Abstract
Background
Research on the role of variations in the mitochondrial genome in pathogenesis of acute lymphoblastic leukemia (ALL) has been unfolding at a rapid rate. Our laboratory has previously described higher number of copies of the mitochondrial genomes per cell in pediatric ALL patients as compared to the healthy controls. In the current study, we evaluated the pattern of mitochondrial genome variations in 20 de-novo pediatric B-ALL cases and seven controls. Quantitative real-time Polymerase Chain Reaction was used for estimation of mitochondrial genomes’ copy number in bone marrow samples of each ALL patient and peripheral blood samples of controls. The complete mitochondrial genomes of all samples were sequenced using the Illumina platform.
Results
Sequencing data analysis using multiple mitochondrial genome databases revealed 325 variants in all 27 samples, out of which 221 variants were previously known while 104 were unassigned, new variants. The 325 variants consisted of 7 loss-of-function variants, 131 synonymous variants, 75 missense variants, and 112 non-coding variants. New, missense variants (n = 21) were identified in genes encoding the electron transport chain complexes with most of them encoding ND4, ND5 of complex I. Missense and loss-of-function variants were found to be deleterious by many predictor databases of pathogenicity. MuTect2 identified true somatic variants present only in tumors between patient-sibling pairs and showed overlap with missense and loss-of-function variants. Online MtDNA-server showed heteroplasmic and homoplasmic variants in mitochondrial genome.
Conclusions
The data suggest that some of these variations might have a deleterious impact on the expression of mitochondrial encoded genes with a possible functional relevance in leukemia.
Similar content being viewed by others
Background
Acute lymphoblastic leukemia (ALL) is a result of uncontrolled, clonal proliferation of lymphoid cells due to arrest at an early stage of differentiation that can invade the bone marrow, blood, and extramedullary sites [32]. In India, 3.6% of new cancer cases were diagnosed as leukemia [3], out of which 75–80% were reported to be ALL [1]. ALL disease phenotype comprises of different subtypes with distinct somatic and genetic alterations, clinical features, and treatment responses [14, 15].
Mitochondria generate adenosine triphosphate (ATP) through oxidative phosphorylation, carried out by the electron transport chain (ETC) complex and also harmonize multiple interconnected aspects of cell proliferation, apoptosis and metabolism [2, 21]. The mitochondria have their own genome, a double-stranded 16.6-kb circular unmethylated DNA, encoding for 13 proteins of ETC complex, 12S rRNA and 16S rRNA, and 22 transfer RNAs. Mitochondrial DNA (mtDNA) is highly susceptible to oxidative stress due to its proximity to reactive oxidative species (ROS) produced in the mitochondrial matrix [33]. The ETC complex is made of multiple proteins encoded both by mtDNA and nuclear DNA (nDNA). A mitochondrion can contain two to eight copies of mtDNA [21], which is constantly turned over to maintain its copy number. The mtDNA changes reported in cancer include mutations, deletions, inversions, and copy number alterations and mutations in mtDNA regions have been reported in all types of human cancers [23, 50]. An analysis of mtDNA mutations in over 1600 tumor types reported over 1900 mitochondrial somatic DNA mutations with great variability in the number of mtDNA mutations observed across cancers with a very low frequency of mtDNA mutations in hematological cancers [24]. We have observed significantly higher mtDNA copy numbers in pediatric ALL patients than in controls and that mtDNA copy numbers could predict survival in ALL cases [20]. Variants in mtDNA have been analyzed in the D-loop and ETC coding regions in ALL [22, 27, 54, 55]. The current study aimed to explore the link between the mtDNA variants and mtDNA copy numbers by whole mitochondrial DNA sequencing to identify the regions encoded by these variants and their associated pathogenicity.
Materials and methods
Patients
The patients recruited in the study were newly diagnosed, untreated pediatric ALL patients. These were patients who visited the outpatient department of Medical Oncology at the B.R.A.I.R.C.H. Cancer Hospital at All India Institute of Medical Sciences (AIIMS), New Delhi between January 2017 and December 2017. The ALL diagnosis was confirmed by morphological, immunophenotyping and cytogenetic analyses on the bone marrow samples.
Sample collection and processing
Samples for sequencing and analysis of mtDNA were selected from bone marrow aspirates. The samples were collected from ALL patients in ethylenediaminetetraacetic acid (EDTA) vacutainers at the time of diagnostic sample collection. Mononuclear cells were obtained by whole blood cell lysis, aliquoted and stored at -80°. Total cellular DNA extraction was done as previously described, using lysis buffer, proteinase K and 20% SDS [8]. DNA was quantified using a Qubit fluorometer and quality was ascertained by the A260/280 ratio. One aliquot was used to estimate the activity of the ETC Complex I.
Mitochondrial DNA copy number estimation
Mitochondrial gene copy number was measured on the Bio-Rad CFX96 Real-Time PCR system for all samples using mitochondrial gene-specific primer sequences from the minor arc [35]. The primer sequences used for mitochondrial gene were Forward: CTAAATAGCCCACACGTTCCC and Reverse: AGAGCTCCCGTGAGTGGTTA and for nuclear gene, β-actin were Forward: TAGCTGTGCTCGCGCTACT and Reverse: TCTCTGCTGGATGACGTGAG. MtDNA copy number, normalized to copies of nuclear gene β-actin per cell, in each subject and control sample was calculated using the formula, 2∧[Ct(β-actin)−Ct(minor arc].
ETC Complex I enzyme activity estimation
Specific enzyme activity of ETC Complex I was estimated in cell pellets from the 20 tumor patients and 7 healthy controls. The activity was estimated using an ELISA-based kit for complex I (ab109721, USA) and protein concentration estimated using BCA Assay Kit (Thermo Scientific Pierce Cat No. 23225) as per the manufacturers’ protocol. Complex I activity was determined by following the oxidation of NADH to NAD+ and the simultaneous reduction of the dye (provided in the kit) as measured by the increase in absorbance at 450 nm in a spectrophotometer.
Whole mitochondrial DNA sequencing
Whole mitochondrial genome sequencing was performed on 27 participants and bone marrow aspirates were collected from 20 ALL patients and peripheral blood samples from two healthy adults and five sibling controls. The patient samples were stratified by mtDNA copy numbers to include 10 high and 10 low mtDNA copy number samples. The mitochondrial genome was amplified in two overlapping fragments using long-range PCR where a two-step PCR protocol was used to capture the target region to prepare the amplicon library for sequencing. PCR primers were designed for the target region, and genomic DNA was enriched for the region of interest by amplicon-based capture. The enriched genomic DNA was ligated with sequencing adapters to produce a library that was sequenced on Illumina HiSeq2500 to generate 2 × 150 bp sequence reads greater than 10,000 X sequencing coverage depth.
Identification of mtDNA variants
Multiple quality checks were done during the sequencing process, described in the Additional file 1: Sect. S1.I. The mtDNA variants, including SNPs and indels, were predicted individually for each sample using HaplotypeCaller and the results were combined using bcftools merge (version 1.9). Also, to distinguish mtDNA somatic variants (SNVs and indels) from germline variants, a case–control analysis was performed using MuTect2 by comparing the sequences of five paired tumor samples of patients with those of matched normal samples (respective siblings) with default parameters. Availability of normal tissue from the patients themselves as a representation of control sample was not an option as ALL is a hematological cancer and the leukemic cells are present everywhere in the blood and bone marrow of ALL patients. In addition, the systemic nature of ALL results in inflammation, which has been seen to affect the integrity of mitochondrial DNA. Hence, we took sibling pairs wherever feasible as the best match option.
Variant annotation
The impacts of rare and common variants on protein structure or function were predicted using SnpEff [4] and ANNOVAR [52]. Various mitochondrial specific databases and predictors were used for predicting the impact of variants on the pathogenicity of the disease, as mentioned in Additional file 1: Sect. S1.II. Additionally, variants were classified as pathogenic, likely pathogenic, of uncertain significance, likely benign or benign using the variant assessment guidelines as specified by the American College of Medical Genetics [39]. Various human mitochondrial genome databases such as, mtDB [18], mtSNP [7], HmtVar [37], PhyloP [36] and relevant literature were used for this classification.
Candidate somatic variants
All common variants reported with a minor allele frequency (MAF) of > 1% in the population-based databases (such as dbSNP-based Aggregate Allele Frequency-ALFA) [44] were not considered since those variants are likely to represent rare polymorphisms or low-penetrant variants rather than pathogenic somatic driver mutations. Known germline variants reported at dbSNP (version 151) were excluded and alterations listed as known somatic variations in COSMIC [6] were retained. All the variants were identified to be either ‘known’, based on their previously reported dbSNP ID, or ‘unreported, new’ due to an unassigned dbSNP ID.
Heteroplasmic and homoplasmic variants
Heteroplasmy is a condition where distinct mtDNA genome sequences for the same gene locus coexist in the same cell. Detection of heteroplasmic and homoplasmic variants and haplogroup classification was performed using publicly available online, mtDNA-server [53].
Statistical analysis
Non-parametric statistical tests were applied using GraphPad Prism 7.0 software (San Diego, California, US). Mann–Whitney U test (two-sample Wilcoxon rank-sum test) was used to look to compare mitochondrial DNA copy numbers and complex I enzyme activities in patients and controls. Analysis of effect of the type of variant (loss-of-function (LOFs)/missense) on the complex I enzyme activities was done using Mann-U Whitney statistical test. A ‘p’ value of < 0.05 was considered to be statistically significant.
Results
Patient characteristics
All 20 pediatric ALL patient samples analyzed in the sequencing analysis were of B-cell origin (Table 1). The mean age of patients was 10.2 years, and 12.8 years for controls. All the patients selected for the mitochondrial genome sequencing had > 70% leukemic lymphoblasts in the bone marrow.
MtDNA copy number analysis
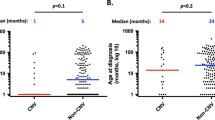
The mtDNA copy numbers in patients were significantly higher than that in healthy controls (p-value = 0.0026, Fig. 1a). The patients were divided into two categories based on their copy numbers i.e., high and low mtDNA copy numbers for comparison of mtDNA genome sequencing data. The categorization was based on the median mtDNA copy number (417.3 copies per cell) in the patient group.
Correlation between complex activity and mtDNA copy number
ELISA-based estimation of ETC Complex I enzyme activity showed a higher activity in the patient samples than controls, which was not statistically significant (p-value = 0.088, Fig. 1b). No significant correlation was found between enzyme activity and mtDNA copy numbers, when analyzed using Spearman’s correlation coefficient (data not shown). However, in our larger cohort, complex activities were found to be significantly higher in tumors and they showed a correlation with mtDNA copy number (unpublished results).
Analysis of variants
Full-length mitochondrial genome sequencing in the patient samples and controls using Illumina HiSeq platform revealed a total of 325 variants in the mitochondrial genome in 20 patients and seven controls that passed the criteria of the minimum genotypic quality score i.e., 20. The 325 variants identified included 104 unreported variants, while the remaining, 221 had a previously known dbSNP ID (database for single nucleotide polymorphism). Out of these 325 variants, there were 94 variants that were overlapping in both tumors and control samples, with 199 uniquely present in ALL patients and 32 base positions present only in controls (Table 2). Region-wise filtering of the unassigned variants from GATK variant calling identified the regions with a disproportionately high number of variants, i.e., the D-loop (n = 17), ND5 (n = 11), ND2 and ND4 regions (n = 10 each).
Based on their impact on subsequent protein levels or function, the 325 variants were classified into low (n = 131) (synonymous), high (n = 7) (loss-of-function, LOF), moderate (n = 75) (missense), and modifier (n = 112) (non-protein coding) variants. The seven LOF variants were observed in 7 out of 20 tumor patients and 2 out of 7 healthy controls. Two LOFs i.e., positions 5186 and 12007 were present in both tumor and sibling samples, while five were exclusively present in tumors. The new LOF variants are given in Table 3; Additional file 1: Table S1 lists the LOFs with a previously known dbSNP ID. Both LOFs in controls were seen in sibling samples. The LOF variants were present in the regions which encoded for ND1, ND2, ND4, and ND5 subunits of complex I and COX1 and COX3 subunits of complex IV of the ETC (Fig. 2a). The complex activities in patients with LOFs in regions encoding for ETC complex I were not significantly different from those patients with no LOFs at all or LOFs present in other complexes.
Out of a total of 75 missense variants mentioned earlier, variant annotation by SnpEff and Annovar found 21 unreported, missense variants encoding for genes of the ETC complexes and tRNAs (Table 4) and 54 missense variants with an assigned dbSNP ID (Additional file 1: Table S2). The 21missense variants, marked in the Circos plot (Fig. 2b), could have a moderate impact on protein-coding genes and could be disease-associated. Among these 21variants, the majority were found in tumor samples (18 ALL patients and 3 siblings) mostly in ND4 and ND5 encoding regions. In comparison of the complex I activities in patients with missense variants in regions encoding for complex I and those patients with no missense variants, no statistically significant difference was found. We observed a particular base position 4769 bp, which is a missense variant to be present in all the patients and this base position has a known dbSNP ID. This base position encodes for ND2, helical region and has been previously reported in multiple diseases such as Schizophrenia, muscle disease, cardiomyopathy, neuropathy.
Differences in mtDNA variants between high and low mtDNA copy number ALL patient samples
LOF variants were present in 4 of 10 ALL patients with high mtDNA copy number and 3 out of 10 with low mtDNA copy number but none of the variants were unique for either group. All ten patients in the high mtDNA copy number group had new, missense variants at different mitochondrial genome sites. There were 11 sites in all that encoded for ND2, ND5 and ND4L of the ETC complex I, COX1 of complex IV and a tRNA. In low mtDNA copy number group, eight out of the ten ALL patients had ten missense variants at various base pair positions, encoding for ND1 of complex I, COX2 and COX3 subunits of complex IV, ATP6 of complex V, and tRNAs. No predilections for any specific regions were observed between the two categories of ALL patients stratified by mtDNA copy number.
Analysis of new, unassigned missense variants using publicly available human mitochondrial genome databases
Deleterious impact and pathogenicity of missense variants that were unassigned by a dbSNP ID were summarized in a variant annotation table. The variants were characterized by pathogenicity predictors, cancer-specific predictors, pathogenicity meta-predictors and database of allelic frequency and phenotype. It was observed that 17 variants with unassigned dbSNP IDs, were classified as either possibly damaging, probably damaging, deleterious, damaging, disease, disease-causing, high impact, pathogenic or being fast-evolving sites based on different predictive software (Additional file 1: Table S3). The most common deleterious effects were seen in missense variants at base positions 5452, 13,022, and 13,535, encoding for complex I and 8594 encoding for complex V. Also, 41 out of 54 missense variants with a known dbSNP ID were listed as pathogenic or deleterious by these tools for predicting pathogenicity in Additional file 1: Table S4.
Analysis of somatic variants
MuTect2 pipeline of GATK compared the five paired samples of ALL patients and their respective siblings to filter out the germ-line variants and list out the true positive somatic variants [6]. Other criteria are listed in Additional file 1: Sect. S1.III. Overall, 93 somatic variants, out of which 6 were known somatic variants as already reported in COSMIC and 87 variants were probable somatic variants. Among these 87, a total of 32 variants had an existing dbSNP ID (Additional file 1: Table S5) and 55 were new probable somatic variants. Table 5 lists out these new probable somatic variants consisting of both missense and LOF variants at various base positions. The new somatic variants predicted by MuTect2 were mapped with the variants called individually by GATK and it was observed that 3 LOFs and 17 missense variants were overlapping between the lists in Tables 3 and 4. The details of somatic variants in the five pairs of ALL patients and their respective siblings are given in Additional file 1: Table S6.
Haplogroup determination of the mtDNA variants in ALL cases and controls
MtDNA-server analyzed the ALL patients and paired sibling controls for assigning haplogroups. The haplogroups assignments were M6, R, U2b1a, U1a1c1d and M35b2.
Analysis of heteroplasmic and homoplasmic variants
MtDNA-server database was used to identify homoplasmic and heteroplasmic variants at different base positions in the sequenced samples. A total of 415 base positions showed various heteroplasmic and homoplasmic variants, of which 64 positions showed heteroplasmy in some patients and homoplasmy in others. There were 58 positions (13.9%) that exclusively showed heteroplasmy and 229 positions had a homoplasmic appearance in the mtDNA genome. We are reporting here, the most common heteroplasmic and homoplasmic mtDNA sites only, from a very long list. The maximum number of homoplasmic variations were found at the mtDNA sites, m.263A > G (D-loop), m.750A > G and m.1438A > G (12S rRNA), m.4769A > G (ND2) and m.8860A > G (ATP6) in all 20 tumor samples and seven controls. m.73A > G (D-loop), m.7028C > T (COX1), m.14766C > T and m.15326A > G (CYTB) were present in 19 patients and seven controls and m.11719G > A (ND4) was found in 18 patients and seven controls. Similarly, the highest number of heteroplasmic variations were found at the base pair position m.3107N > T, present in all patients and controls, encoding for RNR2 (16S rRNA). The presence of homoplasmic and heteroplasmic variants in tumors and controls was also checked in the filtered list of new, missense and LOF variants in all samples (Additional file 1: Sect. S1.IV).
Discussion
The research data presented in this paper describe the deleterious variants that we identified in our study, in addition to previously reported ones, in the mitochondrial genome in lymphoblastic leukemia. Integrated genomic and transcriptomic analyses of large cohorts of childhood ALL have revolutionized our understanding of the genetic basis of ALL by identifying new subtypes, dysregulated pathways, and therapeutic targets within the last decade [41]. Metabolomics and proteomics studies have also been performed in ALL earlier [29]. However, whole mitochondrial DNA sequencing data is limited for ALL and the mitochondrial genome’s specific role in affecting the pathogenesis and prognosis in ALL is still unknown. This is due to factors such as the technical complexity inherent in analyzing the mitochondrial genome, the redundancy of the function of individual units of the mtDNA, and the non-feasibility of acquiring adjacent normal tissue.
Next-generation sequencing (NGS) is being used to detect targetable lesions in patients with ALL and to integrate sequencing data into disease management protocols [17]. The genomic landscape of adult and pediatric BCR-ABL1-like B-lymphoblastic leukemia has been described using parallel DNA and RNA sequencing [42]. A few studies have analyzed the whole mitochondrial genome to identify variants that could be helpful in ALL diagnosis, risk stratification, treatment planning and minimal residual disease (MRD) assessment [5, 25]. A pan-cancer mtDNA study identified the presence of 19 LOF somatic mutations in different regions of ETC complexes in pediatric B-ALL cases [49]. Our work reports novel LOFs not reported previously in B-ALL and, in fact, we have identified distinct LOF variants, with five of them encoding for ND1, ND2, ND4 and ND5 of complex I subunits and the remaining two for COX1 and COX3 of the complex IV region. We observed a higher number of LOFs in tumors (n = 7) as compared to controls (n = 2), suggesting a possible impact on the structure and function of these complexes in leukemia. Aside from previously reported disease-associated ATP6 variants such as m.8860A > G that occur with high frequency in breast cancers [10], we found additional deleterious/disease-related mutations in the ATP6 region in 13 ALL cases studied. Recently, authors have shown the utility of genome sequencing in diagnosis of pediatric patients with suspected mitochondrial disease and identification of nuclear and mtDNA variants [40]. A study performed in MELAS patients carrying the heteroplasmic m.3243A > G mutation showed that high mtDNA copy number and low heteroplasmy levels correlated with less severe disease [11].
An elevated mtDNA copy number might be associated with higher number of variations which might subsequently be associated with the pathogenesis of B-ALL. Previous studies have reported non-neutral missense or nonsense leukemia-specific mutations in mtDNA regions encoding ND1, ND2, ND4L, ND6, COX3, ATP6 and ATP8 genes in pediatric ALL patients indicating a role as a potential diagnostic marker [22, 54, 55]. In comparison to the two recent studies on pediatric B-ALL [22, 49], we observed 21 missense variants at new, unreported sites. Interestingly, many variants were only present in the tumor samples suggesting a likely association with some aspect of cancer. Most of the new variants were in regions encoding for different complex I subunits, of which the greatest number were in ND4 and ND5 regions. Given the role of complex I in maintaining the NAD+/NADH balance and ROS levels, the generation of mitochondrial membrane potential and ATP production, its dysfunction often underlies or contributes to mitochondrial disorders [26]. This hypothesis is supported by studies on mtDNA mutations in other cancers showing that functional disruption of Complex I leads to elevated ROS, manifesting as an increase in metastatic propensity and in the transcription of glycolytic- and metastasis-related genes [16, 24, 57]. We also observed two mutations linked to metastatic potential in other cancers: A13966G and A10398G (Additional file 1: Table S1) with the latter also being a germline polymorphism associated with cancer susceptibility [48]. While recent large-scale pan cancer analysis of mtDNA genomes suggest that the mtDNA mutational load in hematopoietic cancers is low compared to other cancers, the observation of some known and some novel mutations in our dataset indicate that mtDNA changes may contribute to pathogenesis and may have prognostic value [34, 56]. Additionally, the preponderance of variants in Complex I emphasizes its critical role as complex I is the largest complex of the ETC. Its functional alterations could lead to downstream effects on the assembly of mammalian ETC super complexes [31]. No correlation between presence of LOFs and missense variants in regions encoding for complex I and their enzymatic activities of complex I, in ALL patients, indicated that the activity doesn’t depend on the variants, directly, but might affect the functionality of the complexes of ETC.
Appropriate representation for assessing mtDNA mutations requires using paired tissue samples (tumor and normal) for identifying somatic variants [13]. We found 87 probable somatic variants in five case–control pairs, among which 32 had dbSNP IDs but 55 were new somatic variants, most commonly present in ETC complex I. Interestingly, there were overlaps between the new, probable somatic variants and the three LOFs and 17 missense variants present in ALL patients. We found that the ND4 and ND5 regions had the maximum number of new somatic mutations, a fact reported for other cancers [28, 43] and suggested to alter tumor progression [19].
Most pathogenic mtDNA mutations are heteroplasmic; mutant and wild-type (WT) mitochondrial genotypes can coexist, and affected cells will manifest metabolic defects only when the mtDNA mutation levels exceed a critical threshold [47]. Genetic and metabolic plasticity in cancer cells bearing heteroplasmic mutations was shown to allow higher oncogenic/metastatic potential [9]. Studies support the notion that mtDNA mutations occur throughout the tumor course and drift to homoplasmy with appreciable frequency [24, 56]. There appears to be a tissue-specific selection process for mtDNA mutations, advantageous for cancer spread [51]. The LOFs we found were higher in number in tumors and more in the homoplasmic than the heteroplasmic state in ALL samples. LOFs in the control samples were in a heteroplasmic state, which could theoretically maintain a threshold for maintenance of normal mitochondrial function. Deep NGS data from 1916 patients from 24 cancer types showed a positive selection in tumors via a preferential increase in allelic frequencies of non-synonymous mtDNA variants [12]. A direct correlation was seen between the increase in the number of heteroplasmic mtDNA variant and extent of atherosclerotic morphologic phenotype based on ratio of minor and major nucleotide mtDNA variants [45].
We did not observe a correlation between higher mtDNA copy numbers and complex activity, although the overall complex I activity was higher in tumors than controls. These observations indicate that higher mtDNA copy numbers in tumors do not necessarily translate into significantly higher complex activities. Perhaps a cumulative suppressive effect of LOF variants could impair complex function in leukemic cells. A similar observation was made in AML cells where increased mitochondrial mass and activity of the respiratory chain enzymes were not concordant [46]. A study in head and neck cancers, and esophageal squamous cell carcinoma, proposed that increased mtDNA copy number could compensate for the damaged mtDNA to maintain the supply of ATP by mitochondrial respiration to maintain cellular survival [30].
Conclusions
In summary, our data provide some information on novel LOF and missense variants in the ETC complexes in leukemic patients at sites, in addition to those already considered as somatic mutation hotspots in mtDNA genome of cancers. These variants could possibly have disease-associated or disease-modifying effects and deserve further exploration for functional relevance. The loss of function variants may support tumor growth by facilitating the rewiring of mitochondrial function in cancer cells to shift from energy generation to re-routing metabolites for macromolecular biosynthesis. Finally, a lack of correlation between mtDNA copy numbers and complex activity indicates that leukemic cells may maintain complex activities by upregulating copy numbers to reach a critical threshold for mitochondrial function necessary to support tumor growth and survival [38].
The presence of mtDNA variants in B-ALL portends a role in cancer through its impact on protein function and metabolic rewiring. However, the exact role of mitochondrial variants in leukemia requires in-vitro experiments to delineate mechanistic effects.
The major limitation of our study is that although we have found LOF and missense variants in greater numbers in ALL, functional validation of the variants linking them with causation/progression of cancer has not yet been done.
Availability of data and materials
Sequencing data of all the 27 mitochondrial DNA samples have been submitted to the National Center for Biotechnology Information (NCBI) and is available under BioProject accession number PRJNA662870.
Abbreviations
- ALL:
-
Acute lymphoblastic leukemia
- ATP:
-
Adenosine triphosphate
- ETC:
-
Electron transport chain
- MtDNA:
-
Mitochondrial DNA
- ROS:
-
Reactive oxidative species
- nDNA:
-
Nuclear DNA
- AIIMS:
-
All India Institute of Medical Sciences
- EDTA:
-
Ethylenediaminetetraacetic acid
- MAF:
-
Minor allele frequency
- LOF:
-
Loss-of-function
- WBC:
-
Whole blood cell counts
- Hb:
-
Hemoglobin
- NGS:
-
Next-generation sequencing
- MRD:
-
Minimal residual disease
- WT:
-
Wild-type
References
Arora RS, Arora B (2016) Acute leukemia in children: a review of the current Indian data. South Asian J Cancer 5(3):155–160. https://doi.org/10.4103/2278-330X.187591
Basak NP, Banerjee S (2015) Mitochondrial dependency in progression of acute myeloid leukemia. Mitochondrion 21:41–48. https://doi.org/10.1016/j.mito.2015.01.006
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6(2):80–92. https://doi.org/10.4161/fly.19695
Coccaro N, Anelli L, Zagaria A, Specchia G, Albano F (2019) Next-generation sequencing in acute lymphoblastic leukemia. Int J Mol Sci. https://doi.org/10.3390/ijms20122929
Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ (2017) COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res 45(D1):D777–D783. https://doi.org/10.1093/nar/gkw1121
Fuku N, Nishigaki Y, Tanaka M (2005) [Human mitochondrial genome polymorphism database (mtSNP)]. Tanpakushitsu Kakusan Koso. Protein Nucleic Acid Enzyme 50(14 Suppl):1753–1758
Fukuoh A, Cannino G, Gerards M, Buckley S, Kazancioglu S, Scialo F, Lihavainen E, Ribeiro A, Dufour E, Jacobs HT (2014) Screen for mitochondrial DNA copy number maintenance genes reveals essential role for ATP synthase. Mol Syst Biol 10:734. https://doi.org/10.15252/msb.20145117
Gammage PA, Frezza C (2019) Mitochondrial DNA: The overlooked oncogenome? BMC Biol 17(1):53. https://doi.org/10.1186/s12915-019-0668-y
Ghaffarpour M, Mahdian R, Fereidooni F, Kamalidehghan B, Moazami N, Houshmand M (2014) The mitochondrial ATPase6 gene is more susceptible to mutation than the ATPase8 gene in breast cancer patients. Cancer Cell Int 14:21. https://doi.org/10.1186/1475-2867-14-21
Grady JP, Pickett SJ, Ng YS, Alston CL, Blakely EL, Hardy SA, Feeney CL, Bright AA, Schaefer AM, Gorman GS, McNally RJ, Taylor RW, Turnbull DM, McFarland R (2018) MtDNA heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol Med 10(6):e8262. https://doi.org/10.15252/emmm.201708262
Grandhi S, Bosworth C, Maddox W, Sensiba C, Akhavanfard S, Ni Y, LaFramboise T (2017) Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection. Hum Mol Genet 26(15):2912–2922. https://doi.org/10.1093/hmg/ddx172
Hertweck KL, Dasgupta S (2017) The landscape of mtDNA modifications in cancer: a tale of two cities. Front Oncol 7:262. https://doi.org/10.3389/fonc.2017.00262
Hunger SP, Mullighan CG (2015) Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood 125(26):3977–3987. https://doi.org/10.1182/blood-2015-02-580043
Hunger SP, Mullighan CG (2015) Acute lymphoblastic leukemia in children. N Engl J Med 373(16):1541–1552. https://doi.org/10.1056/NEJMra1400972
Imanishi H, Hattori K, Wada R, Ishikawa K, Fukuda S, Takenaga K, Nakada K, Hayashi J (2011) Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PLoS ONE 6(8):e23401. https://doi.org/10.1371/journal.pone.0023401
Inaba H, Azzato EM, Mullighan CG (2017) Integration of next-generation sequencing to treat acute lymphoblastic leukemia with targetable lesions: the St. Jude children’s research hospital approach. Front Pediatr 5:258. https://doi.org/10.3389/fped.2017.00258
Ingman M, Gyllensten U (2006) mtDB: human mitochondrial genome database, a resource for population genetics and medical sciences. Nucleic Acids Res 34:D749-751. https://doi.org/10.1093/nar/gkj010
Iommarini L, Kurelac I, Capristo M, Calvaruso MA, Giorgio V, Bergamini C, Ghelli A, Nanni P, De Giovanni C, Carelli V, Fato R, Lollini PL, Rugolo M, Gasparre G, Porcelli AM (2014) Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum Mol Genet 23(6):1453–1466. https://doi.org/10.1093/hmg/ddt533
Jain A, Bakhshi S, Thakkar H, Gerards M, Singh A (2018) Elevated mitochondrial DNA copy numbers in pediatric acute lymphoblastic leukemia: a potential biomarker for predicting inferior survival. Pediatr Blood Cancer. https://doi.org/10.1002/pbc.26874
Jang Y, Lim K (2018) Recent advances in mitochondria-targeted gene delivery. Molecules 23(9):2316. https://doi.org/10.3390/molecules23092316
Järviaho T, Hurme-Niiranen A, Soini HK, Niinimäki R, Möttönen M, Savolainen E-R, Hinttala R, Harila-Saari A, Uusimaa J (2018) Novel non-neutral mitochondrial DNA mutations found in childhood acute lymphoblastic leukemia. Clin Genet 93(2):275–285. https://doi.org/10.1111/cge.13100
Jiménez-Morales S, Pérez-Amado CJ, Langley E, Hidalgo-Miranda A (2018) Overview of mitochondrial germline variants and mutations in human disease: focus on breast cancer (review). Int J Oncol 53(3):923–936. https://doi.org/10.3892/ijo.2018.4468
Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, Davies HR, Papaemmanuil E, Gundem G, Shlien A, Bolli N, Behjati S, Tarpey PS, Nangalia J, Massie CE, Butler AP, Teague JW, Vassiliou GS, Green AR, Du M-Q, Unnikrishnan A, Pimanda JE et al (2014) Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. https://doi.org/10.7554/eLife.02935
Kodroń A, Ghanim M, Krawczyk KK, Stelmaszczyk-Emmel A, Tońska K, Demkow U, Bartnik E (2017) Mitochondrial DNA in pediatric leukemia patients. Acta Biochim Pol 64(1):183–187. https://doi.org/10.18388/abp.2016_1444
Koshikawa N, Akimoto M, Hayashi J-I, Nagase H, Takenaga K (2017) Association of predicted pathogenic mutations in mitochondrial ND genes with distant metastasis in NSCLC and colon cancer. Sci Rep 7(1):15535. https://doi.org/10.1038/s41598-017-15592-2
Kwok CS-N, Quah TC, Ariffin H, Tay SK-H, Yeoh AE-J (2011) Mitochondrial D-loop polymorphisms and mitochondrial DNA content in childhood acute lymphoblastic leukemia. J Pediatr Hematol Oncol 33(6):e239-244. https://doi.org/10.1097/MPH.0b013e31820a5ece
Larman TC, DePalma SR, Hadjipanayis AG, Cancer Genome Atlas Research Network, Protopopov A, Zhang J, Gabriel SB, Chin L, Seidman CE, Kucherlapati R, Seidman JG (2012) Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A 109(35):14087–14091. https://doi.org/10.1073/pnas.1211502109
Leahy J, Spahis S, Bonneil E, Garofalo C, Grimard G, Morel S, Laverdière C, Krajinovic M, Drouin S, Delvin E, Sinnett D, Marcil V, Levy E (2018) Insight from mitochondrial functions and proteomics to understand cardiometabolic disorders in survivors of acute lymphoblastic leukemia. Metab Clin Exp 85:151–160. https://doi.org/10.1016/j.metabol.2018.03.011
Lin C-S, Lee H-T, Lee M-H, Pan S-C, Ke C-Y, Chiu AW-H, Wei Y-H (2016) Role of mitochondrial DNA copy number alteration in human renal cell carcinoma. Int J Mol Sci. https://doi.org/10.3390/ijms17060814
Lobo-Jarne T, Ugalde C (2018) Respiratory chain supercomplexes: Structures, function and biogenesis. Semin Cell Dev Biol 76:179–190. https://doi.org/10.1016/j.semcdb.2017.07.021
Malard F, Mohty M (2020) Acute lymphoblastic leukaemia. Lancet (Lond, Engl) 395(10230):1146–1162. https://doi.org/10.1016/S0140-6736(19)33018-1
Memon AA, Zöller B, Hedelius A, Wang X, Stenman E, Sundquist J, Sundquist K (2017) Quantification of mitochondrial DNA copy number in suspected cancer patients by a well optimized ddPCR method. Biomol Detect Quantif 13:32–39. https://doi.org/10.1016/j.bdq.2017.08.001
Pérez-Amado CJ, Tovar H, Gómez-Romero L, Beltrán-Anaya FO, Bautista-Piña V, Dominguez-Reyes C, Villegas-Carlos F, Tenorio-Torres A, Alfaro-Ruíz LA, Hidalgo-Miranda A, Jiménez-Morales S (2020) Mitochondrial DNA mutation analysis in breast cancer: shifting from germline heteroplasmy toward homoplasmy in tumors. Front Oncol 10:572954. https://doi.org/10.3389/fonc.2020.572954
Phillips NR, Sprouse ML, Roby RK (2014) Simultaneous quantification of mitochondrial DNA copy number and deletion ratio: a multiplex real-time PCR assay. Sci Rep 4:3887. https://doi.org/10.1038/srep03887
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20(1):110–121. https://doi.org/10.1101/gr.097857.109
Preste R, Vitale O, Clima R, Gasparre G, Attimonelli M (2019) HmtVar: a new resource for human mitochondrial variations and pathogenicity data. Nucleic Acids Res 47(Database issue):D1202–D1210. https://doi.org/10.1093/nar/gky1024
Reznik E, Wang Q, La K, Schultz N, Sander C (2017) Mitochondrial respiratory gene expression is suppressed in many cancers. Elife. https://doi.org/10.7554/eLife.21592
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and Genomics and the association for molecular pathology. Genet Med Off J Am Coll Med Genet 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Riley LG, Cowley MJ, Gayevskiy V, Minoche AE, Puttick C, Thorburn DR, Rius R, Compton AG, Menezes MJ, Bhattacharya K, Coman D, Ellaway C, Alexander IE, Adams L, Kava M, Robinson J, Sue CM, Balasubramaniam S, Christodoulou J (2020) The diagnostic utility of genome sequencing in a pediatric cohort with suspected mitochondrial disease. Genet Med Off J Am Coll Med Genet 22(7):1254–1261. https://doi.org/10.1038/s41436-020-0793-6
Roberts KG (2018) Genetics and prognosis of ALL in children vs adults. Hematol Am Soc Hematol Educ Program 2018(1):137–145
Severson EA, Vergilio J-A, Gay LM, Daniel S, Hemmerich AC, Elvin JA, Britt N, Nahas M, Cohen MB, Brown C, Sathyan P, Rankin A, Miller V, Ross JS, Ramkissoon SH (2019) Genomic landscape of adult and pediatric BCR-ABL1-Like B-lymphoblastic leukemia using parallel DNA and RNA sequencing. Oncologist 24(3):372–374. https://doi.org/10.1634/theoncologist.2018-0272
Shen L, Wei J, Chen T, He J, Qu J, He X, Jiang L, Qu Y, Fang H, Chen G, Lu J, Bai Y (2011) Evaluating mitochondrial DNA in patients with breast cancer and benign breast disease. J Cancer Res Clin Oncol 137(4):669–675. https://doi.org/10.1007/s00432-010-0912-x
Sherry ST, Ward M, Sirotkin K (1999) DbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res 9(8):677–679
Sobenin IA, Zhelankin AV, Khasanova ZB, Sinyov VV, Medvedeva LV, Sagaidak MO, Makeev VJ, Kolmychkova KI, Smirnova AS, Sukhorukov VN, Postnov AY, Grechko AV, Orekhov AN (2019) Heteroplasmic variants of mitochondrial DNA in atherosclerotic lesions of human aortic intima. Biomolecules 9(9):E455. https://doi.org/10.3390/biom9090455
Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, Jhas B, Hurren R, Gronda M, Wang X, Jitkova Y, Sukhai MA, Lin F-H, Maclean N, Laister R, Goard CA, Mullen PJ, Xie S, Penn LZ et al (2015) AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 125(13):2120–2130. https://doi.org/10.1182/blood-2014-08-594408
Stewart JB, Chinnery PF (2015) The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 16(9):530–542. https://doi.org/10.1038/nrg3966
Tengku Baharudin N, Jaafar H, Zainuddin Z (2012) Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular Malaysia. Malays J Med Sci MJMS 19(1):36–42
Triska P, Kaneva K, Merkurjev D, Sohail N, Falk MJ, Triche TJ, Biegel JA, Gai X (2019) Landscape of germline and somatic mitochondrial DNA mutations in pediatric malignancies. Can Res 79(7):1318–1330. https://doi.org/10.1158/0008-5472.CAN-18-2220
van Gisbergen MW, Voets AM, Starmans MHW, de Coo IFM, Yadak R, Hoffmann RF, Boutros PC, Smeets HJM, Dubois L, Lambin P (2015) How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutat Res Rev Mutat Res 764:16–30. https://doi.org/10.1016/j.mrrev.2015.01.001
Wang J, Schmitt ES, Landsverk ML, Zhang VW, Li F-Y, Graham BH, Craigen WJ, Wong L-JC (2012) An integrated approach for classifying mitochondrial DNA variants: one clinical diagnostic laboratory’s experience. Genet Med Off J Am Coll Med Genet 14(6):620–626. https://doi.org/10.1038/gim.2012.4
Wang K, Li M, Hakonarson H (2010) ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. https://doi.org/10.1093/nar/gkq603
Weissensteiner H, Forer L, Fuchsberger C, Schöpf B, Kloss-Brandstätter A, Specht G, Kronenberg F, Schönherr S (2016) mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res 44(W1):W64-69. https://doi.org/10.1093/nar/gkw247
Yacoub HA, Mahmoud WM, El-Baz HAE-D, Eid OM, ELfayoumi, R. I., Elhamidy, S. M., & Mahmoud, M. M. (2014) Novel mutations in the displacement loop of mitochondrial DNA are associated with acute lymphoblastic leukemia: a genetic sequencing study. Asian Paci J Cancer Prev APJCP 15(21):9283–9289. https://doi.org/10.7314/apjcp.2014.15.21.9283
Yacoub HA, Mahmoud WM, El-Baz HA-EE-D, Eid OM, El-Fayoumi RI, Mahmoud MM, Harakeh S, Abuzinadah OH (2014) New haplotypes of the ATP synthase subunit 6 gene of mitochondrial DNA are associated with acute lymphoblastic leukemia in Saudi Arabia. Asian Pac J Cancer Prev APJCP 15(23):10433–10438. https://doi.org/10.7314/apjcp.2014.15.23.10433
Yuan Y, Ju YS, Kim Y, Li J, Wang Y, Yoon CJ, Yang Y, Martincorena I, Creighton CJ, Weinstein JN, Xu Y, Han L, Kim H-L, Nakagawa H, Park K, Campbell PJ, Liang H, PCAWG Consortium (2020) Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet 52(3):342–352. https://doi.org/10.1038/s41588-019-0557-x
Zielonka J, Kalyanaraman B (2008) “ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis”—a critical commentary. Free Radical Biol Med 45(9):1217–1219. https://doi.org/10.1016/j.freeradbiomed.2008.07.025
Acknowledgements
We thank the nursing staff of B. R. Ambedkar Institute Rotary Cancer Hospital (B.R.A.I.R.C.H.), AIIMS, New Delhi, for assistance in the bone marrow sample collection. We are highly grateful to the ALL patients and their guardians who participated in the study. We would like to acknowledge the contribution of MedGenome Labs Ltd., Bangalore, where the mtDNA sequencing was performed under the supervision of Dr. Lakshmi Soundararajan. We are grateful to the ICMR genomics facility at AIIMS, New Delhi, for helping with the analysis of the mitochondrial genome sequencing data.
Funding
This study was financially supported by an Intramural grant from All India Institute of Medical Sciences (AIIMS), New Delhi, India (Grant number A-535) and junior research fellowship from Council of Scientific and Industrial Research (CSIR) (Grant number 0463). Funding from AIIMS had the main role in the design of the study and collection, analysis, and interpretation of data of the manuscript should be declared.
Author information
Authors and Affiliations
Contributions
Author contributions made to the manuscript were as follows: Study design: AS, JKP, SB; Data collection: AJ, RS; Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources: AS, AJ; Data analysis: AJ, AK, HS; Supervision: AS, JKP, SB; Validation, Writing- Original draft preparation, Visualization: AS, AJ, AK; Writing- Reviewing and Editing: AS, AJ, AK, JKP. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was initiated after obtaining ethical clearance by the Institutional Ethics Committee (IECPG/305/27.04.2016). Informed consent or assent was obtained from the guardians of patients and controls prior to enrollment in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Detailed methodology and results of anlaysis of variants.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jain, A., Katiyar, A., Singh, R. et al. Implications of mitochondrial DNA variants in pediatric B-cell acute lymphoblastic leukemia. Egypt J Med Hum Genet 23, 133 (2022). https://doi.org/10.1186/s43042-022-00347-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-022-00347-0