Abstract
Background
To characterize the seminal microbiome associated with normal and abnormal semen parameters, towards the prediction of reproductive health and sperm quality. Despite the association between bacteria and infertility, few studies have looked at the beneficial effects of the seminal microbiome on infertility.
The study comprised semen samples from 69 men with normal spermiograms and 166 men with at least 1 abnormal spermiogram parameter from the Institutional IVF Center between October 2019 and October 2022.
We hypothesized that the composition of the microbiota may affect semen parameters. To determine the composition of uncultured bacteria, the 16S ribosomal RNA (rRNA) gene was amplified using Oxford Nanopore Technology.
Results
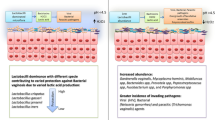
Different groups of bacteria were present in the semen samples of patients with normal semen parameters, such as female factor infertility and abnormal sperm parameters. Bacterial communities differed between samples. However, the relative distribution of Lactobacillus and Prevotella in the normal and abnormal semen groups differed (p = 0.05) and was statistically significant.
In the abnormal semen group, the incidence of Lactobacillus probiotics was lower and the frequency of Prevotella was higher. Additionally, principal component analysis (PCA) revealed differences in the microbial composition of normal and abnormal semen.
Conclusions
In our study, NGS analysis revealed the increased presence of harmful bacteria Prevotella in groups with abnormal semen raises the possibility that certain microbiota may be associated with semen quality and male infertility.
Similar content being viewed by others
Introduction
Microorganisms can be found in almost every environment in nature, including those inhabited by other living things. Microbial communities are present in and on the bodies of all multicellular organisms, and these microbiomes have a profound impact on the biology of their hosts. In the past, pathology was used to study the microbiome. Most studies have focused on the various gut, skin, and oral microbiomes, with relatively little attention paid to the reproductive microbiome [1].
In recent years, research into the diversity of microorganisms in semen samples has focused mainly on specific types of bacteria. Recently, however, new sequencing methods have made it possible to investigate the interactions and effects of entire microbial communities in semen samples [2].
Infertility, which affects 15% of couples attempting to conceive, is thought to be significantly influenced by male factors, either by themselves or in conjunction with female factors. [3]. Despite the fact that modern therapies improve the chances of conceiving for couples who are experiencing male infertility, they frequently neglect the lack of a specific etiological or pathophysiological diagnosis [3, 4].
Chromosomal abnormalities, genetic disorders, hormonal, environmental, physical, or psychological problems can cause male infertility. However, the exact cause of infertility in men is not always known. Male genitourinary (GU) illness is another cause of infertility that affects about 15% of infertile men. Male reproductive processes can be hampered in many ways by infections and the inflammation that follows in the GU tract [5].
Semen samples consistently show enhanced viscosity (seminal hyper-viscosity) in a large proportion of cases (12–29%). This condition is sometimes linked to increased leukocyte counts (leukocytospermia) and can be related to inflammation and genitourinary infections [6].
There is a significant need for study into the causes (and potential preventative treatments) of male infertility. There are several infectious etiologies that account for male factor infertility affecting 15% of couples [7]. Ochsendorf and colleagues indicated that there are so many pathogenic bacteria viral as well as fungal infections and protozoan species that can invade the normal genital-urinary system via sexual transmission, this applies to urine in the canaliculi or hematogenous spread of pregnancy [8, 9]. These diseases of the testicles, testicles, and prostate can cause problems with spermatogenesis and development [10, 11].
Infectious causes, such as urinary tract infections, along with non-infectious factors like exposure to environmental contaminants, man-made materials during intercourse, nicotine products, alcohol, and certain drugs, can contribute to the elevation of seminal leukocytes [12].
Low sperm motility exacerbated by vasectomy, varicocele, autoimmunity, abnormal spermatogenesis, and other non-infectious factors are possible causes of elevated sperm leukocytes [13, 14].
The male reproductive system has received less attention, especially when it comes to studying the 16S ribosomal RNA (16S rRNA) gene to characterize seminal plasma communities. Only seven semen studies with sample sizes ranging from 3 to 96 samples have been done to date, to the best of our knowledge, with half of the studies attempting to find a relationship with male infertility [15,16,17,18,19,20,21]. These examinations generally revealed a wide range of seminal bacteria, but no discernible difference was found between cases associated with infertility and healthy individuals. Yet, a negative relationship between the quality of the semen and the presence of Anaerococcus was found; on the other hand, Lactic acid bacteria, are more common in normal samples and are considered to have a probiotic effect against Pseudomonas and Prevotella [22].
Although next-generation sequencing (NGS) has expanded our toolkit, it also makes the discovery of novel microbes possible without the need for prior knowledge of sequencing information. In up to 45% of instances, the origin of abnormal semen parameters is unknown, therefore a thorough examination of the seminal microbiota should help us better understand male factor infertility [23]. In this study, we performed next-generation sequencing for the characterization of seminal bacterial diversity from 235 male participants according to the presence of normal and abnormal semen parameters respectively.
Materials and methods
Clinical study design and subjects
The study conducted by the Department of Reproductive Medicine and Research between October 2019 and October 2022 included semen samples from 69 men with normal spermiograms and 166 men (20–45 years old) with at least one abnormal spermiogram parameter. This study was approved by the University Hospital institutional ethics committee, with reference number PMU/IEC/089/2019. When their semen sample was used in this investigation, all patients gave their informed consent.
Inclusion criteria
At the time of the sampling, none of the males were receiving antibiotic treatment, and all were in generally good condition with no ongoing urogenital problems or STDs.
Exclusion criteria
Significantly, seminal culture results negative for male accessory gland infections have already ruled out this cohort for a number of sexually transmitted infections, including syphilis, HIV, hepatitis B and C, etc.
Semen collection and spermiograms analysis
After at least three days of abstinence from sexual activity, passing urine, and cleansing hands and genitalia with soap, semen was collected by masturbating. Throughout the whole processing of the semen sample, sterilized laboratory equipment was utilized. On the day of sample collection, a standard spermiogram analysis was carried out. It was then examined after liquefaction at 37 °C for 30 min. Samples by volume, pH, and viscosity, spermatozoa count, total motility were determined using optical microscopy respectively according to WHO 5th edition [24]. All of the samples were collected in sterile Cryo vials with sterile falcon tubes under laminar airflow, and they were promptly frozen at − 80 °C to maintain the variety of the microbiota. Therefore, maintaining stable storage conditions prior to metagenomics sequencing is equally crucial for achieving optimal nucleic acid yields.
Collected samples were stratified into three exclusive phenotypes: asthenospermia (motility < 40%), oligoasthenospermia was defined as sperm concentration (< 15 million/mL), motility (< 40%), and semen hyper-viscosity (thread > 2 cm). As a result, 69 samples were classified as control for normospermia since they did not exhibit any of the phenotypes related to infertility. Based on the results of the spermiogram, individuals were divided into two groups: normal group comprised (Group C = 69) was control with normospermic semen samples, and the abnormal group comprised (Group AT = 43) with asthenospermia, and (Group OA = 67) with oligoasthenospermia, and (Group H = 56) with semen hyper-viscosity respectively.
DNA isolation and quantification
DNA was isolated from semen samples using the DNeasypower soil kit (Qiagen, Hilden, Germany). The amount of isolated DNA was calculated using a Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA).
Ribosomal marker amplification by PCR and sequencing
The 16S Amplicon-Seq V1–V9 hypervariable regions were amplified for sequencing with primers that included the reverse 16S primer 5′-ATCGCCTACCGTGAC-barcode-CGGTTACCTTGTTTACGACTT-3′ and the forward 16S primer 5′-ATCGCCTACCAGGATTGAC-code, both of which contained gene-specific sequences from Oxford Nanopore’s MinION. To amplify the full-length 16S rRNA gene, mix 10 ng DNA template (10 µl), 25 µl Long Amp Taq 2X Master mix (NEB M0287), and 1 µl 16S barcode (barcode 01 is barcode 12) in a 50-µl volume. The 16S Barcoding Kit (SQK-RAB204) includes 14 µl of nuclease-free water with each sample.
The PCR temperature profile was as follows: a 60-s initial denaturation at 95 °C was followed by 25 cycles of 20 s at 95 °C, 30 s at 55 °C, 2 min at 65 °C, and 2 min at 95 °C for the final denaturation. 5′-ATGCCTACCGTGAC-Barcode-AGAGTTTGATMTGGCTCAG-3′ is the forward 16S primer. Using Beckman Coulter AMPure XP beads (High Wycombe, UK), the reverse 16S primer (5′-ATCGCCTACCGTGAC-Barcode-CGGTTACCTTGTTACGACTT-3′ amplicon) was purified at 0.5 × .
Concentrations varied between 10 and 20 ng/μl according to the scenario. Incubate for five minutes at room temperature after combining 5 μl of each pool with 0.5 μl of the SQKRAB204 Sequencing Kit Rapid Adapter (RAP). To establish a pool of amplified samples for sequencing, 1 µl of RAP (Rapid Annealing Primer) was added to the barcoded DNA, yielding a final pool of 100–150 ng and 10 copies.
35.5 μl of running buffer, 25.5 μl of library loading beads, and the generated DNA library (11 μl) were combined to create the mixture. The mixture was placed into SpotON Flow Cells Mk I (R9.4.1) (FLO-MIN106) and subjected to a 12–24-h processing period using MinKNOWTM software version 21.06.0.
Statistical analysis
Alpha and beta diversity, PCA plots, and heatmaps were used to examine the diversity of seminal bacteria. These methods have been shown to be useful for both individual sample and group analyses. Alpha diversity is calculated using the Shanon, Simpson, and Observed Taxa indices. To reduce overrepresentation, pooled data with at least 0.1% or a minimum of 10 reads were used for species categorization, as well as PCA and distance plots using the Bray–Curtis algorithm. The findings of the basic coordinate analysis were displayed in R version 4.0.2. The nonparametric Kruskal–Wallis test was used to compare differences in bacterial load, richness, and diversity. Demographic statistics are shown as interquartile ranges.
Data analysis workflow
The MinKNOW program was used to run the samples. Using Guppy basecaller v3.2.4, fast5 files were base-called following the run. With QCAT v1.0.7, the base called FASTQ files were de-multiplexed using the following command, which also cuts the adapters and barcodes: fastq -b output folder/ –detect-middle –trim -k qcat -f input file RAB204 Following trimming, readings ranging in size from 1200 to 1800 bp were chosen.
For the purpose of identifying species, the demultiplexed and trimmed reads underwent further analysis. The filtered text was read using the SINTAX taxonomic classifier of Usearch v10.0.240, providing higher level taxonomy to parse taxa in a self-contained format for the Ribosomal Repository Project version 16 taxon Read.
Phenotyping methods that group sequences into taxonomic bins based on similarity (taxonomic tracking analysis) were used to provide nanopore sequencing data and determine the composition of the microbiota. Because this strategy does not rely on in silico clustering for consistency, it can withstand many differences sometimes found in OTU-based methods. In the previous publication, it was shown that MinION sequencing based on One Codex analysis can provide rapid and accurate metagenomic analyses and organizations. It was created by limiting taxes.
Results
We evaluated seminal bacterial diversity using 16S rRNA next-generation sequencing (NGS) technologies to define the bacterial communities. Among the 235 males who participated in the study, 69 had normal spermiograms parameters and 166 had one or more abnormal parameters. Table 1 summarizes participant demographics and spermiograms results information.
Nevertheless, some variations in the relative abundance of probiotic Lactobacillus and several species with known pathogenic potential, including Dialister micraaerophilus and prevotella timonensis, were noted. Furthermore, it was shown that the makeup of the bacterial community in aberrant semen differed from controls, indicating that changes in the male urogenital tract microbiota and decreased reproductive success are linked to these conditions.
Bacterial communities' composition at various taxonomic levels
(C group) relative abundance: this sample is mixed/metagenomic. Classification of 38,147 readings using the One Codex database yielded 48.24% (Fig. 1). Relative abundance of (AT = group): this is a mixed/metagenomic sample 69.39% of 15,553 reads were classified. An additional 7.55% of reads were classified, but are non-specific. Lactobacillus crispatus, Streptococcus pneumonia, Lactobacillus delbrueckii, Lactobacillus helveticus, Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus thermophilus were among the highly dominant species (Fig. 2).
Relative abundance of (Group AT): this is a mixed/metagenomic sample 69.39% of 15,553 reads were classified. An additional 7.55% of reads were classified, but are non-specific. Lactobacillus crispatus, Streptococcus pneumonia, Lactobacillus delbrueckii, Lactobacillus helveticus, Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus thermophilus were among the highly dominant species
Relative abundance of (group OA): this sample is mixed/metagenomic. Analyzed Using the One Codex database, 50.57% of 29,626 readings were categorized (Fig. 3). Relative abundance of (group H): this is a mixed/metagenomic sample 100% of 1201 reads were classified using the One Codex database (Fig. 4).
The composition of bacterial communities at various taxonomic levels in our analysis, we detected five prominent strains (> 0.1%) that were shared by all four groups. The most frequent taxa were Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. Fusobacterium phyla were also found in mean quantities of around 1%, but only in groups OA and H (Figs. 3 and 4; Table 2). In this taxonomic category, group H stood out due to the higher proportion of Proteobacteria (27.3%) and the lower frequency of Firmicutes (51.3%).
However, in two-sample or overall comparisons, these group differences were statistically significant (Kruskal–Wallis tests) (Table 2).
When evaluating the difference in proteobacterial rates, borderline values were obtained only between the OA and H groups (Z score P = 0.062) and the AT and H groups (Z score P = 0.072). Group H reached 17.6% Gammaproteobacteria, while other groups ranged from 6.8% to 8.1% (Z score H vs. CP = 0.075, H vs. AT P = 0.062, and H vs. OA P = 0.067). Among Gammaproteobacteria, Enterobacteriaceae, and Pseudomonadales showed a slight increase in group H (around 5%) compared with other groups (0.8 to 3.3%).
Significant P value was obtained from multiple comparisons with Dunn’s test followed by the non-parametric Kruskal–Wallis test; this indicates that the C and AT groups are separated from each other, and the OA and H groups are separated from each other (Table 2). These four categories contained 44 families and 55 genera (more than 0.1%) (Fig. 4; Table 2). The most common bacteria in seminal plasma worldwide are Enterococci (> 23.8%), followed by Staphylococci (> 5.9%).), respectively).
Caulobacteriaceae, Pasteurellaceae (Aggregationella and Haemophilus), and Enterobacteriaceae (Klebsiella and Morganella) are the most abundant Proteobacteria genera. Corynebacteria and Propionibacteria from the phylum Actinobacteriaceae and Flavobacteriaceae from the phylum Bacteroidetes were the most abundant species in each group.
Contrarily, the taxa shown to be more abundant in the OA and H Groups, Pseudomonadaceae (Pseudomonas—Proteobacteria) in the Group H, Aerococcaceae (Aerococcus and Firmicutes), and Gemellaceae (Firmicutes) in the Group AT are those that appear to diverge more between the Groups.
Furthermore, it was shown that control samples had a greater relative abundance of the Lactobacillus genus. compared to abnormal semen sample groups (Figs. 1, 2, 3, and 4). Furthermore, comparison analysis reveals that samples from the AT, OA, and H groups have a higher prevalence of infections Prevotella and Enterococcus faecium (Fig. 5).
Furthermore, compared to normal semen bacterial communities, the abnormal seminal bacterial communities exhibited a statistically significant increase in bacterial diversity, according to alpha-diversity measurements like Shannon’s and Simpson’s diversity indices (Fig. 6, p < 0.01).
Additionally, the Bray–Curtis index (PCA), a measure of beta diversity, revealed a statistically significant difference in bacterial diversity between normal and abnormal semen (p = 0.001) (Fig. 7).
At the genus level, the normal semen group increased the number of the probiotic lactic acid bacteria Lactobacillus while decreasing the abundance of the pathogen Prevotella (p = 0.05) (Fig. 8).
The majority of the seminal microbiota in both normal and abnormal semen samples was made up of Lactobacillus and Prevotella; however, there were notable differences in the relative abundances of these two bacteria between the normal and abnormal samples. As a result, they might have clearly discernible beneficial and unfavorable effects on the quality of semen.
Discussions
This study looked at the microbiological composition of semen from males with both abnormal and normal spermiogram characteristics. Currently, it is unknown if particular bacterial communities could have an impact on sperm function. As there are benefits and drawbacks to every hypervariable region of the 16S rRNA gene, it is currently unclear which should be sequenced.
PCR or culture techniques have been used in earlier research to identify Gardnerella vaginalis in semen or the male genital tract. while two of the seminal microbiota investigations (Weng and colleagues and Hou and colleagues) used the V1–V2 region for sequencing.
Additionally, we used 16S Amplicon-Seq hypervariable regions V1–V9 for sequencing, choosing to use an identical approach, allowing us to compare our results side by side.
We found that there are differences in the overall bacterial load, diversity, and richness of general microbiota profiles. Allowing us to directly compare our findings, measures of alpha-diversity (Shannon and Simpson diversity indices) showed a statistically significant increase in bacterial diversity in abnormal seminal bacterial communities compared to normal seminal bacterial communities (Fig. 6, p < 0.01). However, new research has revealed that fertile male semen has a distinct microbiota [15, 16, 19]. Two of them, Prevotella and Lactobacillus, were analyzed by the enrichment of one genus.
Furthermore, in the total examination of the microbial community in our study, the relative abundance of the Lactobacillus genus was discovered to be higher in the control sample than in abnormal semen, and these results indicate that sperm quality is related to seminal disease. Figures 1, 2, 3, and 4). Lactobacilli are known to have a positive effect on the genital area and have previously been reported in semen samples.
This is consistent with recent research from Taiwan [25], which discovered three distinct types of semen microbial communities, two of which are identical to the ones observed here (a Lactobacillus-dominated group and a Prevotella-dominated group). In addition, [16] also discovered unique clusters of seminal microbiota, with Prevotella and Lactobacillus being two of the most abundant taxa in their research.
Furthermore, comparison analysis reveals a greater incidence of Prevotella and Enterococcus faecium infections in samples in groups AT, OA, and H (Fig. 5).
Urogenital tract infections (UTIs) in men are linked to 8–35% of significant factor infertility in men, is One of the important factors is asymptomatic bacteriospermia [26, 27].
A recent study showed that urinary tract infections are responsible for approximately 15% of male infertility [28]. In vitro fertilization and intrauterine insemination are two methods used to treat infertility and pathogenic bacteria have been found to be associated with these treatments [29].
Reactive oxygen species are released by the reproductive organs in response to pathogenic bacteria such Staphylococcus, Mycoplasma, Chlamydia, and Ureaplasma that create high volumes of white blood cells (ROS) [30,31,32]. These drugs have adverse effects on sperm parameters when consumed in excess [33,34,35].
However, in our study, subtle variations in the relative abundance of particular bacterial taxa were identified by NGS analysis. The genus Prevotella was associated with samples with abnormal spermiograms, especially in the AT and H groups (at least one defective parameter), (Fig. 4). While samples with normal spermiograms were linked to Lactobacillus and Staphylococcus, suggesting that bacteria may have the greatest influence on this parameter. Samples rich in Prevotella had the highest numbers of bacteria, members of the genus linked to vaginosis in women, and abnormal semen group [36, 37].
These microorganisms could alter sperm quality [38]. Reduced sperm viability is associated with microbial infections. [39]. There are different ways in which microorganisms can affect the male reproductive system: (1) some pathogenic bacterial strains found in semen can agglutinate motile spermatozoa, reducing their ability to undergo the acrosome reaction and altering their morphological characteristics [40]. There are many factors associated with male infertility including pre-, testicular, and post-testicular [41].
Inflammation in the male genital tract can be caused by anatomical, viral, immunological, or genetic factors [41, 42]. As a result, poor sperm quality and ultimately male infertility have been associated with an inflammatory state [22]. Microorganisms, in particular, have been shown to impair spermatozoa functions via a variety of mechanisms. Surprisingly, inflammatory cytokines and an increase in the formation of oxygen-reactive species are not the only factors that contribute to this unfavorable effect [43, 44].
Consequently, research on anaerobic bacteria has also been conducted. In this regard, Rehewy and associates observed that infertile people had more viable bacteria when they cultured the semen of both fertile and infertile males [45].
Recently, investigations using next-generation sequencing to detect microbial characteristics in sperm samples that are uniquely connected to infertility have been done [15]. Despite the fact that Hou and colleagues discovered a relationship between Anaerococcus infection and poor sperm quality, may play a role in infertility in men [16].
In the presence of hyper-viscosity and oligoasthenoteratozoospermia, Monteiro and colleagues discovered that the seminal microbiota exhibited a higher abundance of Proteobacteria and a lower abundance of Lactobacillus than the controls [22]. Consistent with these findings, we also observed increased Proteobacteria abundance, indicating that these microbial alterations might be linked to unfavorable consequences. Weng and colleagues studied 96 infertile men and found three distinct groups: Lactobacilli-dominant, Pseudomonas-dominant, and Prevotella-dominant; the last of these was related to sperm quality.
Semen production and sperm function are negatively impacted by Staphylococcus aureus infections. In addition to affecting sperm motility, morphology, and viability, this also impacts semen volume and concentration [15, 17, 46].
It is crucial to ascertain whether a specific microbial profile is connected to a person's reproductive status when discussing male infertility.
To the best of our knowledge, this is the first thorough investigation indicating that abnormal semen may be linked to a particular pathogenic bacterial profile, which can be utilized as a marker to monitor the course of infectious diseases and evaluate reproductive outcomes. As a result, our findings were not wholly surprising and were consistent with previously published literatures.
Conclusions
We intend to describe the role of seminal microbes associated with male infertility. Our findings demonstrated a strong link between sperm health and seminal bacterial communities,
Apart from its potential as a probiotic to preserve the integrity of sperm, Lactobacillus could also be helpful in lessening the negative consequences of Prevotella. However, in our study, NGS analysis revealed an increased presence of harmful Prevotella bacteria in the groups with abnormal sperm, raising the possibility that the male microbiota has a significant impact on male infertility. The different microbiome patterns found in normal and abnormal semen samples in relation to male infertility require confirmation through larger research with larger sample sizes.
Availability of data and materials
All information is accessible from the corresponding authors upon request.
Abbreviations
- GU:
-
Genitourinary
- ESL:
-
Elevated seminal leukocytes
- 16S rRNA:
-
16S ribosomal RNA gene
- NGS:
-
Next-generation sequencing
- WHO:
-
World Health Organization
- AT:
-
Asthenospermia
- OA:
-
Oligoasthenospermia
- H:
-
Hyper-viscosity
- UTIs:
-
Urogenital tract infections
- ROS:
-
Reactive oxygen species
- Et Br:
-
Ethidium bromide
References
Rowe M, Veerus L, Trosvik P, Buckling A, Pizzari T (2020) The reproductive microbiome: an emerging driver of sexual selection, sexual conflict, mating systems, and reproductive isolation. Trends Ecol Evol. 35(3):220–234
Aderaldo J, Teixeira DT, Torres MG, et al (2022) A Shotgun Metagenomic Mining Approach of Human Semen Microbiome. Res Sq. https://doi.org/10.21203/rs.3.rs-1220437/v1
Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, Dohle G et al (2012) European Association of Urology Working Group on Male Infertility. European Association of Urology guidelines on Male Infertility: the 2012 update. Eur Urol. 62(2):324–32
Trussell JC (2013) Optimal diagnosis and medical treatment of male infertility. Semin Reprod Med. 31(4):235–6
Alqawasmeh O, Fok E, Yim H, Li T, Chung J, Chan D (2023) The microbiome and male infertility: looking into the past to move forward. Hum Fertil (Camb). 26(3):450–462
Kumar N, Singh AK (2015) Trends of male factor infertility, an important cause of infertility: A review of literature. J Hum Reprod Sci Oct-Dec 8(4):191–196
Diemer T, Huwe P, Ludwig M, Hauck EW, Weidner W (2003) Urogenital infection and sperm motility. Andrologia. 35(5):283–7
Ochsendorf FR (2008) Sexually transmitted infections: impact on male fertility. Andrologia. 40(2):72–5
Keck C, Gerber-Schäfer C, Clad A, Wilhelm C, Breckwoldt M (1998) Seminal tract infections: impact on male fertility and treatment options. Hum Reprod Update 4(6):891–903
Henkel R, Ludwig M, Schuppe HC, Diemer T, Schill WB, Weidner W (2006) Chronic pelvic pain syndrome/chronic prostatitis affect the acrosome reaction in human spermatozoa. World J Urol. 24(1):39–44
Domes T, Lo KC, Grober ED, Mullen JB, Mazzulli T (2012) Jarvi K (2012) The incidence and effect of bacteriospermia and elevated seminal leukocytes on semen parameters. Fertil Steril 97(5):1050–1055
Close CE, Roberts PL, Berger RE (1990) Cigarettes, alcohol and marijuana are related to pyospermia in infertile men. J Urol. 144(4):900–3
Barratt CL, Bolton AE, Cooke ID (1990) Functional significance of white blood cells in the male and female reproductive tract. Hum Reprod. 5(6):639–48
Simbini T, Umapathy E, Jacobus E, Tendaupenyu G, Mbizvo MT (1998) Study on the origin of seminal leucocytes using split ejaculate technique and the effect of leucocytospermia on sperm characteristics. Urol Int 61(2):95–100
Weng SL, Chiu CM, Lin FM, Huang WC, Liang C, Yang T et al (2014) Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PLoS One. 9(10):e110152
Hou D, Zhou X, Zhong X, Settles ML, Herring J, Wang L et al (2013) Microbiota of the seminal fluid from healthy and infertile men. Fertil Steril. 100(5):1261–9
Chen H, Luo T, Chen T, Wang G (2018) Seminal bacterial composition in patients with obstructive and non-obstructive azoospermia. Exp Ther Med. 15(3):2884–2890
Baud D, Pattaroni C, Vulliemoz N, Castella V, Marsland BJ, Stojanov M (2019) Sperm microbiota and its impact on semen parameters. Frontiers in microbiology. 10:234
Liu CM, Osborne BJ, Hungate BA, Shahabi K, Huibner S, Lester R et al (2014) The semen microbiome and its relationship with local immunology and viral load in HIV infection. PLoS Pathog. 10(7):e1004262
Amato V, Papaleo E, Pasciuta R, Viganò P, Ferrarese R, Clementi N et al (2019) Differential composition of vaginal microbiome, but not of seminal microbiome, is associated with successful intrauterine insemination in couples with idiopathic infertility: a prospective observational study. Open Forum Infect Dis. 7(1):ofz525
Garcia-Segura S, Del Rey J, Closa L, Garcia-Martínez I, Hobeich C, Castel AB et al (2023) Characterization of seminal microbiome of infertile idiopathic patients using third-generation sequencing platform. International journal of molecular sciences. 24(9):7867
Monteiro C, Marques PI, Cavadas B, Damião I, Almeida V, Barros N et al (2018) Characterization of microbiota in male infertility cases uncovers differences in seminal hyperviscosity and oligoasthenoteratozoospermia possibly correlated with increased prevalence of infectious bacteria. Am J Reprod Immunol. 79(6):e12838
Akutsu T, Motani H, Watanabe K, Iwase H, Sakurada K (2012) Detection of bacterial 16S ribosomal RNA genes for forensic identification of vaginal fluid. Leg Med (Tokyo). 14(3):160–2
Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM et al (2010) World Health Organization reference values for human semen characteristics. Hum Reprod Update 16(3):231–245
Birse KD, Kratzer K, Zuend CF, Mutch S, Noël-Romas L, Lamont A et al (2020) The neovaginal microbiome of transgender women post-gender reassignment surgery. Microbiome. 8(1):61
Askienazy-Elbhar M (2005) Infection du tractus génital masculin: le point de vue du bactériologiste [Male genital tract infection: the point of view of the bacteriologist]. Gynecol Obstet Fertil. 33(9):691–7 (French)
Kopa Z, Wenzel J, Papp GK, Haidl G (2005) Role of granulocyte elastase and interleukin-6 in the diagnosis of male genital tract inflammation. Andrologia. 37(5):188–94
Fraczek M, Kurpisz M (2015) Mechanisms of the harmful effects of bacterial semen infection on ejaculated human spermatozoa: potential inflammatory markers in semen. Folia Histochem Cytobiol. 53(3):201–17
Damirayakhian M, Jeyendran RS, Land SA (2006) Significance of semen cultures for men with questionable semen quality. Arch Androl 52(4):239–242
Sasikumar S, Dakshayani D, Sarasa D (2013) An investigation of DNA fragmentation and morphological changes caused by bacteria and fungi in human spermatozoa. Int J Curr Microbiol App Sci 2(4):84–96
Ochsendorf FR (1999) Infections in the male genital tract and reactive oxygen species. Hum Reprod Update Sep-Oct 5(5):399–420
Pasqualotto FF, Sharma RK, Potts JM, Nelson DR, Thomas AJ, Agarwal A (2000) Seminal oxidative stress in patients with chronic prostatitis. Urology. 55(6):881–5
Segnini A, Camejo MI, Proverbio F (2003) Chlamydia trachomatis and sperm lipid peroxidation in infertile men. Asian J Androl. 5(1):47–9
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL et al (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 108 Suppl 1(Suppl 1):4680–7
Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA (2009) Topographical and temporal diversity of the human skin microbiome. Science 324(5931):1190–2
Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ (2010) Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 48(5):1812–9
Srinivasan S, Morgan MT, Liu C, Matsen FA, Hoffman NG, Fiedler TL et al (2013) More than meets the eye: associations of vaginal bacteria with gram stain morphotypes using molecular phylogenetic analysis. PLoS One. 8(10):e78633
Moretti E, Capitani S, Figura N, Pammolli A, Federico MG, Giannerini V et al (2009) The presence of bacteria species in semen and sperm quality. J Assist Reprod Genet. 26(1):47–56
Wolff H (1995) The biologic significance of white blood cells in semen. Fertil Steril. 63(6):1143–57
Sanocka D, Fraczek M, Jedrzejczak P, Szumała-Kakol A, Kurpisz M (2004) Male genital tract infection: an influence of leukocytes and bacteria on semen. J Reprod Immunol. 62(1–2):111–24
Mändar R, Punab M, Korrovits P, Türk S, Ausmees K, Lapp E et al (2017) (2017) Seminal microbiome in men with and without prostatitis. Int J Urol 24(3):211–216
Cariati F, D’Argenio V, Tomaiuolo R (2019) The evolving role of genetic tests in reproductive medicine. J Transl Med. 17(1):267
Sabeti P, Pourmasumi S, Rahiminia T, Akyash F, Talebi AR (2016) Etiologies of sperm oxidative stress. Int J Reprod Biomed. 14(4):231–40
Merino G, Carranza-Lira S, Murrieta S, Rodriguez L, Cuevas E, Morán C (1995) Bacterial infection and semen characteristics in infertile men. Arch Androl 35(1):43–47
Rehewy MS, Hafez ES, Thomas A, Brown WJ (1979) Aerobic and anaerobic bacterial flora in semen from fertile and infertile groups of men. Arch Androl. 2(3):263–8
Vajpeyee M, Tiwari S, Yadav LB, Tank P (2022) Assessment of bacterial diversity associated with assisted reproductive technologies through next-generation sequencing. Middle East Fertil Soc J. 27(1):1–9
Acknowledgements
The authors thank Pacific Medical University and Hospital for the facilities and support provided during the study.
Funding
The financial support was given by the Department of Science and Technology Government of Rajasthan India.
Author information
Authors and Affiliations
Contributions
MV, ST, and LBY wrote and drafted the manuscript. ST and LBY collected the semen samples and clinical information and performed the analysis. MV, ST, and LBY contributed equally to this work. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The institutional ethics committee granted ethical approval, with reference number PMU/IEC/089/2019, participants were given their informed consent before having their semen sample used in the study.
Consent for publication
N/A.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vajpeyee, M., Tiwari, S. & Yadav, L.B. Characterization of seminal microbiome associated with semen parameters using next-generation sequencing. Middle East Fertil Soc J 29, 22 (2024). https://doi.org/10.1186/s43043-024-00181-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43043-024-00181-x