Abstract
Background
Africa has a rich and diverse flora that people use for their food and health needs. This study aims to explore the possibility of using aqueous and/or ethanolic extracts of Alchornea cordifolia leaves as an alternative in the fight against multi-resistant bacteria responsible for gastritis and urinary tract infections.
Results
The results show that 100% of the tested bacilli (E. coli, K. pneumoniae, P. aeruginosa) are resistant to the 10 antibiotic disks tested. This resistance is 66.66% for quinolones and fluoroquinolones. S. aureus was found to be resistant to glycopeptides and aminoglycosides. Three ESBL genes are identified in bacilli against only one type of ESBL gene in cocci. 100% of the bacilli carry the SHV genes, and S. aureus has the Mec A gene. The aqueous extract exerted a bactericidal effect on all the strains with MICs and BMCs varying, respectively, from 0.76 to 50 mg/ml and BMCs from 0.76 to 100 mg/ml. Only 40% of the chemical groups (tannin, flavonoids, mucilages and sterol-terpenes) sought were present in the aqueous extract. The ethanolic extract is not active.
Conclusions
Based on these data, the aqueous extract of A. cordifolia leaves is a good phytomedical candidate for the treatment of gastritis (stomach cramps, watery or bloody diarrhea) and urinary tract infections caused by multi-resistant Gram-negative and Gram-positive bacteria.
Similar content being viewed by others
Background
Just as for food is a basic human need, so is the need for health care. In recent years, in African regions, we have seen an increase in infectious diseases. These diseases are a serious public health problem in developing countries as they are the leading cause of death with an alarming growth rate of antibiotic resistance (Dougnon et al. 2021a). This antibiotic resistance is due to poor medication practices observed in developing countries. The direct impact of these practices is to reduce the therapeutic potential of the drugs available to treat patients with bacterial infection (Cristóbal-Azkarate et al. 2014). Indeed, infections caused by multidrug-resistant bacteria are associated with poorer clinical results and a higher treatment cost than other infections (Lehtinen et al. 2019). This fact may be associated with a risk of exhaustion of therapeutic remedies capable of treating diseases caused by those mutant strains. In order to fight effectively against this bacterial resistance and to limit the cases of therapeutic failures, the search for new, more efficient molecules capable of effectively eradicating the mutant strains responsible for bacterial diseases must be encouraged (Brochot et al. 2017).
In Benin, a country in West Africa, the use of plants for health care is very old and is currently experiencing a revival among the populations. According to the World Health Organization, 80% of the African population still uses traditional medicinal practices for their primary healthcare needs (Ayéna et al. 2021). According to the US Agency for International Development, the importance of medicinal plants is growing due to the sharp increase in global demand for medicinal plants and their products in recent decades and the increasing number of users and the diversity of areas in which they are used (Brochot et al. 2017). Nowadays, it is estimated that at least 25% of all modern medicines are derived, directly or indirectly, from medicinal plants, mainly through the application of modern technologies to traditional knowledge (Saini et al. 2018). It is therefore on the strength of Benin’s rich plant heritage that this study proposes to explore the antibacterial potential of A. cordifolia leaves extracts against multidrug-resistant bacterial strains responsible for human infections.
Methods
Collection and extraction of plant material
Plant material consisting of fresh leaves of A. cordifolia was collected in July 2020 in the wetlands of Pahou, a town located 21 km from Cotonou, Benin. The identification followed by certification was carried out by Professor Hounnankpon YEDOMONHAN, curator of the National Herbarium of the University of Abomey-Calavi, under the reference number YH430/HNB.
After harvesting, the organs were washed with distilled water and dried in the laboratory at a temperature of 25 °C and then pulverized using a RETSCH electric grinder, type SM 100 with a mesh size of 0.5 (Ayéna et al. 2022).
The ethanolic extract was prepared by placing 100 g of A. cordifolia powder in 96° ethanol and stirring continuously for 72 h. The aqueous extract was obtained by infusing 100 g of A. cordifolia powder in 1L of water heated at 75 °C for 30 min. Each macerate was filtered on Whatman No. 3 paper and then dry-evaporated. The extracts were then recovered and stored in a refrigerator at 4 °C before testing. The yields of the extracts were calculated according to the following formula:
where Me is the mass of the extract after evaporation of the solvent and Mv is the mass of the plant material used for extraction.
Phytochemical screening
The qualitative phytochemical screening was performed on the aqueous extract of A. cordifolia leaves. This test is based on differential staining and precipitation reactions according to the method described by Ayéna et al (2022). The main chemical groups investigated in A. cordifolia leaf extract are: alkaloids, tannins (catechin and gallic), phenolic compounds, flavonoids, anthocyanins, leuco-anthocyanins, sterols and tri-terpenes, saponosides, reducing compounds and mucilages.
Evaluation of the antibacterial activity of the extracts
The total aqueous and ethanolic extracts obtained previously were used to prepare solutions with a concentration of 50 mg/ml, which were then filtered through 0.22-µm Millipore membranes and sterilized. The sterility of the extract solutions was checked by inoculating aliquots of each solution onto Mueller–Hinton medium and incubated at 37 °C for 24 h. The absence of bacterial culture indicates that the extract was sterile.
Criteria for selection of bacterial strains
The bacterial strains used in this study were chosen because of their strong involvement in the development of infections, their high degree of pathogenicity and their ability to show resistance to at least three different antibiotics. In addition, they are included in the WHO list of antibiotic-resistant “priority pathogens” (WHO 2017). The clinical strains used are multidrug resistant and possess at least one resistance gene.
Detection of resistance genes
DNA extraction
Young colonies of 18 ± 2 h obtained after culture on Muller–Hinton medium were used for DNA extraction. The young colonies were then added to 200 µl of physiological water, vortexed for about 5 min and centrifuged at 3000 rpm for 15 min. The supernatant was collected, and the pellets were discarded.
Resistance genes were detected following the method described by Koudokpon et al. (2018). A total of 5 genes including 4 genes (TEM, SHV, CTX-M-15 and CTX-M1) in bacilli and 1 gene (mecA) in cocci were detected by PCR method. The reaction volume of the mix prepared was 25 µl and that was introduced into a thermocycler for amplification following the conditions described in Tables 1, 2 and 3. DNA fragments were analyzed by electrophoresis in a 2% agarose gel. Migration was performed, followed by interpretation of the results based on comparison of the migration of the fragments to the marker sizes.
A pre-culture of each bacterial strain was diluted in distilled water to a turbidity of 0.5 McFarland or a concentration of 2.108 CFU/mL. Individual strains were then plated according to the method of Kirby and Bauer on Petri plates containing Mueller–Hinton agar and antibiotic disks (CA-SFM / EUCAST 2020). A chloramphenicol disk loaded at the concentration of 50 mg/ml was used as a positive control. The tests were repeated two times.
Preparation of bacterial suspensions and susceptibility testing
Seven bacterial strains were tested, of which four clinical strains (E. coli, K. pneumoniae, P. aeruginosa, S. aureus) and three reference strains (E. coli ATCC 25922, S. aureus ATCC 25923, P. aeruginosa ATCC 27853) were used as positive controls. Prior to antimicrobial testing, a 24-h pure colony portion from the Mueller–Hinton medium of each strain was emulsified in 5 ml of physiological water to achieve a turbidity of 0.5 on the McFarland scale (CA-SFM/EUCAST 2020).
Antibacterial susceptibility testing
Susceptibility testing of isolates was performed according to the French Society of Microbiology method (CA-SFM 2020). The following antibiotic disks were used to perform the susceptibility test: AMC = Amoxicillin + Clavulanic acid, ATM = Aztreonam, ETP = Ertapenem, AMP = Ampicillin, CIP = Ciprofloxacin, AK = Amikacin, FO = Fosfomycin, NA = Nalidixic acid and PEF = Pefloxacin for bacilli strains. For cocci: CIP = Ciprofloxacin, AK = Amikacin, FO = Fosfomycin, TOB = Tobramycin, E = Erythromycin, AK = Clindamycin, VA = Vancomycin and RP = Rifampicin.
Determination of the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
The MIC and MBC were determined following Dougnon et al. technique (Dougnon et al. 2021b). They were performed based on the susceptibility test. A sample stock solution of each extract was prepared at a concentration of 100 mg/ml in distilled water. Hundred microliters of Mueller–Hinton broth (MHB) was placed in each well of a microplate (1–10 wells). Hundred microliters of the stock extract solution was placed in the first well. After homogenization by aspiration discharge with a micropipette, 200 μL of 100 mg/ml extract solution was obtained. Hundred microliters of this new solution was taken and mixed with the Mueller–Hinton broth contained in the second well, and this half dilution of the well was continued until the 8th well. Finally, 10 μL of the bacterial suspension was added to each well. The 9th and 10th wells represented the positive and negative controls and contained 100 μL of Mueller–Hinton broth + 100 μL of the bacterial suspension and 100 μL of Mueller–Hinton broth, respectively. The microplates were incubated for 24 h at 37 °C.
MICs were estimated by the addition of nitrachlorotetrazolium. All wells with higher MIC concentrations were then plated on Mueller–Hinton agar and placed at 37 °C for 24 h. This allowed the evaluation of the minimum bactericidal concentration which is the lowest concentration of the extract that does not show bacterial colonies. The antibacterial power is considered bactericidal or bacteriostatic according to the ratio R = MBC/MIC. Indeed, according to Dougnon et al. (2021b), if 1 ≤ R ≤ 2, the effect is bactericidal, and if 4 ≤ R ≥ 16, the effect is bacteriostatic.
Data processing and analysis
Data were collected and stored in Excel 2019 spreadsheet, and graphs were made with Excel software. The inhibition diameters are presented as mean ± standard deviation.
Results
The yield of extractions was 46.51% and 51.43% for aqueous and ethanolic extracts, respectively.
Phytochemical composition of the extract
The qualitative phytochemical screening revealed the presence of four major chemical groups (Table 4). These are catechin and gallic tannins, flavonoids, mucilages and sterol-terpenes. The other chemical groups (anthocyanins, leuco-anthocyanins, alkaloids, reducing compounds and saponosides) are absent.
Resistance genes detected
A total of 17 antibiotic families were tested in this study. 100% of the bacilli tested (E. coli, K. pneumoniae, P aeruginosa) are resistant to penicillins and monobactams. This resistance is 66.66% to quinolones and fluoroquinolones. As for S. aureus, it was found resistant to glycopeptides and aminoglycosides. The search for resistance genes shows the existence of three ESBL genes in bacilli against only one type of ESBL gene in cocci. 100% of bacilli carry the SHV genes and S. aureus has the Mec A genes (Table 5).
Antimicrobial activities of extracts inhibition diameters of extract-sensitive strains
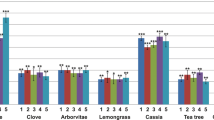
The results of the antimicrobial test reveal that at the concentration of 100 mg/ml, all the extracts combined have proven antimicrobial activities. The aqueous extract inhibited 100% of the bacterial strains with inhibition diameters ranging from 9.33 ± 1.21 to 17 ± 0.89 mm (Table 6). As for MIC and MBC, the results show that they ranged from 0.76 to 50 mg/ml and 0.76 to 100 mg/ml for the aqueous extract, respectively. This extract showed bactericidal power against 75% of the strains (K. pneumoniae, S. aureus and P. aeruginosa). No activity was observed with the ethanolic extract (Fig. 1).
Discussion
The multidrug resistance test showed that 100% of the bacilli were resistant to the combination of Aztreonam and Clavulanic Acid + Amoxicillin compared to 75% to Ampicillin. The S. aureus strain showed absolute resistance (100%) to Vancomycin, Erythromycin and Rifampicin. According to the work of Ischola et al. (2021), the high resistance of the isolated bacterial strains is a concern and calls for a rational use of antibiotics to avoid the transmission of antibiotic resistance from the environment to humans. According to Koudopkon et al. (2018), Enterobacteriaceae easily express resistance to Amoxicillin, Amoxicillin + clavulanic acid and Aztreonam. This high resistance is due to the high frequency of prescription of these antibiotics in medical training (Moussé et al. 2019). For the detection of beta-lactam, four resistance genes (blaTEM, blaSHV, blaCTX-M15 in the bacilli group and Mec A gene in the cocci group) were identified. The resistance genes blaTEM (33.33%), blaSHV (100%), blaCTX-M15 (33.33%) and Mec A (100%) were detected. These results confirm the findings of Abrar et al. (2019) who also report the presence of these genes in clinical strains (Abrar et al. 2019).
The MICs and MBCs of the tested bacteria are recorded in Table 7. Only the aqueous extract of A. cordifolia leaves showed different antibacterial activity against the tested multidrug-resistant strains. The comparison between the inhibition diameters showed that there was no difference between the diameters of E. coli, K. pneumoniae and S. aureus, P. aeruginosa strains at the 0.05 threshold. However, a variation in inhibition diameter was recorded for S. aureus (17 ± 0.89 mm) and P. aeruginosa (17 ± 0.89 mm) strains versus smaller diameters for E. coli (9.33 ± 1.21 mm) and K. pneumoniae (9.33 ± 1.21 mm). This difference in the average diameters of inhibition observed would be due to the presence of resistance genes which are gene 01 for some and gene 02 for others.
The MIC and MBC determined vary, respectively, from 0.76 to 50 mg/ml and from 0.76 to 100 mg/ml. It exerted 100% bactericidal power on the strains studied. According to several researchers, these values indicate that the extract possesses antimicrobial activity of the species toward the studied clinical strains (Djague et al. 2020). The ethanolic extract of the leaves showed no activity. These results are not in line with Fadehan et al. study (2015) who showed that ethanolic extracts of A. cordifolia leaves exhibited antibacterial activity against these strains. Other researchers confirm these results which are not similar to our findings (Boniface et al. 2016; Djimeli et al. 2017). This could be explained by the fact that the active compounds available in the ethanolic extract have no affinity with water. On the other hand, the resistance genes would have strengthened the defensive arsenal of the bacteria, making them more difficult to eliminate. These results would be due to the phytochemical composition of the A. cordifolia leaf extract.
Conclusions
This study on the search for an alternative treatment of infections due to multi-resistant bacteria led to the characterization of the resistance profile of certain bacterial strains and the exploration of the antibacterial potential of aqueous extracts of A. cordifolia leaves. The results show that 100% of the bacilli tested are resistant to the most commonly used antibiotics (penicillins and monobactams). This resistance is 66.66% to quinolones and fluoroquinolones. As for S. aureus, it was found resistant to glycopeptides and aminoglycosides. The detection of resistance genes showed the presence of TEM and CTX-M15, SHV and Mec A resistance genes. Only the aqueous extract had a bactericidal effect on all strains. Regarding the results, aqueous extract of A. cordifolia leaves is a good phytomedical candidate to treat gastritis (stomach cramps, watery or bloody diarrhea) and urinary tract infections caused by multi-resistant Gram-negative and Gram-positive bacteria. However, in addition to the observed antibacterial activity, for a better understanding of the biological potential of this extract, further pharmacological tests such as studying the mechanism of action of the leaf extract should be undertaken.
Availability of data and materials
Data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- PCR:
-
Polymerase chain reaction
- ATCC:
-
American Type Culture Collection
- AMC:
-
Amoxicillin + clavulanic acid
- ATM:
-
Aztreonam
- ETP:
-
Ertapenem
- AMP:
-
Ampicillin
- CIP:
-
Ciprofloxacin
- AK:
-
Amikacin
- FO:
-
Fosfomycin
- NA:
-
Nalidixic acid
- PEF:
-
Pefloxacin for bacilli strains
- CIP:
-
Ciprofloxacin
- AK:
-
Amikacin
- FO:
-
Fosfomycin
- TOB:
-
Tobramycin
- E:
-
Erythromycin
- AK:
-
Clindamycin
- VA:
-
Vancomycin
- RP:
-
Rifampicin for cocci strains
- MIC:
-
Minimum inhibitory concentration
- MBC:
-
Minimum bactericidal concentration
- ESBL:
-
Extended spectrum beta-lactamase
References
Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S (2019) Distribution of blaCTX−M, blaTEM, blaSHV and blaOXA genes in extended-spectrum-β-lactamase-producing clinical isolates: a three-year multi-center study from Lahore. Pak Antimicrob Resist Infect Control 8:80. https://doi.org/10.1186/s13756-019-0536-0
Ayéna AC, Anani K, Dosseh K, Agbonon A, Gbeassor M (2021) Comparative study of antimicrobial, anti-inflammatory, and antioxidant activities of different parts from Pterocarpus Santalinoides l’Her. Ex. DC (Fabaceae). Evid Based Complem Altern Med 2021, Article ID 8938534, 7. https://doi.org/10.1155/2021/8938534
Ayéna ACT, Dosseh K, Idoh K, Agbonon A, Gbeassor M (2022) Comparative physicochemical screening and toxicology of hydroethanol extracts of the parts of Pterocarpus santalinoides l’H´er. ex DC. (Fabaceae) in Wistar Rats. Sci World J, 2022, Article ID 5953094, 7. https://doi.org/10.1155/2022/5953094
Boniface PK, Ferreira SB, Kaiser CR (2016) Recent trends in phytochemistry, ethnobotany and pharmacological significance of Alchornea cordifolia (Schumach. & Thonn.) Muell. Arg J Ethnopharmacol 191:216–244. https://doi.org/10.1016/j.jep.2016.06.021
Brochot A, Guilbot A, Haddioui L, Roques C (2017) Antibacterial, antifungal, and antiviral effects of three essential oil blends. Microbiol Open 6:e00459. https://doi.org/10.1002/mbo3.459
CA-SFM (2020) Comité de l’antibiogramme de la Société Française de Microbiologie
Cristóbal-Azkarate J, Dunn JC, Day JMW, Amábile-Cuevas CF (2014) Resistance to antibiotics of clinical relevance in the fecal microbiota of mexican wildlife. PLoS ONE 9:e107719. https://doi.org/10.1371/journal.pone.0107719
Djague F, Lunga PK, Toghueo KRM, Melogmo DYK, Fekam BF (2020) Garcinia kola (Heckel) and Alchornea cordifolia (Schumach. & Thonn.) Müll. Arg. from Cameroon possess potential antisalmonellal and antioxidant properties. PLoS One 15(8), article e0237076
Djimeli MN, Fodouop SPC, Njateng GSS, Fokunang C, Tala DS et al (2017) Antibacterial activities and toxicological study of the aqueous extract from leaves of Alchornea cordifolia (Euphorbiaceae). BMC Complem Altern Med 17:349. https://doi.org/10.1186/s12906-017-1854-5
Dougnon V, Agbodjento E, Hounsa E, Legba BB, Deguenon, et al (2021) An ethnobotanical survey of seventeen plants species used against diarrhoea and other diseases in southern Benin (West Africa). J Biol Res Boll Della Soc Ital Biol Sper. https://doi.org/10.4081/jbr.2021.9486
Dougnon V, Hounsa E, Agbodjento E, Keilah LP, Legba BB et al. (2021b). Percentage destabilization effect of some West African medicinal plants on the outer membrane of various bacteria involved in infectious diarrhea. BioMed Res Int Article ID 4134713, 12. https://doi.org/10.1155/2021b/4134713
Fadehan G, Boamah D, Adotei D, Edoh Edoh et al (2015) Screening of Ageratum conyzoides Linn and Alchornea cordifolia (Schumach & Thonn) extracts for antibacterial activity. Eur J Med Plants 10(4):1–7. https://doi.org/10.9734/EJMP/2015/20739
Ichola OD, Dougnon VT, Koudokpon CH, Agbankpe AJ, Deguenon E et al. (2021) Assessment of the bacterial pollution and detection of antibiotic resistance genes in Benin: case of the hydrographic channel complex Cotonou-Nokoue Lake. J Environ Public Health 2021. https://doi.org/10.1155/2021/6664816
Koudokpon H, Dougnon V, Hadjadj L, Kissira I, Fanou B et al (2018) First sequence analysis of genes mediating extended-spectrum beta-lactamase (ESBL) bla-TEM, SHV- and CTX-M production in isolates of enterobacteriaceae in Southern Benin. Int J Infect. https://doi.org/10.5812/iji.83194
Lehtinen S, Blanquart F, Lipsitch M, Fraser C (2019) On the evolutionary ecology of multidrug resistance in bacteria. PLoS Pathog 15:e1007763. https://doi.org/10.1371/journal.ppat.1007763
Moussé W, Sina H, Mama-Sirou IA, Anago E, Dah-Nouvlessounon D et al (2019) Antibiotic resistance and production of extended spectrum β-lactamases by clinical gram-negative bacteria in Benin. J Adv Microbiol. https://doi.org/10.9734/jamb/2019/v18i230158
Saini S, Tulla K, Maker AV et al (2018) Therapeutic advances in anaplastic thyroid cancer: a current perspective. Mol Cancer 17:154. https://doi.org/10.1186/s12943-018-0903-0
Acknowledgements
The authors thank all the staff of the Research Unit in Applied Microbiology and Pharmacology of Natural Substances of the Polytechnic School of Abomey-Calavi for their support throughout the research period.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
FA and VD designed the study. VA and ACA collected and interpreted the data. FA, ACA and VA wrote the manuscript. MM, JRK and LBM reviewed the draft manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adounkpe, F., Ayena, A.C., Aholoukpe, V. et al. Use of the leaves of Alchornea cordifolia (Schumach. & Thonn.) Müll (Euphorbiaceae) and prospects for treatment of infections due to multidrug-resistant bacteria. Bull Natl Res Cent 46, 132 (2022). https://doi.org/10.1186/s42269-022-00821-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-022-00821-0