against insects, herbivores, and microbial pathogens [64]. However, glucosinolates have recently attracted great research attention because of their potential health benefits in degenerative diseases such as cancer, cardiovascular, and neurodegenerative diseases [11, 83].
Enhanced glucosinolates accumulation in plants has been related to high expression levels of genes encoding the glucosinolate biosynthesis enzymes or transcription factors associated with the regulation of these genes, such as methylthioalkylmalate synthase 1 (MAM1), myeloblastosis transcription factor 34 (MYB34), myeloblastosis transcription factor 51 (MYB51) and SUPERROOT 1 (SUR1) [51, 92]. MAM1 and SUR1 are aliphatic glucosinolate biosynthetic genes, whereas MYB34 and MYB51 regulate genes encoding enzymes in the indolic glucosinolate biosynthetic pathway in E. sativa [49]. Glucosinolates biosynthesis occurs in three successive steps: amino acid elongation for aliphatic GSLs, core structure formation, and side chain modification. In the first step, methionine is de-aminated to a 2-oxo-acid, which in turn is condensed with acetyl-CoA. A subsequent decarboxylation step leads to the net gain of one methyl group, and this elongation cycle can be repeated two to six times [23]. These reactions are catalyzed by enzymes belonging to the methylthioalkylmalate synthases (MAMs) family [36]. In the second step, the resulting chain elongated products are oxidized to the corresponding aldoximes by cytochrome P450 enzymes [72]. A C-S lyase (SUR1) catalyzes the cleavage of the S-alkylthiohydroximates and the formation of thiohydroximates [71]. Then, thiohydroximates are glucosylated for the formation of desulfoglucosinolates. The core structure formation is terminated by the sulfation reactions of desulfo-GSLs [80]. The final step of GSL biosynthesis includes the side chain modification reactions, which are responsible for the differences in biological activity of GSL by-products found in plants [52]. Transcriptional regulation of GSL biosynthetic genes is reportedly under the control of MYB-family transcription factors [96].
On the other hand, kaempferol is a dominant flavonoid found generally in vegetables and fruits. E. sativa has been reported as one of the major sources of kaempferol [70, 87]. Dietary kaempferol has attracted extensive attention because of the beneficial effects on human health [13, 15, 16, 35, 42]. The first enzyme in flavonoid biosynthesis, chalcone synthase, is tightly regulated in response to a variety of environmental and developmental stimuli [22, 98, 104].
Plants exposed to different metals have been found to stimulate essential enzymes in the phenylpropanoid pathway, such as phenylalanine ammonia-lyase (PAL) and chalcone synthase [94]. Recently, total phenolic compounds: tannins and flavonoids, in roots of red radish were affected by foliar application of ZnO and FeO nanoparticles [65]. Besides their role in improving the effectiveness and bioavailability of new fertilizers, nanofertilizers are used to reduce the amount of these materials lost to the surrounding environment [90]. The most interesting metal nanoparticles are silver nanoparticles (AgNPs) due to their strong biological activity [38, 60]. They affect plants at many different levels [69, 106]. Their positive effects include stimulation of germination [79], growth invigoration [43], and increment of biomass accumulation [37]. In wheat plants, the application of AgNPs enhanced the agro-morphological criteria including photosynthesis and yield [86].
Microalgae are ubiquitous photosynthesizing microscopic phototrophs: autotrophs or heterotrophs that grow in a wide range of aquatic habitats [14]. They provide organic matter for plant growth [82] and improve soil structure and porosity through polysaccharides and mucilage secretions [47]. Also, microalgae have been reported to associate with higher plants in a symbiotic relationship [34]. Moreover, they were used as a biofertilizer for a variety of crops, tomato [77], cucumber, eggplant and lettuce [29], spinach [28], wheat [88, 93] and maize [26]. Furthermore, using microalgae in crops has been shown to increase biomass, fruit quality and vegetable yield. Chlorella vulgaris as a biofertilizer has been found to be cost-effective in ameliorating soil nutrients for increasing the productivity of Hibiscus esculentus [3].
Currently, both microalgae and nanoparticles have been reported as promising tools for many biotechnological applications via enhancing production of diverse phytochemicals, especially medicinal natural products [43, 50, 68]. The main goal of this study is to investigate the impact of AgNPs and Chlorella sp. MF1 suspension on expression of five key genes in the biosynthetic pathway of two highly important groups of secondary metabolites (glucosinolates and kaempferol) in E. sativa plants as well as to assess their effect on glucosinolates and kaempferol content. In addition, amplification and sequencing of chalcone synthase (CHS) PCR fragments were conducted to investigate the potential sequence variation in CHS PCR products of E. sativa plants treated with high level of AgNPs compared with control.
2 Methods
2.1 Cultivation of Chlorella
The marine microalga Chlorella sp. MF1 was procured from the Taxonomy and Biodiversity Lab of Aquatic Biota, National Institute of Oceanography and Fisheries, Egypt. It was cultivated in BG11 broth medium [89]. Before autoclaving, the pH of the medium was adjusted to 7.5. Then, microalga was incubated for 15 days at room temperature with light/dark condition of 16:8 h and gently regular agitation in sterile 1000-mL Erlenmeyer flasks containing 500 mL medium and 10% inoculum. Chlorella biomass was collected by centrifugation at 4500 rpm for 10 min at the end of the logarithmic phase, washed twice with distilled water and dried in an oven at 80 °C for 16 h.
2.2 Biosynthesis and characterization of AgNPs
Algal extracts were prepared by powdering the dried algal biomass with a pestle and mortar, then suspending 10 mg in 100 mL of double distilled water and filtering with Whatman No.1 filter paper. Finally, the filtrate was utilized for biosynthesis of AgNPs by mixing of 10 mL AgNO3 solution (10 mM) and 100 mL of algal extract, the reduction reaction of silver nitrate was started. Then, the reaction mixture was heated at 60 °C for 1 h. Nanoparticles formation was observed using color alteration. The produced AgNPs were centrifuged at 15,000 rpm for 20 min at 4 °C, and the pellet was washed several times with dist. H2O and finally with 90% ethanol to get pure AgNPs powder. The UV–Vis Spectrophotometer (Lambda 25, PerkinElmer, UK) was used to characterize AgNPs. The wavelength ranged between 300 and 800 nm. FTIR Spectroscope (Bruker Optics, GmBH, Germany) was used to measure the FTIR spectra of AgNPs using KBr pellet. In addition, the morphology of AgNPs was examined using a TEM (Jeol, Japan), and the XRD pattern was evaluated using a Diffractometer (Xpert-Pro, England) at room temperature using 30 mA and 40 kV. The peaks were recorded at 2θ range of 20°–80°.
2.3 Plant treatments
Seeds of E. sativa Mill. (Sementi Da Orto Prato Flore Andria, Milan, Italy) were surface sterilized with 10% commercial bleach [5% (w/v) sodium hypochlorite] for 10 min and rinsed three times with distilled water before sowing. Three principal treatments were carried out as follows: Controls: 20 seeds were sown in a plastic pot, filled with 2 kg of sieved air-dried clay and sand (2:1) soil and left to grow in the greenhouse of the Botany and Microbiology Department, Faculty of Science, Tanta University, Egypt. Soaking treatments: 200 seeds were soaked for 4 h in 10 mL of 5, 10, 20, and 40 mg L−1 AgNPs or 1, 2, 3 and 4 g L−1 Chlorella suspension and then, seeds of each treatment were sown in 3 pots as described above. Foliar treatments: 200 seeds were sown in pots as described in controls. After 14 days, leaves were sprayed twice a week with 5, 10, 20, and 40 mg L−1 AgNPs or 1, 2, 3, and 4 g L−1 Chlorella suspension. After 35 days, leaves were gathered and frozen in liquid nitrogen, then stored at − 80 °C for further analysis. For both treatments and controls, three replicates were used.
2.4 Measurement of plant Ag content
The mixed acid digestion method was used for determination of Ag content in E. sativa-treated plants (sprayed or soaked) according to Allen et al. [5]. An amount (0.1 g) of plant dry powder was mixed in a 150-mL digestion flask with 69% Nitric acid and 30% H2O2 (5:2 v/v). The mixture was heated gently until the disappearance of nitric acid fumes, and the whole mixture turned into a pale-yellow clear solution. Silver was evaluated by using Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES), PerkinElmer/Optima 7000 apparatus at the Central Lab of Tanta University.
2.5 Gene expression analysis
Quantitative real-time PCR (qRT-PCR) was used to assess the mRNA transcript levels of the five studied genes. To extract total RNA, 50–100 mg of leaves were homogenized in liquid nitrogen and then extracted using the TRIzol™ Reagent (Invitrogen, Germany) according to the manufacturer's instructions. A DNase I treatment was performed to eliminate DNA contamination. Then, 1 μg of total RNA was utilized as a template for cDNA synthesis using the QuantiTects Reverse Transcription Kit (Qiagen, USA). cDNA aliquots were used as a template for qRT-PCR analysis using gene-specific primers and β-actin as a house-keeping gene (Table 1). qRT-PCR was conducted to assess gene expression using Rotor-Gene Q (Qiagen, USA). cDNA amplicons were amplified with Maxima SYBR Green/Fluorescein qPCR Master Mix according to the manufacturer’s protocol. The following procedure for reactions was applied: 1 cycle of 50 °C 2 min, 95 °C for 10 min and 40 cycles of 95 °C for 15 s, 60 °C for 60 s. Relative gene expression was estimated as fold change using 2−∆∆ct. The house-keeping gene was used for normalization and determination of the relative amount of gene expression [63].
2.6 HPLC analysis of glucosinolates and kaempferol
2.6.1 Glucosinolates extraction and quantification
For glucosinolates extraction, leaf samples were ground in liquid nitrogen and pulverized into a fine powder. Isolation of glucosinolates was conducted according to Kliebenstein et al. [52]. Twenty milligrams of powdered leaf material were used for glucosinolates extraction. The sephadex/sulfatase protocol was followed with slight changes. To extract glucosinolates, the following mix was used: 400 μL of methanol, 10 μL of 0.3 M lead acetate, 120 μL of water and 12 μL of glucotropaeolin as internal standard were added. 10 μL of water and 10 μL of sulfatase (Sigma-Aldrich, St. Louis) solution were added to each column to desulfate glucosinolates and then incubated overnight at room temperature. Desulfoglucosinolates were eluted with 100 μL of 60% (v/v) methanol and twice with 100 μL of distilled water. This experiment was carried out five times for each sample. Then, the extracts were utilized for the separation and identification of the glucosinolates.
For quantification of glucosinolates, 20 μL of the glucosinolates extract were used. The chromatographic analyses were conducted using a high-performance liquid chromatography (HPLC; Waters, Milford, MA) equipped with a 2695 Waters Separations Module and a 996 Waters photodiode array detector. The HPLC column was a C18 Waters Symmetry Shield RP18 (5 μm particle size, 150 × 4.8 mm i.d.) and protected with a C18 guard column (Waters). The oven temperature was pre-heated to 30 °C. The glucosinolates were identified at 229 nm and separated by using the following program with a flow rate of 0.8 mL min−1: three min. at 100% H2O, 23 min. gradient from 0 to 25% (v/v) acetonitrile, 1 min. at 25% (v/v) acetonitrile, 9 min. gradient from 25 to 0% (v/v) acetonitrile and a final 4 min. at 100% H2O.
2.6.2 Kaempferol extraction and quantification
Frozen ground samples (2.0 g) were mixed with 80 mL of methanol/water (62.5:27.5, v/v) in a 500-mL flask with a round bottom and spiked with an internal standard of kaempferol (Kaempferol (3,5,7-trihydroxy-2-(4-hydroxyphenyl)-4H1-benzopyran-4-one, 95%, Sigma-Aldrich St. Louis, MO, USA) to a final concentration of 0.005 mg mL−1 and then refluxed for 1.5 h at 100 °C [53]. After cooling, a 2.0 mL aliquot was centrifuged and a sub-aliquot of 200 mL of hydrolysate was mixed with 200 mL of methanol. After that, the mixture was sonicated for 10 min. before being filtered through 0.45 mm Millex-FH (PTFE) SLFH 013 NL (Millipore, Bedford, MA, USA) filters and quantified as described in the case of glucosinolates.
Glucosinolate and kaempferol contents are given as μg per gram dry weight (DW) leaf powder calculated from HPLC peak areas using response factor computed for standard at 229 nm [52].
2.7 Chalcone synthase (CHS) amplification and sequencing
In order to evaluate the influence of seed soaking and foliar spraying of 40 mg L−1 AgNPs on amplified CHS PCR products of E. sativa, PCR was used to amplify the CHS gene sequence using gene-specific primers (Table 1). The A. thaliana CHS gene sequence (GenBank Accession No.: AY612787.1) was used to design these two primer sets. The primer set (CHSF1R1 and CHSF2R2) was designed to amplify 488 and 252 bp, respectively. First, total genomic DNA was isolated from AgNPs-treated and control E. sativa plants using Thermo Scientific GeneJET plant genomic DNA purification mini Kit (K0792/Thermo Fisher, USA) following the manufacturer’s instructions.
DreamTaq PCR Master Mix (2×) (K1071/Thermo Fisher USA) was used for PCR amplification. The thermal cycler conditions were as follows: denaturation at 94 °C for 30 s followed by annealing at 55 °C for 30 s. and extension at 72 °C for 30 s. There was an initial delay of 15 min at 95 °C at the beginning of the first cycle and a 10 min delay at 72 °C at the end of the last cycle as a post extension step. PCR products were resolved on 1.5% agarose using TBE as running buffer and at 80 V for 100 min. The expected PCR bands were extracted from agarose gel and purified with E.Z.N.A.® Gel Extraction Kit (V-spin) (Omega Bio-TEK, USA) Microspin filters and quantified spectrophotometrically. The sequencing of the purified PCR bands was conducted using the ABI PRISM® 3100 Genetic Analyzer (Micron-Corp, Korea).
2.8 Chalcone synthase (CHS) sequence analysis
The sequences of all PCR products were compared against sequences in the NCBI database using BLASTn software. Sequence alignment of E. sativa and sequences showing high similarity was conducted with MEGAX (version10.1.8) software. The phylogenetic tree was obtained automatically by applying neighbor-joining algorithms to the matrix of pairwise distances estimated using the Jukes–Cantor model. The phylogenetic tree was generated automatically using a matrix of pairwise distances by applying neighbor-joining algorithms. 1000 was used as the bootstrap value.
2.9 Statistical analysis
All results were presented as the mean of 3 replicates ± standard error (SE). Differences among treatments for all measured variables were tested by one-way analysis of variance (ANOVA), followed by Tukey’s HSD post hoc test when significant differences were found (p < 0.05). To test whether an equal effect of AgNPs and Chlorella sp. suspension on glucosinolates and kaempferol content was observed in a test combination, Chi-square test was performed. JMP statistical analysis software ver. 13.2.0 (Statistical Discovery From SAS©, USA) was used.
3 Results
3.1 Characterization of biosynthesized AgNPs
3.1.1 Ultraviolet–visible spectrophotometer analysis
One of the most frequently used approaches for structural characterization of nanoparticles is UV–visible spectroscopy. The absorption spectrum of biosynthesized AgNPs revealed that AgNPs have a strongly symmetric single band absorption with a peak limit at 449 nm signifying Ag nanoparticles (Fig. 1a).
3.1.2 Transmission electron microscopy (TEM)
To complement the above optical and spectroscopic characterization and assign size and morphology of the produced silver nanoparticles, TEM was carried out. The TEM images showed that the particles exhibit small size disparity. The spherical shape of AgNPs was observed and their size ranged from 1.45 to 5.08 nm with a mean of 3.19 ± 0.14 (Fig. 1b).
3.1.3 Fourier-transform infrared (FTIR) spectroscopy
FTIR spectroscopy was employed to identify the functional groups responsible for the biosynthesis of silver nanoparticles. The FTIR spectra of AgNPs are illustrated in Fig. 2. FTIR spectra showed a shift in the following peaks: 3488.72 (due to O–H stretching of phenol or carboxylic acids), 2059.8–2800.64 (due to C–H stretching of alkane), 1635.64 (due to N–H stretching mode in amide group which is commonly found in the protein), 1319.31 (due to N=O symmetry stretching typical of the nitro compounds) and 694.37–648.08 (due to C–H stretching of alkane).
3.2 Plant silver content
The accumulation of silver (Ag) in E. sativa-treated plants was measured using an ICP-OES apparatus. As shown in Table 2, no Ag (0.0 μg g−1) was detected in control plants. On the contrary, Ag content increased gradually with increasing AgNPs level in E. sativa plants (spraying or soaking). This means that there is a positive relationship between the dose (spraying or soaking) of AgNPs used and Ag content in E. sativa-treated plants. Also, results indicate that spraying of E. sativa plants with AgNPs significantly increased Ag content, especially in plants sprayed with the high (40 mg L−1) AgNPs.
3.3 Gene expression analysis
In this study, the expression levels of four key genes in the biosynthesis pathway of glucosinolates in E. sativa (methylthioalkylmalate synthase 1 (MAM1), myeloblastosis transcription factor 34 (MYB34), myeloblastosis transcription factor 51 (MYB51), and SUPERROOT 1 (SUR1)) were determined in response to treatment with AgNPs and Chlorella sp. suspension. In addition, the expression level of chalcone synthase (CHS) that is involved in biosynthesis of kaempferol was examined. The results revealed that the key genes (CHS, MAM1, MYB34, MYB51 and SUR1) responded differently to various treatments of AgNPs and Chlorella sp. suspension and their expression levels were significantly different from control plants, exhibiting the highest expression level of these genes at 40 mg L−1 AgNPs and 4 g L−1 Chlorella sp. suspension (seed soaking and foliar spray) (Fig. 3).
Genes expression analysis of five glucosinolates and kaempferol-related genes in Eruca sativa plants treated either soaking or spraying with different concentrations of AgNPs and Chlorella sp. suspension. Different letters indicate statistically significant difference at P < 0.05. a Chalcone synthase CHS, b methylthioalkylmalate synthases 1 (MAM1), c myeloblastosis transcription factor 34 (MYB34), d myeloblastosis transcription factor 51 (MYB51), E: SUPERROOT 1 (SUR1). Sp. Nano- and So. Nano-: spraying or soaking with AgNPs (5, 10, 20, 40 mg L−1), Sp. Alga- and So. Alga-: spraying or soaking with Chlorella sp. suspension (1, 2, 3, 4 g L−1)
The higher upregulation of MAM1, MYB34, MYB51 and SUR1 genes was observed in plants whose seeds were soaked with 40 mg L−1 AgNPs than with 4 g L−1 Chlorella sp. suspension. The highest fold change of these genes was found in plants whose seeds were soaked with 40 mg L−1 AgNPs was 2.48-, three-, 1.9- and 2.48-fold, respectively (Fig. 3b–d). Similarly, the highest fold change was observed in the case of plants whose leaves were sprayed with 40 mg L−1 AgNPs was 1.46-, three-, 0.74- and 1.29-fold, respectively.
On the other hand, the highest fold change (2.03-fold) of the CHS gene was observed in plants whose leaves were sprayed with 40 mg L−1 AgNPs (Fig. 3a). However, the highest CHS expression was found in plants whose seeds were soaked with 4 g L−1 Chlorella sp. suspension (Fig. 3a).
3.4 HPLC analysis of glucosinolates and kaempferol
Data in Table 3 and Fig. 4 showed that treating E. sativa plants with AgNPs and Chlorella sp. suspension (soaking and spraying) led to alteration of glucosinolates and kaempferol content compared with the control plants. The highest glucosinolates and kaempferol contents were detected in plants sprayed with AgNPs (40 mg L−1) and with Chlorella sp. suspension (4 g L−1), 81.92 μg g−1 DW and 80.70 μg g−1 DW, respectively (Table 3). Also, foliar spray with Chlorella sp. suspension (4 g L−1), seeds soaking with AgNPs (40 mg L−1) and seeds soaking with Chlorella sp. suspension (4 g L−1) exhibited high glucosinolates and kaempferol contents compared with the control plants (Fig. 4). Chi-square test revealed that glucosinolates and kaempferol content depends on the type of treatment (Table 3). In other words, there is a significant variation in the concentration of glucosinolates and kaempferol among different treatments (p value < 0.0001).
HPLC chromatograms of glucosinolates and kaempferol showing their contents in Eruca sativa a control plants, b AgNPs (40 mg L−1) seeds-soaked plants, c AgNPs (40 mg L−1) leaves-sprayed plants, d Chlorella sp. suspension (4 g L−1) seeds-soaked plants, e Chlorella sp. suspension (4 g L−1) leaves-sprayed plants
3.5 Sequencing of Eruca sativa CHS gene fragments
In order to evaluate the effect of AgNPs on DNA of E. sativa CHS, specific primers were designed and used to amplify E. sativa CHS DNA fragments. E. sativa-specific CHS gene bands were eluted from agarose gel and were sequenced. The length of amplified DNA fragment from untreated E. sativa CHS (control) was 674 bp, while the length of CHS gene amplicon from AgNPs (40 mg L−1)-soaked plants was 677 bp. Finally, the length of the CHS gene fragment in the case of spraying AgNPs (40 mg L−1) was 673 bp. The sequences of all 6 PCR amplicons were compared against sequences in the NCBI database using BLASTn software.
The results of NCBI BLASTn showed that all CHS sequences were strongly conserved and shared high sequence identity (> 99%) with A. thaliana CHS gene sequences. Also, NCBI BLAST showed that the two amplified sequences were highly similar to A. thaliana chalcone synthase (CHS) gene, partial cds (Accession no.: AY612787) with 99.77% sequence identity and 98% query coverage. The CHS sequences of untreated (control) and treated E. sativa (soaking AgNPs and spraying AgNPs treatments at 40 mg L−1) were aligned with CHS sequences of A. thaliana and other Arabidopsis species using MEGAX (version 10.1.8) software. The multiple sequence alignment (MSA) analysis indicated that there are several single nucleotide polymorphisms (SNPs) and Indel (insertion and deletion) variations mainly in the CHS fragment sequence of E. sativa plants sprayed with AgNPs (40 mg L−1) compared with control (untreated E. sativa), AgNPs (40 mg L−1)-soaked E. sativa plants and Arabidopsis CHS gene sequences.
The phylogenetic analyses revealed that the CHS fragment sequences of untreated E. sativa (control) were closely related to the A. thaliana CHS gene (AY612787.1), while the CHS fragment sequence of E. sativa plant sprayed with AgNPs (40 mg L−1) and the CHS fragment sequence of E. sativa plant soaked with AgNPs (40 mg L−1) were grouped together into a separate major cluster (Fig. 5).
4 Discussion
Glucosinolates (GSLs) and kaempferol are two highly important groups of secondary metabolites, which are found at high levels in Brassica species [6]. In this study, application of abiotic and biotic elicitors (AgNPs and Chlorella sp. suspension) was found to increase glucosinolates and kaempferol in E. sativa plants sprayed with 40 mg L−1 AgNPs and 4 g L−1 Chlorella sp. suspension. Such elicitors have modified plant primary and secondary metabolites, as reported by Marslin et al. [67], Chanda et al. [14] and Anjum et al. [6]. Nanoelicitation was found to be a powerful technique for increasing secondary metabolite synthesis, which can boost metabolism and induce physiological responses in plants [76].
In the present study, biosynthesis of silver nanoparticles (AgNPs) using suspension of the marine chlorophyte Chlorella sp. MF1 was performed. For characterization and confirmation of biosynthesized AgNPs, UV–Vis spectroscopy, TEM and FTIR analyses were utilized. UV–Vis spectroscopy is regularly used to investigate AgNPs surface plasmon resonance (SPR). In this study, the UV–Vis spectrum revealed a single strong and large SPR peak at 449 nm (Fig. 1), indicating that AgNPs were synthesized successfully. Similarly, various studies have identified a surface plasmon resonance peak in the range of 410–450 nm as an indicator of AgNPs biosynthesis [21, 100, 105]. Also, TEM images of AgNPs biosynthesized in the present study indicated that AgNPs are spherical and have a diameter that ranged between 1.45 and 5.08 nm (Fig. 1). According to Pirtarighat et al. [81], spherical nanoparticles have only a single SPR band, and the number of peaks increases as the diversity of particle shapes increases. However, various forms of biologically synthesized nanoparticles, including rectangular, cubic, and spherical, have been observed in a study by Soleimani and Habibi-Pirkoohi [95].
In addition, FTIR analysis was used in the present study to classify potential biomolecules present in the algal extract, which are involved in AgNPs biosynthesis, stabilization and capping, according to Moldovan et al. [73]. The results showed the existence of various functional groups of phytochemicals such as amides, phenols and carboxylic groups in algal water extract, which were involved in the reduction of silver ions to AgNPs as well as their stabilization (Fig. 2). The presence of amides indicated that AgNPs were capped and protected by the proteins present in the algal extract. This result is in accordance with Rajeshkumar et al. [84] and Abdul Razack et al. [1], who suggested that proteins are the major constituents in the algal extract involved in reducing Ag ions into nanoparticles, capping and stabilizing AgNPs by electrostatic forces. Moreover, negatively charged groups present in the algal extract, such as carboxylate (COO−) and hydroxyl groups (OH−), have a high tendency to bind Ag ions, contributing to both reduction (AgNPs biosynthesis), and stabilization of Ag ions [4, 48].
In this study, HPLC results exhibited higher content of glucosinolates and kaempferol in plants foliar-sprayed with AgNPs (40 mg L−1) and Chlorella sp (4 g L−1) than with the seed soaking method (Table 3 and Fig. 4). Similarly, the foliar application of AgNPs (20 and 40 ppm) increased total phenolics and carotenoids in oakleaf lettuce (Lactuca sativa L. var. foliosa Bremer) seedlings [45], while ZnO and FeO green synthesized nanoparticles increased total tannins and flavonoids in roots of red radish [65]. Moreover, foliar application of TiO2 nanoparticles has significantly elevated carotenoids and phenolic compounds in the maize crop [74]. Furthermore, CuONPs-treated seedlings of Brassica rapa ssp. rapa also had substantially higher levels of glucosinolates and phenolic compounds than control seedlings [18].
Also, in this study, the expression levels of four key genes in the glucosinolate biosynthetic pathway (MAM1, MYB34, MYB51 and SUR1) increased significantly in response to treatment with AgNPs or Chlorella sp. suspension. The highest expression level of these genes was observed when seeds were soaked with 40 mg L−1 AgNPs or 4 g L−1 Chlorella sp. suspension. In addition, the fold change was higher in the case of AgNPs treatment, 2.48-, three-, 1.9- and 2.48-fold, respectively (Fig. 3b–e). It has been reported that high expression levels of genes encoding glucosinolate biosynthesis pathway enzymes or transcription factors involved in their regulation, such as MAM1, MYB34, MYB51 and SUR1, have been linked to glucosinolate accumulation in plants [51, 92], which is similar to the finding of this study. In addition, MAM1 and SUR1 are aliphatic glucosinolate biosynthetic genes, whereas MYB34 and MYB51 regulate genes encoding enzymes in the indolic glucosinolate biosynthetic pathway in E. sativa [49]. As described above, there was an upregulation of the four key genes in the glucosinolate biosynthetic pathway in this study, which is in agreement with the finding of Thiruvengadam et al. [99], who detected that expression of aliphatic GSLs (BrMYB28 and BrMYB29) and indolic GSLs (BrMYB34 and BrMYB51) as well as aliphatic GSL biosynthetic genes (St5C and SUR1) transcripts was upregulated in plants treated with high concentrations (5 and 10 mg L−1) of AgNPs.
Similarly, the expression of aliphatic GSLs (BrMYB28 and BrMYB29) and indolic GSLs (BrMYB34 and BrMYB122) transcripts of Chinese cabbage was upregulated in CuONPs-elicited hairy roots [17]. Moreover, higher concentrations of CuONPs (250 and 500 mg L−1) increased the expression of aliphatic GSL (BrMYB28 and BrMYB29) and indolic GSL (BrMYB34 and BrMYB51) transcripts in Brassica rapa ssp. rapa seedlings [18]. Recently, Ramezani et al. [85] discovered that AgNPs function as elicitors and strongly increase the transcription of key genes (UGT85C2 (UDP-glycosyltransferases), KAH (Kaurenoic acid-13 hydroxylase), UGT74G1 and UGT76G1) of Stevia rebaudiana B. responsible for secondary metabolites involved in the pathway of glycosides biosynthesis. Similar to the finding of this study, microalgal extracts from Chlorella vulgaris and Scenedesmus quadricauda had bio-stimulant effects on the expression of sugar beet genes involved in various biological pathways and processes, such as primary and secondary metabolites [10]. Recently, Farid et al. [30] found that crude polysaccharides from Chlorella sorokiniana had an important stimulatory effect on PAL gene function, suggesting an upregulation of main genes in the phenolics pathway.
In contrast to the results of glucosinolate genes, plants treated with 40 mg L−1 AgNPs (foliar spray) as 4 g L−1 algal suspension (foliar spray) did not induce a high expression of CHS, 2.03- and 0.74-fold, respectively (Fig. 3a). It is well known that CHS is the first enzyme in the biosynthetic pathway of all flavonoids [98, 104]. As mentioned above, the results of this study indicated that CHS gene expression levels were upregulated in most treatments. It has been shown that CHS expression causes accumulation of flavonoids such as the natural flavonol kaempferol [12], which is in accordance with the finding of this study. Similarly, AgNPs upregulated the key genes of the flavonoid and anthocyanin biosynthetic pathway in A. thaliana as reported by Garcia-Sanchez et al. [31]. Moreover, the total phenolic and flavonoid contents, as well as the expression levels of their genes (PAL, CHI and FLS), were significantly high in CuONPs-elicited hairy roots [17].
The precise mechanism by which nanoparticles (NPs) regulate plant secondary metabolism is not yet fully understood. The upregulation of mitogen-activated protein kinase (MAPK) cascades, reactive oxygen species (ROS) and cytoplasmic Ca+2 efflux was thought to be increased as the plant's early responses to NPs treatment [46, 54]. In general, MAPK phosphorylation and activation of downstream transcription factors generally lead to the transcriptional reprogramming of secondary metabolism in plants [103].
In order to investigate the uptake and accumulation of AgNPs in E. sativa plants (soaked or sprayed with different AgNP levels), Ag content was measured in their leaves. The results showed a positive relationship between the dose of AgNPs and Ag content. However, the highest Ag content was 1.22 µg g−1 in plants sprayed with 40 mg L−1 (Table 2). This result suggests that AgNPs uptake in the case of foliar application through stomatal pores is higher than that penetrated into endosperm and embryonic tissues through the seed coat. Similar findings have been observed by Larue et al. [59] in Lactuca sativa, Hong et al. [41] in Cucumis sativus, Cocozza et al. [19] in tree plants and Noori et al. [75] in tomato. This finding explains the higher glucosinolates content in plants sprayed with AgNPs compared to soaked plants (Table 3 and Fig. 4) due to higher Ag bioaccumulation, which is similar to the finding of Li et al. [62].
It was found that treatment of E. sativa plants with AgNPs showed no phytotoxicity, where E. sativa plants sprayed with high levels of AgNPs (40 or 20 mg L−1) exhibited the highest significant increase in both fresh weight and dry weight, indicating that the application of silver nanoparticles enhanced plant vegetative growth [91]. Regarding the potential risk of toxicity to humans who will feed on plants treated with nanoparticles, results in Table 2 revealed that the highest Ag content was 1.22 µg g−1 in plants sprayed with 40 mg L−1, which is a very small amount compared with the maximum daily intake of silver, which should be less than 350 µg according to the US Environmental Protection Agency (EPA) reference dose [27]. Also, the European Food Safety Authority (EFSA) has considered a total life-time oral intake of 10 g Ag (0.39 mg/day/person) as the human “no observed adverse effect level” [2].
To detect if AgNPs induced sequence variation in CHS sequences of AgNPs-treated plants (sprayed or soaked) compared to control and CHS sequence of A. thaliana, multiple sequence alignment (MSA) analysis was performed. MSA results reflected high sequence similarity between CHS A. thaliana and CHS E. sativa sequences of control and treated plants (seed soaking and foliar spray treatments). Besides, MSA analysis showed several single nucleotide polymorphisms (SNPs) and Indel (insertion and deletion) variations in the E. sativa CHS fragment sequence of plants that were foliar sprayed with AgNPs (40 mg L−1) compared to control plants (untreated E. sativa). Treatments with NPs have affected DNA integrity, proteins and DNA expression in plants [8, 58, 101], which is in accordance with the findings of this study. Cu and CdNPs acted as conventional mutagens and affected chromosomal attributes [56]. They induced several types of aberrations in Lathyrus sativus L. [33]. Treatment with silver and gold NPs resulted in some specific chromosomal abnormalities in Allium cepa roots [24].
It has been shown that chemical interaction of NPs with DNA during interphase can influence DNA replication and transcription of DNA [20]. In addition, silver ions have been discovered to bind to DNA in a localized binding mode, which means that the metal ions are in direct contact with the DNA structure, causing a significant shift in DNA conformation [44]. CeO2NP treatment of soybean (Glycine max L.) resulted in the appearance of new bands in a random amplified polymorphic DNA pattern indicating NP-induced mutations [39]. The phylogenetic tree showed that the sequence of CHS of E. sativa control was closely related to the A. thaliana CHS gene, while the sequence of CHS fragment of E. sativa plants sprayed with AgNPs (40 mg L−1) was clustered into a separate major cluster (Fig. 5) indicating the variation that was induced by high AgNPs level in the CHS sequence of the treated plants.
5 Conclusions
The biosynthesized silver nanoparticles (AgNPs) were spherical and had a slight size disparity. Glucosinolates and kaempferol key genes responded differently when plants were treated with AgNPs and Chlorella sp. suspension. However, upregulation of studied genes was observed and the highest expression level of these genes as well as glucosinolates and kaempferol content was at 40 mg L−1 AgNPs and 4 g L−1 Chlorella suspension (seed soaking or foliar spray). E. sativa plants sprayed with AgNPs (40 mg L−1) showed several single nucleotide polymorphisms (SNPs) and Indel (insertion and deletion) variations in the CHS fragment sequence. Therefore, more attention should be paid when using AgNPs.
Availability of data and material
Not applicable.
Abbreviations
- AgNPs:
-
Silver nanoparticles
- CdNPs:
-
Cadmium nanoparticles
- CeO2NPs:
-
Cerium dioxide nanoparticles
- CHS :
-
Chalcone synthase
- CuONPs:
-
Copper oxide nanoparticles
- FTIR:
-
Fourier-transform infrared
- GSLs:
-
Glucosinolates
- HPLC:
-
High-performance liquid chromatography
- Indels:
-
Insertions and deletions
- MAM1 :
-
Methylthioalkylmalate synthase 1
- MAPK:
-
Mitogen-activated protein kinase
- MSA:
-
Multiple sequence alignment
- MYB34 :
-
Myeloblastosis transcription factor 34
- MYB51 :
-
Myeloblastosis transcription factor 51
- NPs:
-
Nanoparticles
- PAL :
-
Phenylalanine ammonia-lyase
- qRT-PCR:
-
Quantitative real-time PCR
- RA S :
-
R osmarinic acid synthase
- ROS:
-
Reactive oxygen species
- SNPs:
-
Single nucleotide polymorphisms
- SUR1 :
-
SUPERROOT 1
- TEM:
-
Transmission electron microscopy
- UV–Vis:
-
Ultraviolet–visible
References
Abdul Razack S, Duraiarasanb S, Manic V (2016) Biosynthesis of silver nanoparticle and its application in cell wall disruption to release carbohydrate and lipid from C. vulgaris for biofuel production. Biotechnol Rep 11:70–76
Aguilar F, Autrup H, Barlow S, Castle L, Crebelli R, Dekant W, Toldra F (2007) Scientific opinion of the panel on food additives, flavourings, processing aids and materials in contact with food (AFC) on a request related to a 16th list of substances for food contact materials. Eur Food Saf Auth J 555-563:1-31
Agwa OK, Ogugbue CJ, Williams EE (2017) Field evidence of Chlorella vulgaris potentials as a biofertilizer for Hibiscus esculentus. Int J Agric Res 12(4):181–189
Ajitha B, Reddy YAK, Reddy PS, Jeon HJ, Ahn CW (2016) Role of capping agents in controlling silver nanoparticles size, antibacterial activity and potential application as optical hydrogen peroxide sensor. R Soc Chem Adv 6(42):36171–36179
Allen SE, Grimshaw HM, Parkinson JA, Quarmby C (1974) Chemical analysis of ecological materials. Blackwell Scientific, Oxford
Anjum S, Anjum I, Hano C, Kousar S (2019) Advances in nanomaterials as novel elicitors of pharmacologically active plant specialized metabolites: current status and future outlooks. R Soc Chem RSC Adv 9(69):40404–40423
Arts ICW, Hollman PCH (2005) Polyphenols and disease risk in epidemiologic studies. Am J Clin Nutr 81:317S-325S
Atha DH, Wang H, Petersen EJ, Cleveland D, Holbrook RD, Jaruga P, Dizdaroglu M, Xing B, Nelson BC (2012) Copper oxide nanoparticle mediated DNA damage in terrestrial plant models. Environ Sci Technol 46:1819–1827
Baoming T, Dandan S, Yuli L, Haiyan S, Hua L, Xin Z, Bonan W, Zhenqiang P (2011) Analysis of the RNAi targeting FAD2 gene on oleic acid composition in transgenic plants of Brassica napus. Afr J Microbiol Res 5(7):817–822
Barone V, Baglieri A, Stevanato P, Broccanello C, Bertoldo G, Bertaggia M, Cagnin M, Pizzeghello D, Moliterni VMC, Mandolino G, Fornasier F, Squartini A, Nardi S, Concheri G (2018) Root morphological and molecular responses induced by microalgae extracts in sugar beet (Beta vulgaris L.). J Appl Phycol 30:1061–1071
Björkman M, Klingen I, Birch ANE, Bones AM, Bruce TJA, Johansen TJ, Meadow R, Mølmann J, Seljåsen R, Smart LE, Stewart D (2011) Phytochemicals of Brassicaceae in plant protection and human health influences of climate, environment and agronomic practice. Phytochemistry 72(7):538–556
Blanco E, Sabetta W, Danzi D, Negro D, Passeri V, Lisi A, Paolocci F, Sonnante G (2018) Isolation and characterization of the flavonol regulator CcMYB12 from the globe artichoke [Cynara cardunculus var. scolymus (L.) Fiori]. Front Plant Sci 9:941
Calderón-Montaño JM, Burgos-Morón E, Pérez-Guerrero C, López-Lázaro M (2011) A review on the dietary flavonoid kaempferol. Mini Rev Med Chem 11(4):298–344
Chanda M, Merghoub N, El-Arroussi H (2019) Microalgae polysaccharides: the new sustainable bioactive products for the development of plant bio-stimulants? World J Microbiol Biotechnol 35:177
Chandramohan G, Al-Numair KS, Alsaif MA, Veeramani C (2015) Antidiabetic effect of kaempferol a flavonoid compound, on streptozotocin-induced diabetic rats with special reference to glycoprotein components. Prog Nutr 17:50–57
Chen AY, Chen YC (2013) A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem 138:2099–2107
Chung IM, Rekha K, Rajakumar G, Thiruvengadam M (2018) Influence of silver nanoparticles on the enhancement and transcriptional changes of glucosinolates and phenolic compounds in genetically transformed root cultures of Brassica rapa ssp. Rapa. Bioprocess Biosyst Eng 41(11):1665–1677
Chung IM, Rekha K, Venkidasamy B, Thiruvengadam M (2019) Effect of copper oxide nanoparticles on the physiology, bioactive molecules, and transcriptional changes in Brassica rapa ssp. rapa seedlings. Water Air Soil Pollut 230(2):48
Cocozza C, Perone A, Giordano C, Salvatici MC, Pignattelli S, Raio A, Schaub M, Sever K, Innes JL, Tognetti R, Cherubini P (2019) Silver nanoparticles enter the tree stem faster through leaves than through roots. Tree Physiol 39:1251–1261
Collins AR, Kumar A, Dhawam A, Stone V, Dusinska M (2013) Mechanisms of genotoxicity. Review of recent in vitro and in vivo studies with engineered nanoparticles. Nanotoxicology 52:1–70
Dada AO, Adekola FA, Dada FE, Adelani-Akande AT, Bello MO, Okonkwo CR, Inyinbor AA, Oluyori AP, Olayanju A, Ajanaku KO, Adetunji CO (2019) Silver nanoparticle synthesis by Acalypha wilkesiana extract: phytochemical screening, characterization, influence of operational parameters, and preliminary antibacterial testing. Heliyon 5(10):e02517
Dao TTH, Linthorst HJM, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10:397–412
de Kraker JW, Gershenzon J (2011) From amino acid to glucosinolate biosynthesis: protein sequence changes in the evolution of methylthioalkylmalate synthase in Arabidopsis. Plant Cell 23:38–53
Debnath P, Mondal A, Hajra A, Das C, Mondal NK (2018) Cytogenetic effects of silver and gold nanoparticles on Allium cepa roots. J Genet Eng Biotechnol 16(2):519–526
Delouee SAW, Urooj A (2007) Antioxidant properties of various solvent extracts of mulberry (Morus indica L.) leaves. Food Chem 102:1233–1240
Dineshkumar R, Subramanian J, Gopalsamy J, Jayasingam P, Arumugam A, Kannadasan S, Sampathkumar P (2019) The impact of using microalgae as biofertilizer in maize (Zea mays L.). Waste Biomass Valoriz 10(5):1101–1110
Dolara P (2014) Occurrence, exposure, effects, recommended intake and possible dietary use of selected trace compounds (aluminium, bismuth, cobalt, gold, lithium, nickel, silver). Int J Food Sci Nutr 65(8):911–924
El-din SM, Hassan SM (2016) The promotive effect of different concentrations of marine algae on spinach plants (Spinacia oleracea L.). Egypt J Hortic 43:109–122
Elhafiz A, Gaur AESS, Osman NHM, Lakshmi TR (2015) Chlorella vulgaris and Chlorella pyrenoidosa live cells appear to be promising sustainable biofertilizer to grow rice, lettuce, cucumber and eggplant in the UAE soils. Recent Res Sci Technol 7:14–21
Farid R, Mutale-joan C, Redouane B, Najib EM, Abderahime A, Laila S, Hicham EA (2019) Effect of microalgae polysaccharides on biochemical and metabolomics pathways related to plant defense in Solanum lycopersicum. Appl Biochem Biotechnol 188(1):225–240
Garcia-Sanchez S, Bernales I, Cristobal S (2015) Early response to nanoparticles in the Arabidopsis transcriptome compromises plant defence and root-hair development through salicylic acid signalling. BMC Genom 16:341–349
Garg G, Sharma V (2014) Eruca sativa (L.): botanical description, crop improvement, and medicinal properties. J Herbs Spices Med Plants 20(2):171–182
Ghosh B, Datta AK, Pramanik A, Kumbhakar DV, Das D, Paul R, Biswas J (2017) Mutagenic effectivity of cadmium sulphide and copper oxide nanoparticles on some physiological and cytological attributes of Lathyrus sativus L. Cytologia 82(3):267–271
Gorelova OA (2006) Communication of cyanobacteria with plant partners during association formation. Microbiology 75(4):465–469
Granado-Serrano AB, Martin MA, Bravo L, Goya L, Ramos S (2006) Quercetin induces apoptosis via caspase activation, regulation of Bcl-2, and inhibition of PI-3-kinase/Akt and ERK pathways in a human hepatoma cell line (HepG2). J Nutr 136:2715–2721
Graser G, Schneider B, Oldham NJ, Gershenzon J (2000) The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch Biochem Biophys 378:411–419
Gupta SD, Agarwal A, Pradhan S (2018) Phytostimulatory effect of silver nanoparticles (AgNPs) on rice seedling growth: an insight from antioxidative enzyme activities and gene expression patterns. Ecotoxicol Environ Saf 161:624–633
Haider A, Kang IK (2015) Preparation of silver nanoparticles and their industrial and biomedical applications: a comprehensive review. Adv Mater Sci Eng 2015:165–257
Halder S, Mandal A, Das D, Gupta S, Chattopadhyay AP, Datta AK (2015) Copper nanoparticle induced macromutation in Macrotyloma uniflorum (Lam.) Verdc. (Leguminosae): a pioneer report. Genet Resour Crop Evol 62(2):165–175
Hellín P, Novo-Uzal E, Pedreño MA, Cava J, Garrido I, Molina MV, Fenoll J, Flores P (2018) Impact of plant biostimulants on the production of glucosinolates in broccoli. Acta Hortic 1194:99–104
Hong J, Peralta-Videa JR, Rico C, Sahi S, Viveros MN, Bartonjo J, Zhao L, Gardea-Torresdey JL (2014) Evidence of translocation and physiological impacts of foliar applied CeO2 nanoparticles on cucumber (Cucumis sativus) plants. Environ Sci Technol 48(8):4376–4385
Imran M, Salehi B, Sharifi-Rad J, Aslam Gondal T, Saeed F, Imran A, Shahbaz M, Tsouh Fokou PV, Umair Arshad M, Khan H, Guerreiro SG, Martins N, Estevinho LM (2019) Kaempferol: a key emphasis to its anticancer potential. Molecules (Basel, Switzerland) 24(12):2277
Jasim B, Thomas R, Mathew J, Radhakrishnan EK (2017) Plant growth and diosgenin enhancement effect of silver nanoparticles in Fenugreek (Trigonella foenum-graecum L.). Saudi Pharm J 25:443–447
Jiang WY, Ran SY (2018) Two-stage DNA compaction induced by silver ions suggests a cooperative binding mechanism. J Chem Phys 148(20):205102
Jurkow R, Pokluda R, Sękara A, Kalisz A (2020) Impact of foliar application of some metal nanoparticles on antioxidant system in oakleaf lettuce seedlings. BMC Plant Biol 20:290
Karakaş Ö (2020) Effect of silver nanoparticles on production of indole alkaloids in Isatis constricta. Iran J Sci Technol Trans A Sci 44:621–627
Karthikeyan N, Prasanna R, Nain L, Kaushik BD (2007) Evaluating the potential of plant growth promoting cyanobacteria as inoculants for wheat. Eur J Soil Biol 43(1):23–30
Kashyap M, Samadhiya K, Ghosh A, Anand V, Shirage PM, Bala K (2019) Screening of microalgae for biosynthesis and optimization of Ag/AgCl nano hybrids having antibacterial effect. R Soc Chem RSC Adv 9(44):25583–25591
Katsarou D, Omirou M, Liadaki K, Tsikou D, Delis C, Garaqounis C, Krokida A, Zambounis A, Papadopoulou KK (2016) Glucosinolate biosynthesis in Eruca sativa. Plant Physiol Biochem 9:452–466
Khan MI, Shin JH, Kim JD (2018) The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb Cell Factories 17:36
Kim YB, Li X, Kim SJ, Kim HH, Lee J, Kim H, Park SU (2013) MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 18:8682–8695
Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126:811–825
Koh E, Wimalasiri KMS, Chassy AW, Mitchell AE (2009) Content of ascorbic acid, quercetin, kaempferol and total phenolics in commercial broccoli. J Food Compos Anal 22(7-8):637–643
Kohan-Baghkheirati E, Geisler-Lee J (2015) Gene expression, protein function and pathways of Arabidopsis thaliana responding to silver nanoparticles in comparison to silver ions, cold, salt, drought, and heat. Nanomaterials 5:436–467
Kołodziejski D, Piekarska A, Hanschen FS, Pilipczuk T, Tietz F, Kusznierewicz B, Bartoszek A (2019) Relationship between conversion rate of glucosinolates to isothiocyanates/indoles and genotoxicity of individual parts of Brassica vegetables. Eur Food Res Technol 245(2):383–400
Kumbhakar DV, Datta AK, Mandal A, Das D, Gupta S, Ghosh B, Dey S (2016) Effectivity of copper and cadmium sulphide nanoparticles in mitotic and meiotic cells of Nigella sativa L. (black cumin)-can nanoparticles act as mutagenic agents? J Exp Nanosci 11(11):823–839
Kurutas EB (2015) The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: current state. Nutr J 15(1):1–22
Landa P, Prerostova S, Petrova S, Knirsch V, Vankova R, Vanek T (2015) The transcriptomic response of Arabidopsis thaliana to zinc oxide: a comparison of the impact of nanoparticle, bulk, and ionic zinc. Environ Sci Technol 49(24):14537–14545
Larue C, Castillo-Michel H, Sobanska S, Cécillon L, Bureau S, Barthès V, Ouerdane L, Carrière M, Sarret G (2014) Foliar exposure of the crop Lactuca sativa to silver nanoparticles: evidence for internalization and changes in Ag speciation. J Hazard Mater 264:98–106
Lee SH, Jun BH (2019) Silver nanoparticles: synthesis and application for nanomedicine. Int J Mol Sci 20(4):865
Le Gall G, Metzdorff SB, Pedersen J, Bennett RN, Colquhoun IJ (2005) Metabolite profiling of Arabidopsis thaliana (L.) plants transformed with an antisense chalcone synthase gene. Metabolomics 1:181–198
Li CC, Dang F, Li M, Zhu M, Zhong H, Hintelmann H, Zhou DM (2017) Effects of exposure pathways on the accumulation and phytotoxicity of silver nanoparticles in soybean and rice. Nanotoxicology 11:699–709
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Maag D, Erb M, Köllner TG, Gershenzon J (2015) Defensive weapons and defense signals in plants: some metabolites serve both roles. BioEssays 37(2):167–174
Mahmoud AWM, Abdelaziz SM, El-Mogy MM, Abdeldaym EA (2019) Effect of foliar ZnO and FeO nanoparticles application on growth and nutritional quality of red radish and assessment of their accumulation on human health. Agriculture (Pol’nohospodárstvo) 65(1):16–29
Manach C, Scalbert A, Morand C, Rémésy C, Jimenez L (2004) Polyphenols—food sources and bioavailability. Am J Clin Nutr 79:727–747
Marslin G, Sheeba CJ, Franklin G (2017) Nanoparticles alter secondary metabolismin plants via ROS burst. Front Plant Sci 8:832
Martínez-Ballesta M, Gil-Izquierdo Á, García-Viguera C, Domínguez-Perles R (2018) Nanoparticles and controlled delivery for bioactive compounds: outlining challenges for new “smart-foods” for health. Foods (Basel, Switzerland) 7(5):72
Mehmood AB (2018) Overview of the application of silver nanoparticles to improve growth of crop plants. IET Nanobiotechnol 12:701–705
Michael HN, Shafik RE, Rasmy GE (2011) Studies on the chemical constituents of fresh leaf of Eruca sativa extract and its biological activity as anticancer agent in vitro. J Med Plants Res 5(7):1184–1191
Mikkelsen MD, Naur P, Halkier BA (2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37(5):770–777
Mikkelsen MD, Petersen BL, Olsen CE, Halkier BA (2002) Biosynthesis and metabolic engineering of glucosinolates. Amino Acids 22:279–295
Moldovan B, Sincari V, Perde-Schrepler M, David L (2018) Biosynthesis of silver nanoparticles using Ligustrum ovalifolium fruits and their cytotoxic effects. Nanomaterials 8(8):627
Morteza E, Moaveni P, Farahani HA, Kiyani M (2013) Study of photosynthetic pigments changes of maize (Zea mays L.) under nano TiO2 spraying at various growth stages. Springer Plus 2(1):247
Noori A, Ngo A, Gutierrez P, Theberge S, White JC (2020) Silver nanoparticle detection and accumulation in tomato (Lycopersicon esculentum). J Nanopart Res 22:131
Nourozi E, Hosseini B, Maleki R, Abdollahi Mandoulakani B (2019) Iron oxide nanoparticles: a novel elicitor to enhance anticancer flavonoid production and gene expression in Dracocephalum kotschyi hairy-root cultures. J Sci Food Agric 99(14):6418–6430
Özdemir S, Sukatar A, Oztekin GB (2016) Production of Chlorella vulgaris and its effects on plant growth, yield and fruit quality of organic tomato grown in greenhouse as biofertilizer. J Agric Sci 22:597–605
Pandey KB, Rizvi SI (2009) Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev 2(5):270–278
Parveen A, Rao S (2015) Effect of nanosilver on seed germination and seedling growth in Pennisetum glaucum. J Clust Sci 26:693–701
Piotrowski M, Schemenewitz A, Lopukhina A, Muller A, Janowitz T, Weiler EW, Oecking C (2004) Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J Biol Chem 279:50717–50725
Pirtarighat S, Ghannadnia M, Baghshahi S (2019) Green synthesis of silver NPs using the plant extract of Salvia spinosa grown in vitro and their antibacterial activity assessment. J Nanostruct Chem 9(1):1–9
Grzesik M, Romanowska-Duda Z (2014) Improvements in Germination, Growth, and Metabolic Activity of Corn Seedlings by Grain Conditioning and Root Application with Cyanobacteria and Microalgae. Pol J Environ Stud 23(4):1147–1153
Quirante-Moya S, García-Ibañez P, Quirante-Moya F, Villaño D, Moreno DA (2020) The role of brassica bioactives on human health: Are we studying it the right way? Molecules 25(7):1591
Rajeshkumar S, Malarkodi C, Paulkumar K, Vanaja M, Gnanajobitha G, Annadurai G (2014) Algae mediated green fabrication of silver nanoparticles and examination of its antifungal activity against clinical pathogens. Int J Met 2014:8
Ramezani M, Asghari S, Gerami M, Ramezani F, Karimi Abdolmaleki M (2019) Effect of silver nanoparticle treatment on the expression of key genes involved in glycosides biosynthetic pathway in Stevia rebaudiana B. Plant. Sugar Tech 22:518–527
Razzaq A, Ammara R, Jhanzab HM, Mahmood T, Hafeez A, Hussain S (2016) A novel nanomaterial to enhance growth and yield of wheat. J Nanosci Technol 2(1):55–58
Ren J, Lu Y, Qian Y, Chen B, Wu T, Ji G (2019) Recent progress regarding kaempferol for the treatment of various diseases. Exp Ther Med 18(4):2759–2776
Renuka N, Prasanna R, Sood A, Ahluwalia AS, Bansal R, Babu S, Singh R, Shivay YS, Nain L (2016) Exploring the efficacy of wastewater-grown microalgal biomass as a biofertilizer for wheat. Environ Sci Pollut Res 23(7):6608–6620
Rippka R, Deruelles J, Waterbury JB, Herdman M, Stainer RY (1979) Generic assignment, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111(1):1–61
Salama DM, Osman SA, Abd El-Aziz ME, Abd Elwahed MSA, Shaaban EA (2019) Effect of zinc oxide nanoparticles on the growth, genomic DNA, production and the quality of common dry bean (Phaseolus vulgaris). Biocatal Agric Biotechnol 18:101083
Sayed-Ahmed HI, Gaafar RM, Elsherif DE, Haider A, El-Shanshoury AR (2020) Effect of silver nanoparticles and Chlorella sp. suspension on vegetative growth of Eruca sativa. Egypt J Exp Biol (Botany) 16(1):27–33
Seo MS, Kim JS (2017) Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae. Molecules 22(9):1549
Shaaban MM (2001) Green microalgae water extracts as foliar feeding to wheat plants. Pak J Biol Sci 4:628–632
Singh S, Parihar P, Singh R, Singh VP, Prasad SM (2016) Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and genomics. Front Plant Sci 6:1143
Soleimani M, Habibi-Pirkoohi M (2017) Biosynthesis of silver nanoparticles using Chlorella vulgaris and evaluation of the antibacterial efficacy against Staphylococcus aureus. Avicenna J Med Biotechnol 9(3):120–125
Sonderby IE, Burow M, Rowe HC, Kliebenstein DJ, Halkier BA (2010) A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol 153:348–363
Spadafora ND, Amaro AL, Pereira MJ, Müller CT, Pintado M, Rogers HJ (2016) Multi-trait analysis of post-harvest storage in rocket salad (Diplotaxis tenuifolia) links sensorial, volatile and nutritional data. Food Chem 211:114–123
Sun W, Meng X, Liang L, Jiang W, Huang Y, He J, Hu H, Almqvist J, Gao X, Wang L (2015) Molecular and biochemical analysis of chalcone synthase from freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE 10(3):e0119054
Thiruvengadam M, Gurunathan S, Chung IM (2015) Physiological, metabolic, and transcriptional effects of biologically-synthesized silver nanoparticles in turnip (Brassica rapa ssp. rapa L.). Protoplasma 252:1031–1046
Torabfam M, Malmiri HJ (2018) Microwave-enhanced silver nanoparticle synthesis using chitosan biopolymer: optimization of the process conditions and evaluation of their characteristics. Green Process Synthesis 7:530–537
Tripathi DK, Singh S, Singh S, Pandey R, Singh VP, Sharma NC, Prasad SM, Dubey NK, Chauhan DK (2017) An overview on manufactured nanoparticles in plants: uptake, translocation, accumulation and phytotoxicity. Plant Physiol Biochem 110:2–12
Tungmunnithum D, Thongboonyou A, Pholboon A, Yangsabai A (2018) Flavonoids and other phenolic compounds from medicinal plants for pharmaceutical and medical aspects: an overview. Medicines 5(3):93
Vasconsuelo A, Boland R (2007) Molecular aspects of the early stages of elicitation of secondary metabolites in plants. Plant Sci 172:861–875
Waki T, Mameda R, Nakano T, Yamada S, Terashita M, Ito K, Tenma N, Li Y, Fujino N, Uno K, Yamashita S, Aoki Y, Denessiouk K, Kawai Y, Sugawara S, Saito K, Yonekura-Sakakibara K, Morita Y, Hoshino A, Takahashi S, Nakayama T (2020) A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat Commun 11(1):1–14
Yasin S, Liu L, Yao J (2013) Biosynthesis of silver nanoparticles by bamboo leaves extract and their antimicrobial activity. J Fiber Bioeng Inform 6(6):77–84
Zuverza-Mena N, Martínez-Fernández D, Du W, Hernandez-Viezcas JA, Bonilla-Bird N, López-Moreno ML, Komarek M, Peralta-Videa JR, Gardea-Torresdey JL (2017) Exposure of engineered nanomaterials to plants: Insights into the physiological and biochemical responses-a review. Plant Physiol Biochem 110:236–264
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
RG, AE and AH conceived the research idea and designed the experiments. DE performed the experiments, DE and RG analyzed the data. RG, DE and HSA wrote and revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sayed Ahmed, H.I., Elsherif, D.E., El-Shanshory, A.R. et al. Silver nanoparticles and Chlorella treatments induced glucosinolates and kaempferol key biosynthetic genes in Eruca sativa. Beni-Suef Univ J Basic Appl Sci 10, 52 (2021). https://doi.org/10.1186/s43088-021-00139-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43088-021-00139-2