Abstract
Background
To assess bacterial diversity in infertile couples with their biochemical pregnancy outcomes. Using a retrospective case-control study design, participants were recruited for collection of vaginal swab, follicular fluid, endometrial fluid, and semen samples. The microbial composition was analyzed by 16S ribosomal RNA (rRNA) gene amplification with (MinION) Oxford Nanopore Ltd.
Results
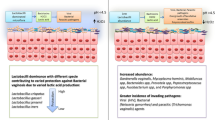
Our findings revealed that age and endometrial thickness had a significant impact on the pregnancy success rate of pregnant (P) and non-pregnant (NP) patients receiving IVF, with high levels of luteinizing hormone, estrogen, and progesterone in the P group. In addition, the partial least squares discriminant analysis (PLS-DA) revealed a difference in microbial composition between the P and NP groups, as well as a higher microbial abundance in non-pregnant patients compared to pregnant patients. After comparison between pregnant patients and non-pregnant patients, pregnant patients had a higher abundance of Firmicutes and Proteobacteria and a lower abundance of Actinobacteria, Fusobacterium, and Bacteroidetes at the phylum level. Non-pregnant patients had a lower abundance of the probiotics lactobacillus and a higher abundance of infections Prevotella and Gardnerella at the genus level. As a result, a disordered microbiota in non-pregnant patients, characterized by a decrease in probiotics and an increase in pathogens, could be used as a possible marker for a higher IVF failure rate.
Conclusion
Alteration of the microbiota of the reproductive tract or the presence of certain microbes, regardless of the degree of pathogenicity that can affect fertilization, as well as implantation and subsequent embryonic development. This could result in failed fertility treatments and a lower live birth (LBR) rate.
Similar content being viewed by others
Background
Infertility is an increasingly common problem around the world. There is a microbiome dependent host disease relationship. Infertility affects between 8% and 12% of couples of childbearing age worldwide [1]. However, in some regions of the world, infertility rates are much higher, reaching 30% in some populations [2]. The microbiome associated with the reproductive tract makes up 9% of the human microbiota, which includes the flora in the vagina, endometrial follicular fluid in women, and seminal fluid in men. Many reports showed the higher live birth rate (90–95%) after embryo transfer in the presence of predominant lactobacilli as vaginal microflora while being negatively affected by pathogenic bacteria [3]. The vaginal flora consists of about 30 species of microbes, which mainly live in the mucosa of the lateral wall in the vagina and some also in the cervix [4]. One of them, lactobacilli, is the most common microbe that plays a very important role in vaginal health [5, 6].
Lactobacillus can secrete a large amount of lactic acid in the anaerobic respiration of glycogen, which has the ability to make the vaginal environment acidic and maintain vaginal pH, and this acidic environment helps inhibit the spread of pathogenic bacteria. Lactobacillus has a high absorptivity of the vaginal mucosa, and therefore it can reduce the adsorption of other pathogenic bacteria to the vaginal mucosa and prevent the long-term presence of the pathogenic bacteria in the vagina [7, 8]. The presence of lactobacilli in the vagina is a vaginal microbiome necessary for maintaining vaginal health and protecting the reproductive system from harmful microorganisms [9]. Follicular fluid microbiome which can affect the outcome of IVF treatment due to the presence of Lactobacillus species and associated with improved embryo quality, leading to extremely high embryo transfer and pregnancy rates. Endometrium, originally considered a sterile site, and the presence of microbiota and Lactobalillus species has shown that it improves the implantation rate [10].
Infertility is caused by male factors (alone or in combination with female factors) in about 40% to 50% of infertile couples. Unfortunately, male infertility is still classified as idiopathic in a high percentage of cases [11,12,13]. The presence of pathogenic bacteria, namely Anaerococcus in the semen is negatively associated with its quality.
The aim of this present research was to find out whether the microbiota obtained by metagenomics analysis and present in vaginal swab, follicular fluid, endometrial fluid and semen are associated with poor treatment outcomes in assisted reproduction. Using next-generation sequencing, 16S ribosomal RNA (rRNA) gene sequence analysis can be used to identify bacteria without the need for culture in a way that does not depend on the subjective opinion of the investigator.
Therefore, a total of 255 IVF patients were enrolled in this study, and the researchers used a high-throughput sequencing technique to compare the patients' microbial diversity with the successes and failures of IVF outcomes.
Materials and methods
Ethics statement
The institutional ethics committee of Pacific Medical University has approved the use of human subjects for this study dated 30th June 2018 with the reference number (PMU/IEC/085/2018). All patients gave informed consent to the use of their vaginal swab, follicular fluid, endometrial fluid, and semen samples in this study.
Recruitment of participants
The inclusion criteria were as follows:
From July 2018 to December 2020, participants were recruited who met all of the following enrollment criteria: primary infertility patients attending Pacific IVF Center Udaipur India, no history of gynecological and abdominal surgery, having the normal sonographic texture of ovaries, and with no signs of hyperandrogenemia. Premenopausal Indian women who were not pregnant, non-clinical symptoms strongly suggestive of BV, schedule hormone replacement embryo transfer cycle with frozen embryos, and those who were provided written informed consent. The metagenomics study included vaginal swabs, follicular fluid, endometrial fluid, and semen samples.
The exclusion criteria were as follows:
Who had a female partner older than 40 years, more than 3 failed embryo transfers, and abnormal uterocervical anatomy, known to have vaginal or systemic infections, pelvic inflammatory disease or infected sperm. Known history of HIV-1/2, hepatitis B/C, syphilis, or genital chlamydia infection; known history of diabetes; known history of cervical/vaginal surgery; known history of use of intrauterine device; known history of antibiotic, steroid, or vaginal suppository use within the previous 2 weeks.
Sample collection
All vaginal samples and follicular fluid and semen samples were collected on the day of oocyte pickup, endometrial fluid samples were collected on the day of embryo transfer at day 3.
In order to keep the sample away from contamination, it is important to maintain an appropriate sample environment during sample collection, as changes in temperature, humidity, or other factors can alter or contaminate samples. In addition, we found that the proximity of different samples could lead to cross-contamination, which could later lead to incorrect results. In addition, sample collection time was minimized and aseptic laboratory resources were used, including gloves, masks and hats and appropriate sterile catheters in the embryology laboratory premises. All samples were collected in sterile Cryo vials under the laminar airflow using autoclaved sterile catheters and falcon tubes help reduce contamination and we performed rapid cooling at 80 °C to preserve the diversity of microbiota produced by a dry storage at 4 °C is changed significantly. Therefore, it is equally important to maintain consistent storage conditions to achieve optimal nucleic acid yields prior to metagenomics sequencing.
Vaginal swabs were collected from the vaginal midpoint and posterior fornix sample using a sterile swab to examine an area on the lateral wall of the vagina approximately midway between the introitus and the cervix. To avoid contamination from cervical mucus, the swab was gently pressed into the sidewall of the vagina and rotated four times to thoroughly coat the swab. Samples of follicular fluid were collected during oocyte retrieval in the embryology laboratory using an appropriate sterile protocol in a laminar air flow. After the embryo transfer was performed, we collected endometrial samples from the material remaining on the transfer catheter in a laminar airflow and transferred them to a Petri dish. The sterile 1.8 ml nest tube was opened in laminar air flow and the endometrial specimen was transferred to the tube and tightly capped. Semen samples were collected from male patients during the semen preparation procedure for the IVF cycle. All samples were frozen at − 80 °C (< 30 days) as soon as possible for further analysis. The entire analysis process is described in Fig. 1.
Based on the IVF pregnancy result, the samples were divided into two groups: those who had a successful pregnancy (P) and those who did not have a pregnancy (NP). All patients were given access to their medical records to learn more about their reproductive history and outcomes.
Reproductive outcomes analysis
Pregnancy outcome was determined fifteen days after embryo transfer by detecting b-human chorionic gonadotropin (b-HCG) in serum. The patients were divided into two groups, pregnancy group (P) and non-pregnancy group (NP) based on pregnancy outcome of IVF and their baseline condition, age, baseline hormone levels, number of embryos, and endometrial thickness were compared in Table 1.
DNA extraction and quantification
Bacterial DNA was extracted from samples according to the manufacturer’s instructions using the commercially available DNeasypower soil kit (Qiagen, Germany. Extracted DNA quantity was assessed using Nanodrop spectrophotometer (Thermo).
PCR amplification of ribosomal marker and sequencing
PCR mixture for the full-length 16S rRNA gene (50 μl total volume) contained 10 ng of DNA template (10 μl), 25 μl Long Amp Taq 2X master mix (NEB M0287), 1 μl 16S Barcode (Barcode 01 to Barcode 12 each for 1 sample separately) and 14 μl nuclease-free water as suggested in 16S Barcoding Kit (SQK-RAB204). The PCR thermal profile consisted of an initial denaturation of 60 s at 95 °C, followed by 25 cycles of 20 s at 95 °C, 30 s at 55 °C, 2 min at 65 °C, and a final step of 5 min at 65 °C. Forward 16S primer: 5′-ATCGCCTACCGTGAC-barcode-AGAGTTTGATCMTGGCTCAG-3′ Reverse 16S primer: 5′-ATCGCCTACCGTGAC-barcode-CGGTTACCTTGTTACGACTT-3′ The amplicons were cleaned-up with the AMPure XP beads (Beckman Coulter) using a 0.5× ratio. For each sample quantity was assessed using Nanodrop (Thermo Fisher Scientific) spectrophotometer. Success of 16S amplification was assessed in 1% agarose gels stained with ethidium bromide (EtBr). Samples that showed well-defined ≈ 1500 bp bands were mixed at proportional volumes. The pools were then purified using Beckman Coulter’s AMPure XP beads and resuspended in 10 mM Tris-HCl pH 8.0 with 50 mM NaCl, as directed by ONT. Nanodrop One (Thermo Fisher Scientific) was used to assess concentration of the purified pools. Wherever required, concentrations were adjusted to 10–20 ng/μl. Finally, 5 μl of each pool were mixed with 0.5 μl of the rapid adapter (RAP) of the SQKRAB204sequencing kit and incubated for 5 min at room temperature. By adding 1 l of RAP (rapid annealing primer) to the barcoded DNA, the different barcoded samples were mixed in an equimolar ratio to generate a final pool (100–150 ng in 10 l) for the sequencing library. The prepared library (11 μl of DNA library) was mixed with Library Loading beads (25.5 μl) and Running Buffer with fuel mix (35.5 μl) and further loaded on SpotON Flow Cells Mk I (R9.4.1) (FLO-MIN106) and sequenced for 12–24 h using the MinKNOW™ software version 21.06.0 (Oxford Nanopore Technologies Ltd.).
Statistical analysis
The statistical analysis is also being performed, where the alpha and beta diversity, PCA plots, heat map, etc. has been presented as applicable for individual samples (the same has been presented in figures footnotes) as well as within a group. Alpha diversity: based on Shannon, Simpson, and Observed taxa indices. Beta diversity: PCoA, PCA, and distance plots based on Bray-Curtis algorithm as well as grouped data was included to minimum 0.1% or a minimum of 10 reads classifying species to reduce overrepresentation. The findings of the principle coordinate analysis were shown using R version 4.0.2. Statistical analysis was performed using SPSS (version 25.0; IBM). Statistical significance was determined using Student's test, one-way or two-way ANOVA and annotated using international convention for statistical presentation. For continuous variables that satisfy a normal distribution, such as age and data, are reported as SD mean.
Results
Patient enrollment and baseline characteristics
The patient’s enrollment procedure is described in Fig. 2. Total 255, patients undergoing IVF and patients were enrolled this study between July 2018 and December 2020, 52 patients were excluded due to a lack of follow-up and 6 patients excluded for data missing. Total 197 patients were included in the study. In the end, further divided them into pregnancy group P (N = 99) and non-pregnancy group NP group (N = 98) respectively. Different endometrial preparation programs were chosen based on the patient’s physical condition including NC (natural cycle), COH (controlled ovarian hyper stimulation), HRT (hormone replacement therapy), D + HRT (downregulating hormone replacement therapy), and C + HRT (constant hormone replacement therapy).
All of the participants had primary infertility with regular menstrual cycles and no other medical issues. As shown in Table 1, age (p = 0.00) and endometrium thickness (p = 0.02) had a strong relationship related to pregnancy outcome. All the hormonal level were assessed at the start of the cycle (Table 1).
Microbial compositions in couples with positive and negative IVF clinical outcome
The microbial diversity of the P group and NP groups was compared using 16S ribosomal RNA (rRNA) gene amplification with (MinION) Oxford Nanopore Ltd. Our results indicated that a NP group had a higher Shannon index and Simpson index than P group.
Alpha-diversity and a comparison of the microbial community between the pregnancy (P) and non-pregnancy (NP) group
Vaginal swabs, follicular fluid, endometrial fluid, and semen samples collected from 197 enrolled infertile patients to learn more about the factors that influence IVF success. These samples were divided into group P (N = 99) and NP group (N = 98) respectively based on pregnancy status. The microbial diversity of the P and NP groups was compared using high-throughput sequences. Our results show that a total of 2,624,974 filtered clean reads (4,525.81 reads/samples) and 3,413 OTUs were obtained from all samples (data not shown). As shown in Fig. 3A, B, higher Shannon and Simpson index were obtained in the NP group compared to the P group. In Fig. 3C, the Scalar-Venn results indicated that there were 452 OTUs and 465 OTUs in P group and NP group, and 423 common OTUs were observed, which occupied 93.58% (423/452) and 90.97% (423/465) in P group and NP group, respectively. In addition, the partial least squares discriminant analysis (PLS-DA) result indicated that most dots in P groups scatted far away from that in NP groups (Fig. 3D).
In the P and NP groups, alpha-diversity and comparative study of bacterial diversity were performed. The Shannon index (A), the Simpson index, (B) the Scalar-Venn, (C) the PLS-DA (D). P, pregnancy success after embryo transfer; NP, pregnancy failure after embryo transfer; *means P < 0.05, p < 0.05 which indicates significant difference
Microbial community comparison between the P and NP groups at the phylum and genus levels
Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, and Bacteroidetes are the most common predominant in the P group (80.77, 9.52, 9.18, 0.04, and 0.40%, respectively) and the NP group (72, 61, 12.41, 8.01, and 3.75%). It was a population that accounted for 99.91 and 99.83%, respectively, of the total number of sequences in these two groups Compared with the NP group, a higher abundance of Firmicutes (80.77 vs. 72.61%) and Proteobacteria (9.18 vs. 8.01%), and a lower abundance of Actinobacteria (9.52 vs. 12.41%, p = 0.03), Fusobacteria (0.04 vs. 8.01%, p = 0.02), and Bacteroidetes (0.40 vs. 3.75%, p = 0.03) were observed in P group (Fig. 4).
At the phylum level, a comparison of the microbial communities of the P and NP groups. A Phylum-level cumulative bar charts of the main taxa in P and NP samples Firmicutes (B), Actinobacteria (C), Proteobacteria (D), Fusobacteria (E), Bacteroidetes (F). P, pregnancy success after embryo transfer; NP, pregnancy failure after embryo transfer; *means P < 0.05, p < 0.05 indicates significant difference
At the genus level, pregnancy increased the abundance of probiotic lactic acid bacteria (74.61 vs 63.09%) and decreased the abundance of the pathogen Gardnerella (6.03 vs 7.24%, p = 0.03), Atopobium (0.84 vs 4.14%, p = 0.02), Sneathia (0.03 vs. 3.75%, p = 0.02), and Prevotella (0.38 vs. 3, 02%, p = 0.04) (Fig. 5).
At the genus level, the microbial community of the P and NP groups was compared. A In the P and NP samples, cumulative bar charts of the main taxa at the genus level. Lactobacillus (B), Gardnerella (C), Atopbium (D), Sneathia (E), Prevotella (F). P, pregnancy success after embryo transfer; NP, pregnancy failure after embryo transfer; *means P < 0.05, p < 0.05 indicates significant difference
Discussion
In our study, we find that a decrease in Lactobacillus and an increase in Gardnerella, Atopobium, and Prevotella had a strong association with IVF failure [14].
These links were significantly stronger after the genera Gardnerella, Atopobium, and Prevotella became overgrown, as patients with a high incidence of Gardnerella, Atopobium, and Prevotella either failed to conceive or underwent abortion after embryo transfer. Therefore, it is important to describe the importance of the vaginal microbiota in IVF and demonstrate its potential as a biomarker for predicting IVF outcomes [15].
This study was conducted to search for an additional etiology for the absence of pregnancy after excluding all possible genetic, hormonal, overt infections, and other organic causes of infertility. All of these subjects appeared to be physically and mentally fit for conception [16, 17]. Fetal development from fertilization is not a sterile process. We found an abundance of bacteria, and normal and abnormal flora in our patient population. Culture yield is inferior in samples from the reproductive tract, so next-generation sequencing, a culture-independent technique, can identify and quantify bacterial DNA signature many times over.
The presence of non-Lactobacillus species such as Bacillus, Staphylococcus, Streptococcus, Acinetobacter, Enterococci, Prevotella, Sneathia, and Gardenella in our patient population at a certain titer individually or collectively exceeding a threshold value may indicate the positive impact of Lactobacillus species on pregnancy in an otherwise normal internal milieu, thereby reducing the live baby rate through various mechanisms [18].
Biofilm properties such as Enterococci, Bacillus species, Gardenella vaginalis, Stenotrophomonas maltophilia, Prevotella amnis, and Sneathia present in our samples help them to remain dormant in the reproductive tract, escape antibiotics such as metronidazole, and trigger biofilm dispersion in certain triggers such as stress and inflammation to generate local milieu [19]. A bacterial biofilm is a structured community of bacteria attached to a biological tissue or inert surface. A biofilm can be entrapped in a mucus substance: an extracellular polymeric (EPS) self-produced matrix [20]. This inflammation can affect gametogenesis, fertilization and implantation, and further embryonic development. Traditionally, follicular fluid, endometrial fluid, and semen samples are considered relatively sterile in conventional culture techniques, but metagenomics analysis has revealed the presence of distinct bacterial populations. These can contribute to infertility and failed fertility treatments. The vaginal microbiota of the woman of reproductive age consists largely of at least five different communities [21]. Lactic acid producing Lactobacillus species dominates four of these common state types (CSTs). While the fifth is generally anaerobic and strictly anaerobic, it is sometimes associated with vaginal symptoms. CST-IV does not contain large numbers of lactobacilli but consists of a polymicrobial mix of strict and facultative anaerobes including Gardnerella, Atopobium, Mobiluncus, Prevotella, and other Clostridiales taxa [22,23,24,25]. More than 90% of the vaginal microbiota consists of Lactobacillus species and further up in the endometrium and follicular fluid it makes up the predominant microbiota in people with a higher live birth rate. When this balance is disrupted, it can lead to infertility, as shown in this study. Semen samples and follicular fluid showed the presence of Bacillus species in abundance in our patient pool. Interestingly, microorganisms such as Bacillus subtilis, Pseudomonas aeruginosa, and Escherichia coli are isolated in the cervix area of infertile human females, which inhibit motility and agglutinate human sperm due to their elastase-positive activity [26]. Bacillus species, Propionibacterium species, Streptococcus species, Actinomyces species, Staphylococcus species, and some of the Bifidobacterium species of bacteria that have been linked to poor IVF outcomes. Our results show that bacterial species in follicular fluids, as well as follicular fluids themselves, can have both positive and negative effects on IVF outcomes [27]. The normal flora of the reproductive tract includes a number of lactobacilli species that promote a healthy and supportive environment for the embryo in the pre- and peri-conceptual periods. This can promote a supportive environment for implantation through the development of lactic acid hydrogen peroxide (H2O2), bacteriocins, antibiotics, harmful hydroxyl radicals, and probiotics [28, 29]. In our study, Lactobacillus was predominant in vaginal microbes, but other bacterial species dominated the follicular fluid, endometrium, and semen samples, revealing their role in infertility through direct and indirect mechanisms. In the presence of bacterial infections, much of the focus in recent years on intrauterine inflammation has been on the cytokine network. This signaling system is believed to play an important role from conception to implantation [30]. Embryo cytokine receptor expression allows cytokines and growth factors secreted into maternal fallopian tubes and uterine epithelial cells to influence the proper development and adaptation of the embryo to its microenvironment [30].
Granulocyte-macrophage colony-stimulating factor (GM-CSF) and colony-stimulating factor (CSF-1) growth factors are leukemia inhibitory factor (LIF), heparin-binding EGF-like growth factor (HB-EGF), insulin-like growth factor (IGF-1, IGF-2), and the cytokines IL-4, IL-10, and IL-11 which are essential for normal blastocyst production while other growth factors are critical for normal blastocyst development such as B. The male reproductive tumor necrosis factor tract. This bacterial infection can affect sperm activity in the male reproductive system and in the adrenal glands [31].
Staphylococcus aureus infections severely affecting the development of semen and sperm activity. This affects sperm volume and sperm concentration as well as sperm motility, morphology and viability [32]. As a result, there may be a causal relationship between staph infection and male infertility. 20.6% of S. aureus is registered, according to a previous study, S. aureus infection was found in semen samples from men with fertility problems. More directly, S. aureus infection has been found to be closely associated with low semen quality and reduced sperm motility [33]. This may play a role in what is known as idiopathic infertility. Although this bacterial flora is relatively harmless, on the one hand it increasingly exceeds certain threshold values and on the other hand it reduces the lactobacilli population and can lead to infertility. We need larger studies to provide more detailed information on this and the possible use of probiotics or antibiotics in the infertility management toolkit.
In conclusion, this is the first NGS-based study of the entire reproductive microbiota to show the presence of diverse bacterial populations in our population and possible implications for fertility potential and ART treatment outcome. Certain bacteria can affect fertility outcome more than others, making them an avenue for further research.
Availability of data and materials
All data available upon reasonable request.
Abbreviations
- ART:
-
Assisted reproductive technologies
- GU:
-
Genitourinary
- ESL:
-
Elevated seminal leukocytes
- NGS:
-
Next-generation sequencing
- IVF:
-
In vitro fertilization
- TEM:
-
Transmission electron microscopy
- PCR:
-
Polymerase chain reaction
- STD:
-
Sexually transmitted disease
- LBR:
-
Live birth rate
- BV:
-
Bacterial vaginosis
- GM-CSF:
-
Granulocyte-macrophage colony-stimulating factor
- CSF:
-
Colony-stimulating factor
- LIF:
-
Leukaemia inhibitory factor
- PTB:
-
Preterm birth
- TLR:
-
Toll-like receptors
- MMP-8:
-
Matrix metalloproteinase 8
- EMMPRIN:
-
Extracellular matrix metalloproteinase inducer
- OTU:
-
Operational taxonomic unit
- b-HCG:
-
b-Human chorionic gonadotropin
- IGF:
-
Insulin-like growth factor
- P:
-
Pregnancy group
- NP:
-
Non-pregnancy group
References
Ombelet W, Cooke I, Dyer S, Serour G, Devroey P (2008) Infertility and the provision of infertility medical services in developing countries. Hum Reprod Update 14(6):605–602
Nachtigall RD (2006) International disparities in access to infertility services. Fertil Steril 85(4):871–875
Moreno I, Simon C (2019) Deciphering the effect of reproductive tract microbiota on human reproduction. Reprod Med Biol 18(1):40–50
Song JH, Zheng JJ, Zhang HY, Tuo Y, Song SF (2011) Identification and analysis of vaginal lactobacilli in patients with bacterial vaginosis patients and healthy women in nationality of pastoral area. Zhonghua fu Chan ke za zhi 46(1):41–44
Xiao BB, Liao QP (2012) Analysis of diversity of vaginal microbiota in healthy Chinese women by using DNA-fingerprinting. Beijing da xue bao Yi xue ban= J Peking Univ Health Sci 44(2):281–287
Zhang D (2010) A study of vaginal flora. J Pract Obstetrics Gynecol 26(2):89–90
Chee WJ, Chew SY, Than LT (2020) Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microbial cell factories 19(1):1–24
Xiao CY, Zhang HW (2012) Analysis on the composition of 2066 cases with vagina microecological imbalance. Int J Obstetrics Gynecol 39(2):191–193
Van De Wijgert JH, Borgdorff H, Verhelst R, Crucitti T, Francis S, Verstraelen H et al (2014) The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One 9(8):e105998
Moreno I, Codoñer FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazán J (2016) Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 215(6):684–703
Kamischke A, Nieschlag E (1999) Analysis of medical treatment of male infertility. Hum Reprod 14(suppl_1):1–23
Carr BR (2013) Optimal diagnosis and medical treatment of male infertility. In: Seminars in reproductive medicine (Vol. 31, no. 04, ). Thieme Medical Publishers, pp 231–232
Rittenberg V, El-Touchy T (2010) Medical treatment of male infertility. Hum Fertil 13(4):208–216
Kong Y, Liu Z, Shang Q, Gao Y, Li X, Zheng C et al (2020) The disordered vaginal microbiota is a potential indicator for a higher failure of in vitro fertilization. Front Med 7:217
Amabebe E, Anumba DO (2018) The vaginal microenvironment: the physiologic role of lactobacilli. Front Med 5:181
Nuriel-Ohayon M, Neuman H, Koren O (2016) Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7:1031
Einenkel R, Zygmunt M, Muzzio DO (2019) Microorganisms in the healthy upper reproductive tract: from denial to beneficial assignments for reproductive biology. Reprod Biol 19(2):113–118
Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P et al (2007) Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J 1(2):121–133
Machado A, Cerca N (2015) Influence of biofilm formation by Gardnerella vaginalis and other anaerobes on bacterial vaginosis. J Infect Dis 212(12):1856–1861
Adegoke AA, Stenström TA, Okoh AI (2017) Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol 8:2276
Fredricks DN, Fiedler TL, Marrazzo JM (2005) Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 353(18):1899–1911
Campos AC, Freitas-Junior R, Ribeiro LF, Paulinelli RR, Reis C (2008) Prevalence of vulvovaginitis and bacterial vaginosis in patients with koilocytosis. Sao Paulo Med J 126(6):333–336
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL et al (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci 108(Supplement 1):4680–4687
Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X et al (2012) Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4(132):132ra52
KAUR M, Tripathi KK, Bansal MR, Jain PK, Gupta KG (1988) Bacteriology of the cervix in cases of infertility: effect on human and animal spermatozoa and role of elastase. Am J Reprod Immunol Microbiol 17(1):14–17
Pelzer ES, Allan JA, Waterhouse MA, Ross T, Beagley KW, Knox CL (2013) Microorganisms within human follicular fluid: effects on IVF. PLoS One 8(3):e59062
Hawes SE, Hillier SL, Benedetti J, Stevens CE, Koutsky LA, Wølner-Hanssen P et al (1996) Hydrogen peroxide—producing lactobacilli and acquisition of vaginal infections. J Infect Dis 174(5):1058–1063
Aroutcheva AA, Simoes JA, Behbakht K, Faro S (2001) Gardnerella vaginalis isolated from patients with bacterial vaginosis and from patients with healthy vaginal ecosystems. Clin Infect Dis 33(7):1022–1027
Ng SC, Hart AL, Kamm MA, Stagg AJ, Knight SC (2009) Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis 15(2):300–310
Hillier SL, Krohn MA, Rabe LK, Klebanoff SJ, Eschenbach DA (1993) The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women. Clin Infect Dis 16(Supplement_4):S273–S281
Robertson SA, Chin PY, Glynn DJ, Thompson JG (2011) Peri-conceptual cytokines–setting the trajectory for embryo implantation, pregnancy and beyond. Am J Reprod Immunol 66:2–10
Farsimadan M, Motamedifar M (2020) Bacterial infection of the male reproductive system causing infertility. J Reprod Immunol 142:103183
Onemu SO, Ibeh IN (2001) Studies on the significance of positive bacterial semen cultures in male fertility in Nigeria. Int J Fertil Women's Med 46(4):210–214
Acknowledgements
The authors acknowledge the Pacific Medical University and Hospital, for providing facilities and support during the study. We sincerely thank Dr. Shivoham Singh for Statistical analysis.
Funding
The study has been funded by the Department of Science and Technology Government of Rajasthan India.
Author information
Authors and Affiliations
Contributions
MV, ST, and LBY designed and wrote the manuscript. MV, ST, LBY, and PT revised the article. All authors have read and approved the manuscript. MV, ST, and LBY, contributed equally. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Institutional ethics committee of Pacific Medical University has approved the use of human subjects for this study dated 30th June 2018 with the reference number (PMU/IEC/085/2018). All patients gave informed consent to the use of their vaginal swab, follicular fluid, endometrial fluid, and semen samples in this study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vajpeyee, M., Tiwari, S., Yadav, L.B. et al. Assessment of bacterial diversity associated with assisted reproductive technologies through next-generation sequencing. Middle East Fertil Soc J 27, 28 (2022). https://doi.org/10.1186/s43043-022-00117-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43043-022-00117-3