Abstract
Background
Breast cancer is the most common malignancy in women throughout the world. Mitochondria play important roles in cellular energy production, free radical generation and apoptosis. Identification of mitochondrial DNA mutations and/or polymorphisms as cancer biomarkers is rapidly developing in molecular oncology research.
Methods
In this study, the DNA alterations of the mitochondrial ATPase 6 and 8 genes were investigated in 49 breast cancer patients using PCR amplification and direct DNA sequencing on mtDNA. A possible association between these variants and tumorigenesis was assessed. Furthermore, the impact of non-synonymous substitutions on the amino acid sequence was evaluated using the PolyPhen-2 software.
Results
Twenty eight distinct somatic mitochondrial DNA variants were detected in tumor tissues but not in the corresponding adjacent non-tumor tissues. Among these variants, 9 were observed for the first time in breast cancer patients. The mtDNA variants of A8384 (T7A), T8567C (I14T), G8572A (G16S), A9041G (H172R) and G9055A (A177T) showed the most significant effects probably due to damaging changes to the resulting protein. Furthermore, non-synonymous amino acid changing variants were more frequent in the ATPase6 gene compared to the ATPase8 gene.
Conclusion
Our results showed that the ATPase6 gene is more susceptible to variations in breast cancer and may play an important role in tumorigenesis by changing the energy metabolism level in cancer cells.
Similar content being viewed by others
Introduction
Breast cancer is a major public health problem in women throughout the world. In 2008, it was the most common cause of death in women (458,000 deaths) worldwide [1]. Breast cancer is also the most common cancer and the fifth cause of mortality due to malignancies among Iranian women [1, 2].
The human mitochondrial genome consists of a circular double-stranded DNA of 16,569 base pairs, including genes encoding for the electron transport chain (complexes I-IV), ATP synthase or complex V in oxidative phosphorylation as well as a displacement loop region, 2 ribosomal RNAs (16 and 23) and 22 transfer RNAs [3, 4]. Variants of ATPase subunit 6 (8366–8572) and ATPase subunit 8 (8527–9207) in mitochondrial DNA (mtDNA) has been reported in different types of cancers, including breast, colon and ovarian [5–7].
The mitochondrion plays a critical role in cellular energy production [8], carcinogenesis and tumor progression, and could be a prognostic marker in different cancer types [5, 9–15]. To date, various types of mtDNA alterations, including point variants, large deletion and copy number changes have been reported in breast, colon and ovarian cancers [5, 16]. There is strong evidence that mtDNA alterations can enhance oxidative stress and the risk of tumor development as well as tumor initiation, proliferation [17], metastasis [18–20] and resistance of cancer cells to apoptosis [21].
Therefore, this study was undertaken to evaluate mitochondrial ATPase6 and 8 alterations in tumor and adjacent non-tumor tissues in breast cancer patients. We also investigated the correlation between the variants in these genes and the clinico-pathological features in these breast cancer patients.
Materials and Methods
Tumor tissue collection
Forty-nine breast cancer patients (34–75 years of age with a median age of 52.43 years) took part in this study. The patients were referred to the National Cancer Institute (NCI) at Imam Khomeini Hospital Complex, Tehran, Iran, from Oct. 2007 to Oct. 2009. Tumor tissue and adjacent non-tumor tissue samples were obtained from the Iranian National Tumor Bank (INTB) at NCI. Each specimen was immediately frozen following resection and stored at −80°C until DNA extraction. The pathologic changes in tumor samples were confirmed by two expert pathologists as adenocarcinomas according to the American Joint Committee on Cancer [22]. None of the patients received chemotherapy or radiotherapy treatment before they underwent surgery. All patients were informed on the aim of the study and signed an informed consent approved by the INTB Ethical Committee for the genetic analysis.
DNA extraction and PCR
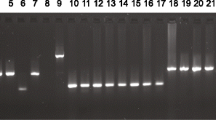
In order to identify the alterations in the mtDNA ATPase6 and ATPase8 genes, PCR-sequencing was performed as described previously with some modifications [23] Total genomic DNA was extracted from fresh tumor samples containing at least 90% neoplastic cells, as well as their adjacent non-tumor tissues, using the QIAamp Mini Kit (USA). The sequences of the primers were as follows: F-ATPase: 5′- CTACGGTCAATGCTCTGAAA -3′ (Accession No. NC_012920.1, 8161–8180). R-ATPase: 5′-TACTATATGATAGGCATGTGA-3′ (9219–9239). PCR amplification was performed using a ready-to-use PCR master mix (Sinaclon LTD, Tehran, Iran) in a final volume of 50 μl containing 5 ng of genomic DNA and 0.10 μM of each primer in a MJ Mini Gradient Thermal Cycler PTC-1148 (Bio-Rad, USA). PCR amplification was carried out with the following program: a 5-min pre-PCR incubation step at 95°C, 35 cycles of 95°C for 60 s, annealing temperature at 55°C for 1 min and 72°C for 2 min, and a final extension of 72°C for 10 min. The amplified fragment (1078 bp) was observed on 1.5% agarose gel.
Sequencing analysis
The PCR products were sequenced using the previously reported primers [23] on a ABI Prism 3700 automated sequencer (Applied Biosystems, USA). Sequence analysis was carried out using the FinchTV 1.4 software (Geospiza, Inc., USA). The sequences were compared to the human mtDNA reference sequence (Gene Bank ID: NC_012920.1) using the BLAST sequence analysis tool (NCBI, Bethesda, USA). The Mitomap database was used to identify mitochondrial genome sequence variants.
Prediction of pathogenicity by protein modeling analysis
The impact of non-synonymous (coding) substitutions in the resulting protein was assessed using PolyPhen-2 (v. 2.2.2) software, a tool for predicting the possible impact of an amino acid substitution variant on the structure and function of the corresponding protein, which is interpreted as benign and damaging effects [24].
Statistical analysis
The correlation between each alteration in the ATPase6 and ATPase8 genes in tumor samples and their adjacent normal tissue were analyzed by Fisher’s exact test using statistical package SPSS (v.16.1). The correlation between the groups was considered statistically significant if the p- value was less than 0.05. Additionally, for each variant the odds ratio (OR) and 95% confidence interval (95% CI) were calculated in order to determine its association to the increased risk in breast cancer patients. The association between mtDNA alteration and clinico-pathological characteristics of breast cancer patients with more than one missense mutation was evaluated using One-way ANOVA analysis.
Results
In this study, the complete sequences of the ATPase6 and 8 genes of 49 tumor tissues and adjacent non-tumor tissues were analyzed in a cohort of breast cancer patients. The clinico-pathological characteristics of the patients are summarized in Table 1. From 49 breast cancer cases, 28 mtDNA variants were found in tumor tissues, which were not present in their adjacent normal tissues. From 28 variants, 23 (82.14%) were found in the ATPase6 gene and the remaining 5 sequence variants were detected in the ATPase8 gene. All cases showed variants in the ATPase6 gene, whereas only 8.16% (4 of 49) cases had variants in the ATPase8 gene. Among 28 mtDNA alterations, 26 were at the homoplasmic state and the remaining 2 variants were at the heteroplasmic state (Table 2). However, there was no significant correlation (P > 0.05) between the ATPase6 and 8 gene variants and the clinico-pathological characteristics of the patients (Table 1). Our results indicated that the A8860G variant was detected in 100% of tumor tissue samples compared to adjacent non-tumor tissues, showing that this alteration may significantly increase breast cancer risk (P < 0.05). However, the patients’ survival was shorter in cases with more than one mtDNA non-synomous ATPase variant compared to the patients with only one mtDNA non-synonymous ATPase variant (A8860G ) (p =0.051, Table 1).
Furthermore, the damaging impact of an amino acid substitution on the structure and function of the ATPase6 and 8 proteins was predicted using PolyPhen-2 software (Table 3). The mtDNA variants A8384 (T7A), T8567C (I14T), G8572A (G16S), A9041G (H172R) and G9055A (A177T) showed significant effects on the resulting protein. However, there was no significant association between mtDNA alterations and the clinico-pathological characteristics of breast cancer patients.
Discussion
The identification of mitochondrial DNA mutations and/or polymorphism patterns is rapidly developing in the field of molecular oncology. A large number of somatic mutations in the mitochondrial genome have been recently reported in different types of cancer,luding breast, colon and ovarian cancers [5, 6]. These molecular markers may have potential implication in cancer research.
Mitochondrial complex V genes play an important role in ATP production [25] and the apoptosis pathways [5]. The contribution of mtDNA complex V variants in cell transformation, elevated ROS production, and tumor progression has been described previously [26]. Moreover, efficient programmed cell death needs the molecular machinery of ATP synthase [27].
The ATPase6 gene, one of the complex V genes, contributes to mtDNA maintenance [25]. Furthermore, the ATPase8 variants have been detected in rat and human bladder cancer cells developed through chemically-induced carcinogenesis [28]. In a meta-analysis study carried out by Lu et al. a total of 55 variants, comprising 34 missense variants, 20 silent variants and 1 nonsense variant, were found in the ATPase6 gene and a total of 9 variants, including 2 missense variants and 7 silent variants, were detected in the ATPase8 gene [6].
In our study, among 28 distinct somatic variants, 18 were missense variants. Six variants have been previously reported in breast cancer [29–32] and 9 variants were new, including 4 missense and 5 silent variants which were observed for the first time in breast cancer patients. However, 17 variants were previously reported in other types of cancers and diseases (Table 2). In addition, more non-synonymous amino acid changing variants were found in the ATPase6 gene in comparison with the ATPase8 gene (Table 2). Our findings suggest that in breast cancer patients, the ATPase6 gene might be more susceptible to mutation in comparison to the ATPase8 gene. Shidara et al. and Kirches reported that ATPase6 gene variants may enhance cancer progression by preventing apoptosis pathways [6, 33].
The functional role of ATPase6/8 variants in tumorigenesis is debatable; however, some of these variants are located in structurally and functionally important regions of the proteins. For instance, the A8860G alteration in ATPase6 has been reported as a polymorphism in different studies [29, 31, 34–40]. The frequency of this polymorphism has been reported to be from 79–91.66% in breast cancer patients [30, 31], 75-100% in other types of cancers [38–40] and 92.85%-100% in neurodegenerative diseases [37, 41–43]. Our results indicated that the A8860G variant was present in 100% of tumor tissue samples. Although this variant is located in a poorly conserved protein region with no impact on protein structure based on PolyPhen-2 software (Table 3), the variation may still contribute to other mtDNA and nDNA mutations.
The frequency of the G9055A variation has been reported as either 10.5% [28] or 18.6% [30] in breast cancer patients, indicating that it may increase the risk of breast cancer progression (OR: 3.03, 95% CI: 1.63-5.63, P = 0.0004) [32, 44]. This variation is located in a conserved protein region with damaging impact on protein structure (Table 3). Furthermore, the frequency of this polymorphism has been reported as 10% in pancreatic cancer [45] and as 57% and 100% in tubular and villous adenomas, respectively [40]. Moreover, the high frequency of this variation has been shown in non-muscle invasive bladder cancer [46]. In addition, this polymorphism has been reported as a protective factor (OR: 0.46, 95% CI: 0.22-0.91, P = 0.03) in Caucasian women with Parkinson’s disease [47]. From these results, we propose that this mtDNA variation is unfavorable for neurodegenerative disorders, while having a protective effect on cancer. According to our results, the frequency of this variation was 6.12% (3 of 49) in tumor samples.
A study by Petros et al. indicated that T8993G in ATPase6 can contribute to tumor growth in nude mice [48]. Another study showed that cybrids with a T8993G or T9176 ATPase6 mutation in nude mice can contribute to tumor development by preventing apoptosis in the early stages of tumor growth [10]. However, we detected none of these mutations in breast cancer patients.
Based on our results, the existence of more than one missense variants in some cases with different clinico-pathological features (Table 4) suggests a synergistic effect of different mtDNA variations on carcinogenesis.
In conclusion, the high frequency of ATPase6 gene alterations in breast cancer proposes that mitochondrial gene variants may play an important role in tumorigenesis, changing the energy metabolism in cancer cells, and may be suggested as molecular biomarkers in breast cancer.
References
Most frequent cancers; Women. [updated; 15/5/2013;cited];available from;http://globocan.iarc.fr/factsheets/cancers/breast.asp
Movahedi MHS, Khayamzadeh M, Moradi A, Ghanbari-Motlagh A, Mirzaei H, Esmail-Akbari M: Survival rate of breast cancer based on geographical variation in Iran, a national study. Iran Red Crescent Med J. 2012, 14 (12): 798-804. 10.5812/ircmj.3631.
Anderson SBA, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG: Sequence and organization of the human mitochondrial genome. Nature. 1981, 290: 457-464. 10.1038/290457a0.
Wallace DC: a mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annul Rev Gene. 2005, 39: 359-408. 10.1146/annurev.genet.39.110304.095751.
Czarnecka AMKW, Krawczyk T, Scinska A, Kukwa A, Cappello F: Mitochondrial DNA mutations in cancer from bench to bedside. Front Biosci. 2010, 15: 437-463. 10.2741/3629.
Lu JSL, Bai Y: Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Research. 2009, 19: 802-815. 10.1038/cr.2009.69.
William C, Copeland JTW, Johnson FM, Penta JS: Mitochondrial DNA Alterations in Cancer. Cancer Investigation. 2002, 20 (4): 554-566.
Chatterjee ADS, Sidransky D: Mitochondrial subversion in cancer. Cancer Prev Res. 2011, 4: 638-649. 10.1158/1940-6207.CAPR-10-0326.
William C, Copeland JTW, Johnson FM, Penta JS: Mitochondrial DNA Alterations in Cancer. Cancer Investigation. 2002, 20 (4): 12-
Shidara YYK, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S: Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005, 65 (5): 1655-1663. 10.1158/0008-5472.CAN-04-2012.
Amuthan GBG, Zhang SY, Klein-Szanto A, Vijayasarathy C: Avadhani NG.; 20(8), 1910–20.: Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. Embo J. 2001, 20 (8): 10-
Yu MZY, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R: Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007, 59 (7): 450-456. 10.1080/15216540701509955.
Matsuyama WNM, Wakimoto J, Hirotsu Y, Kawabata M, Osame M: Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Hum Mutat. 2003, 21 (4): 441-443. 10.1002/humu.10196.
Uzawa KBT, Uchida F, Yamatoji M, Kasamatsu A, Sakamoto Y, Ogawara K, Shiiba M, Bukawa H, Tanzawa H: Circulating tumor-derived mutant mitochondrial DNA: a predictive biomarker of clinical prognosis in human squamous cell carcinoma. Oncotarget. 2012, 3 (7): 670-677.
Kulawiec MSJ, Ericson NG, Wanagat J, Bielas JH: Generation, function, and prognostic utility of somatic mitochondrial DNA mutations in cancer. Environ Mol Mutagen. 2010, 51 (5): 427-439.
Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J: A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009, 18 (9): 1578-1589. 10.1093/hmg/ddp069.
Desler CMM, Singh KK, Rasmussen LJ: The Importance of Mitochondrial DNA in Aging and Cancer. Journal of Aging Research. 2011, -doi:10.4061/2011/407536
Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J-I: ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008, 320 (5876): 661-664. 10.1126/science.1156906.
Dasgupta SKR, Westra WH, Califano JA, Ha PK, Sidransky D, Koch WM: Mitochondrial DNA mutation in normal margins and tumors of recurrent head and neck squamous cell carcinoma patients. Cancer Prev Res. 2010, 3: 1205-1211. 10.1158/1940-6207.CAPR-10-0018.
Ishikawa KIH, Takenaga K, Hayashi J: Regulation of metastasis; mitochondrial DNA mutations have appeared on stage. J Bioenerg Biomembr. 2012, 44 (6): 639-10.1007/s10863-012-9468-6.
Kulawiec MOK, Singh KK: Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009, 8 (14): 1378-1385. 10.4161/cbt.8.14.8751.
Compton CC, Byrd DR, Garcia-Aguilar J, Kurtzman SH, Olawaiye A, Washington MK: AJCC Cancer Staging Atlas: A Companion to the Seventh Editions of the AJCC Cancer Staging Manual and Handbook (Greene, AJCC Cancer Staging Atlas). 2012, New York: Springer
Houshmand MKS, Etemad Ahari S, Moin M, Bahar M, Zamani A: Investigation of tRNA and ATPase 6/8 gene mutations in Iranian ataxia telangiectasia patients. Arch Med Sci. 2011, 7 (3): 523-527.
Adzhubei IASS, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods. 2010, 7 (4): 248-249. 10.1038/nmeth0410-248.
Jonckheere An ISJAM, Rodenburg Richard JT: Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. 2012, 35: 221-225.
Amuthan GBG, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG: Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. Embo J. 2001, 20 (8): 1910-1920. 10.1093/emboj/20.8.1910.
Matsuyama SXQ, Velours J, Reed JC: The Mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol Cell. 1998, 1 (3): 327-336. 10.1016/S1097-2765(00)80033-7.
Chen GFCF, Hong BF, Chan LW, Chan PS: Mitochondrial DNA mutations in chemical carcinogeninduced rat bladder and human bladder cancer. Oncol Rep. 2004, 12 (2): 463-471.
Zhu WQW, Bradley P, Wessel A, Puckett CL, Sauter ER: Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid Carcinogenesis. Carcinogenesis. 2005, 26 (1): 145-152.
Tan DJBR, Wong LJ: Comprehensive Scanning of Somatic Mitochondrial DNA Mutations in Breast Cancer. Cancer Res. 2002, 62 (4): 972-976.
Czarnecka AMKA, Krawczyk T, Zdrozny M, Arnold RS, Bartnik E, Petros JA: Mitochondrial NADH-dehydrogenase polymorphisms as sporadic breast cancer risk factor. Oncol Rep. 2010, 23 (2): 531-535.
Bai RK LS, Covarrubias D, Liu A, Wong LJ: mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007, 67 (10): 4683-4687.
Kirches E: Mitochondrial and nuclear genes of mitochondrial components in cancer. Curr Genomics. 2009, 10 (4): 281-292. 10.2174/138920209788488517.
Sawabe M, Tanaka M, Chida K, Arai T, Nishigaki Y, Fuku N, Mieno MN, Kuchiba A, Tanaka N: Mitochondrial haplogroups A and M7a confer a genetic risk for coronary atherosclerosis in the Japanese elderly: an autopsy study of 1,536 patients. Journal of Atherosclerosis and Thrombosis. 2011, 18 (2): 166-175. 10.5551/jat.6742.
DeHaan C, Habibi-Nazhad B, Yan E, Salloum N, Parliament M, Allalunis-Turner J: Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells. Mol Cancer. 2004, 3: 19-10.1186/1476-4598-3-19.
Taylor RWBM, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, Greaves LC, Kirkwood TB, Turnbull DM: Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003, 112 (9): 1351-1360. 10.1172/JCI19435.
Houshmand MMM, Kuchekian N, Noohi F, Nozar G, Zamani A: Is 8860 variation a rare polymorphism or associated as a secondary effect in HCM disease?. Arch Med Sci. 2011, 7 (2): 242-426.
Aikhionbare FO KM, Carey D, Okoli J, Go R: Is cumulative frequency of mitochondrial DNA variants a biomarker for colorectal tumor progression?. Mol Cancer. 2004, 3: 30-10.1186/1476-4598-3-30.
Aikhionbare FO MS, Kumaresan K, Zavareh M, Olatinwo M, Odunsi K, Partridge E: Mitochondrial DNA sequence variants in epithelial ovarian tumor subtypes and stages. J Carcinog. 2007, 6: 1-10.1186/1477-3163-6-1.
Mehrabi SAJ, Adams G, Grizzle W, Yao X, Aikhionbare FO: A sequence variant in colorectal adenomatous polyps. Diagn Pathol. 2010, 5: 66-10.1186/1746-1596-5-66.
Fauser SLJ, Besch D, Leo-Kottler B: Sequence analysis of the complete mitochondrial genome in patients with Leber’s hereditary optic neuropathy lacking the three most common pathogenic DNA mutations. Biochem Biophys Res Commun. 2002, 295: 342-347. 10.1016/S0006-291X(02)00672-1.
Mostafaie N, Rossmanith W, Hombauer H, Dechat T, Raffelsberger T, Bauer K, Worofka B, Kittl E, Hofmann J, Hejtman M: Mitochondrial genotype and risk for Alzheimer’s disease: Cross-sectional data from the Vienna-Transdanube-Aging “VITA” study. J Neural Transm. 2004, 111 (9): 1155-1165.
Kirk RFR, Amos W, Cooper G, Rubinsztein JS, Walsh C, Paykel ES, Rubinsztein DC: Mitochondrial genetic analyses suggest selection against maternal lineages in bipolar affective disorder. Am J Hum Genet. 1999, 65: 508-518. 10.1086/302507.
Tan DJCJ, Chen WL, Agress LJ, Yeh KT, Wang B, Wong LJ: Novel heteroplasmic frameshift and missense somatic mitochondrial DNA mutations in oral cancer of betel quid chewers. Genes Chromosomes Cancer. 2003, 37: 186-194. 10.1002/gcc.10217.
Wang LBW, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, Petersen GM: Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007, 17 (7): 1455-1459.
Guney AIED, Tavukcu HH, Koc G, Kirac D, Ulucan K, Javadova D, Turkeri L: Detection of mitochondrial DNA mutations in nonmuscle invasive bladder cancer. Genet Test Mol Biomarkers. 2012, 16 (7): 672-679. 10.1089/gtmb.2011.0227.
van der Walt JM NK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Goetz CG, Small GW, Mastaglia F, Tajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM: Mitochondrial Polymorphisms Significantly Reduce the Risk of Parkinson Disease. Am J Hum Genet. 2003, 72 (4): 804-812. 10.1086/373937.
Petros JA BA, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC: mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005, 102 (3): 719-724. 10.1073/pnas.0408894102.
Acknowledgements
Tumor tissues and adjacent non-tumor tissues samples were provided by the Iran National Tumor Bank (INTB) which is funded by the Cancer Institute of Tehran University for Cancer Research. This work was also supported by funding from Iranian Research Organization for Science and Technology (IROST) and National Institute for Genetic Engineering & Biotechnology (NIGEB), Tehran, Iran.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MG carried out the experimental procedures, participated in the sequence alignment and drafted the manuscript. RM, FF, and NM participated in the coordination of the study. BK wrote his constructive comments and edited the manuscript. MH conceived the project and supervised the study. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ghaffarpour, M., Mahdian, R., Fereidooni, F. et al. The mitochondrial ATPase6 gene is more susceptible to mutation than the ATPase8 gene in breast cancer patients. Cancer Cell Int 14, 21 (2014). https://doi.org/10.1186/1475-2867-14-21
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2867-14-21