Abstract
Background
Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), known as fall armyworm (FAW), is a wide-scale invasion pest that resulted in crop yield loss and certainly caused critical economic damage. Therefore, several control strategies such as the application of entomopathogen agent to control the population can be applied. The study aimed to designate and identify the candidate entomopathogens fungi (EPF) from South Sulawesi to control FAW.
Results
The research was conducted in several stages: field exploration, isolation, purification, bioassay, and morphological or genetical identification of selected fungi. The identification found out that: Sarocladium strictum, Talaromyces purpureogenus, and Aspergillus terreus had significant mortality percentages and incubation time in killing FAW. The highest mortality percentage was obtained in the case of the A. terreus (MLN8) isolate with an average mortality of 83.33% (2 days after incubation); A. terreus (4b) with an average mortality of 76.67% (4 days after incubation); both S. strictum (3) and T. purpureogenus (2B) required 3 and 5 days to control FAW, respectively, with 73% mortality percentage.
Conclusion
Ultimately, the exploration of several areas in South Sulawesi discovered potential EPF to suppress the FAW population biologically. Therefore, the identification of those EPFs contributes to FAW strategies control and the development of biopesticides.
Similar content being viewed by others
Background
Maize is a primary cereal grain widely consumed worldwide, alongside wheat and rice. On a global scale, maize along with wheat and rice is important elements in the human diet. Therefore, maize is important agriculturally because of the diverse roles it plays in food security and nutrition throughout the world.
The outbreak of fall armyworm (FAW) Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) affected the stability of food systems, livelihoods, ecosystem biodiversity, and local, regional, and global trade in many countries (Kasoma et al. 2021). Montezano et al. (2018) reported that FAW is a polyphagous pest that can attack approximately 353 plant species from 76 families including the Poaceae, Asteraceae, and Fabaceae. Below the economic threshold, leaves damaged by FAW are still capable of regenerating. In an outbreak, these lepidopterans will destroy the growing point’s effect on plant (Day et al. 2017). Moreover, it is difficult to control this pest due to the rapid migration ability with a wide geographic area coverage (up to 500 km) and it can be found all year round (Prasanna et al. 2018). Because of those reasons, there has been a growing need for sustainable and environmentally friendly pest management strategies.
Jing et al. (2021) reported that FAW has been resistant to insecticides from the carbamates, organophosphates, and pyrethroids groups in the USA. Zhang et al. (2021) reported that in China FAW has been resistant to insecticides from the organophosphate and pyrethroid groups. Excessive insecticide use on FAW infestation leads to the development of resistance, contributes to an increase in the greenhouse effect, and impacts human health (Tudi et al. 2021). Therefore, it is important to minimize the use of insecticides of FAW and develop sustainable integrated pest management program that significantly use biological control (Day et al. 2017).
Biological control agents generally do not attack non-target organisms or cause resistance in the pest insect. Some microorganisms cause disease insects or other arthropods. Insect pathogens enter the insect's body in two ways. Insect host can ingest the pathogens during the feeding process (known as passive entry) typical of bacteria, viruses, and protozoa. The second way is for the pathogen to enter through natural holes by penetrating the cuticle (known active entry), which is the typical method of fungi. Entomopathogenic fungi (EPFs) have emerged as promising biocontrol agents due to their potential to selectively target and control the populations of agricultural pests, while posing minimal risks to non-target organisms and the environment (Islam et al. 2021).
This research endeavored to address the critical issue of FAW infestations in Indonesia by focusing on the selection and molecular identification of EPFs capable of effectively combating this pest. The systematic screening of entomopathogens and their molecular identification are essential steps toward developing efficient and sustainable biological control strategies tailored to the Indonesian. The present research explores the selection process of identifying efficacious and selective EPFs to use in sustainable pest management programs for FAW. The results of this research aimed to provide biological control agents, and efficacious EPFs that can be used in pest management programs for FAW in Indonesia.
Methods
The research began with a survey and sample collection of FAW infected with entomopathogens in the maize fields. Entomopathogens were isolated from collected cadavers and cultured in purified cultures. Bioassays were conducted to determine the efficacy of each culture. For those entomopathogen cultures that were efficacious, the entomopathogens were identified using morphological and molecular methods such as the polymerase chain reaction method (Fig. 1).
Infected insect survey and collection
The study was carried out between October 2022 and March 2023 at several locations in both the highlands and lowlands. The locations chosen were Bajeng sub-district, Gowa regency (coordinates: − 5.309644, + 119.510651, 27.2 MASL), Tombolo Pao sub-district, Gowa regency (coordinates: − 5.243806, + 119.939263, 1517 MASL), and Bantimurung sub-district, Maros regency (coordinates: − 5.035665, + 119.687881, 21.9 MASL).
Determination of infected FAW in the field was based on morphological characteristics such as ill-looking larvae that moved slowly, or larva-covered white hyphae. The field samples were collected in insect boxes (8 × 8 × 4 cm) and kept in the freezer (± − 5 °C) if those samples were not cultured immediately. There were two methods for culturing the samples. First, the inoculation procedures began with cutting the samples into small pieces (1 × 1 cm) using tweezers. Next, the samples were cleaned and sterilized by soaking in 70% alcohol for 5 min and rinsed twice with distilled water. The samples were ready for inoculation in sterilized Potato Dextrose Agar (PDA) media. The second method was the spread plate method; 10 g of clean and sterile samples was grinded using mortar and pestle and diluted in 10 ml distilled water. After that, 1 ml from the first dilution was diluted in 9 ml of distilled water. The dilution was carried out 7 times for better micro-biome density. Furthermore, 1 ml of the last dilution was spread in sterile PDA media. Both methods of inoculation were placed at 26 °C for 2 to 5 days and checked for purification.
Preparation of insects
The larvae were collected from the maize field in the Maros regency and then reared in the laboratory of Center for Standard Testing of Cereal Plant Instruments, Maros, South Sulawesi. To maintain the larval population for the bioassay, healthy larvae were collected from the field. The larvae were fed with fresh young maize leaves, and the larvae were checked daily for food. When the larvae reached the pupal stage, the pupae were placed in a breeding container (modified jar) (30 cm diameter × 27 cm height). Once the pupae emerged, the adults were fed on the sugar solution and placed young maize plant (14 DAP) for oviposition. The egg masses were collected and allowed to hatch. The first instar larvae (neonates) were provided with maize leaves as food. The larvae reached the second instar about 3–4 days after hatching. The second instar larvae were used for the bioassay. The not-used larvae were kept maintaining FAW colony.
Isolates preparation for bioassay
To isolate fungal species for bioassay, 28 isolates are cultured on PDA media containing 100 mg/l chloramphenicol (Sigma Aldrich, USA). The Petri dishes containing the isolates are incubated for 14 days under aerobic conditions at 26 °C. After the conidia had grown and filled the Petri dishes, it was harvested and transferred to an Erlenmeyer flask (100 ml) in sterile condition. Next, fill the flask with distilled water 100 ml and 0.05% Tween 80 and homogenize by stirring. Then, the solution was filtered to separate the hyphae and standardized with a density of 109 hyphae/ml of solution using a hemocytometer. The solutions are transferred into a sprayer for application.
Bioassay test of entomopathogenic fungi (EPF)
EPF bioassay test is the strategy to screen the isolates which is effective in controlling the larval stage of FAW. In the bioassay, the 2nd instar larvae were placed in round plastic containers (25 ml) with fresh maize as the diet. Next, larvae were sprayed with EPF solution until wet but no puddles inside. The test used a completely randomized design of 28 treatments with 3 replications. Each replication contained 10 observation units with 1 larva for each unit. Observations were made by recording the incubation time and percentage of larval mortality for 18 days after EPF solution applied. The incubation period was observed by recording the initial time of death in the test larvae. Mortality percentage was observed by counting the number of dead larvae for each treatment.
Morphological characterization and molecular identification of EPF candidates
Entomopathogenic fungi (EPF) are identified using morphological and molecular approaches. Morphological identification of EPF was carried out based on standard guidelines. Identification at the genus level was carried out on the morphology of conidia, hyphae, conidiophores, and colony color (Alexopoulos et al. 1996; Humber 1997). Molecular identification of EPF isolates was carried out by suspending pure isolates aged 14 days in PDA media. The EPF suspension was extracted following the Zymo Research Quick-DNA Fungal/Bacterial Miniprep Kit procedure.
DNA amplification was carried out following the method used by Mirsam et al. (2022). The results of DNA extraction were followed by DNA amplification using the PCR method. The PCR was carried out by mixing 12.5 µl KAPA Taq ReadyMix PCR, 8.5 µl ddH2O, 1 µl forward primer, 1 µl reverse primer, and 2 µl DNA template. The primers used are universal primers, namely primers ITS1 (50-TCCGTAGGTGAACCTGCGG-30) and ITS4 (50TCCTCCGCTTATTGATATGC-30) for fungi. The PCR cycle consists of an initiation or pre-denaturation step at 95 °C in 3 min, followed by 35 cycles of a denaturation step at 95 °C in 15 s, an annealing step at 55 °C in 30 s, an extension step at 72 °C in 1 min, and an extension step final in 3 min at 72 °C. The PCR product was then electrophoresed at a voltage of 110 V for 50 min. The electrophoresis results were then visualized by a UV trans-illuminator for taking pictures using a camera.
The amplification products were sent to FirstBase (Malaysia) for nucleotide sequence analysis. The sequence results were analyzed using the Basic Local Alignment Search Tool (BLAST) with an optimization program to obtain DNA base sequences available at the National Center for Biotechnology Information (NCBI). The nucleotide sequence results obtained were then analyzed using multiple alignments in ClustalW Bioedit Sequence Alignment Editor Version 7.2 software. Relationships among isolates were constructed using Molecular Evolutionary Genetic Analysis Software version 10.0 (MEGAX) with bootstrapping 1000 times.
Statistical analysis
Mortality percentage is calculated using the formula:
M: mortality percentage; A: number of dead samples; B: number of samples observed.
The observational data were statistically analyzed the variance, and then, for significant difference “least significant difference” (LSD) at 95% confidence level was calculated using ANOVA.
Results
EPF virulence testing
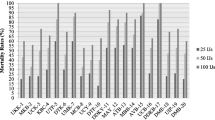
The 27 candidates of entomopathogenic fungi from FAW in Gowa and Maros regency, South Sulawesi Province, demonstrated varying incubation periods (Table 1) and virulence (Fig. 2) in the bioassay test. Four isolates were identified in the bioassays as warranting additional investigation. The isolates included isolates MLN8, 4b, 3, and 2b. The four isolates showed mixed results in the incubation period with an average 2–5 days post-inoculation (Table 1). MLN8 had the highest mortality (83.33%) of the four isolates, but this mortality was non-significantly different (P < 0.05) than the mortality imparted by 4b (76.67%; Fig. 2).
Morphological characterization of EPF
The morphological characteristics of selected microbiomes are presented in Table 2. Identification under the microscope refers to several references indicating a similar morphology from previous research based on the color, shape of the colony, conidial form, and color. The microscopic and macroscopic morphological appearance of the selected isolates is shown in Fig. 3.
Molecular Identification (selected isolates)
The polymerase chain reaction (PCR) results using the primer pair ITS1-ITS4 showed DNA amplification measuring ± 600 bp in the EPF isolates. This shows the existence of the ITS gene which is commonly found in fungal groups. The fungus group has a DNA band size with a size range of ± 600 bp in PCR amplification using primers ITS1-ITS4 (Yuan et al. 2023) (Fig. 4).
a Results of 1% agarose gel electrophoresis of 4 EPF isolates amplified using the primer pair ITS1(F)-ITS4(R) using a 1-kb DNA marker. b Phylogenetic tree of 4 EPF isolates based on neighbor-joining analysis with distance calculations implemented in the Blast and Mega X programs; the genetic distance scale value describes the average number of nucleotide differences in each isolate
The sequence analysis results using the BLAST program showed that four EPF isolates were identified as three different species with various homologies. Phylogenetic analysis based on ITS sequences showed that Isolate 2b had the same class tree location value as the fungus Talaromyces purpureogenus accession OM373024 from China, accession KX359607 from Austria, and accession OR421153 from Morocco with a bootstrap analysis value of 99%. Isolate 3 has the same class location value as the fungus Sarocladium stricum accession OR268327 from China, accession AY138486 from the USA, and accession KY022425 from China with bootstrap analysis values in the range of 46–100%. Isolates 4b and MLN8 have the same class location values as the fungus Aspergillus terreus accession OK465110 from China, accession JQ697519 from China, accession ON935604 from South Korea, and accession KT310979 from China with bootstrap value of 99%.
Discussion
Entomopathogens caused disease followed by insects’ death. The death of Spodoptera caterpillars due to biotic factors is an interesting topic to discuss, especially death due to pathogen infection and the type of pathogen that infects. Various groups of fungi are known to have great potential as EPF; only a small number have been characterized morphologically and identified at the molecular level. EPF found in this survey of FAW were T. purpureogenus, S. strictum and A. terreus. Although not commonly found, other isolates have been reported from Spodoptera species.
Talaromyces sp. is a member of the Ascomycetes which has been known as an antagonist of plant pathogens. Recently, several studies have been conducted on the potential of Talaromyces as a biocontrol agent for several insects (Nicoletti and Becchimanzi 2021). Talaromyces flavus is found in Galleria mellonella larvae in China (Sun and Liu 2008), Prays oleae in Portugal (Oliveira et al. 2012), and several insects from the order Blattodea, Coleoptera; Diptera, Hemiptera, Hymenoptera, Lepidoptera; Orthoptera; Thysanoptera; Trichoptera (Nicoletti and Becchimanzi 2021). There are several species of Talaromyces sp. which are known to have virulence against insects, including T. pulveris (Ziganshina et al. 2018); T. atroroseus, T. pinophilus (Hadj Taieb et al. 2020); T. flavus (Hernández et al. 2007); and T. diversus (Herlinda et al. 2020). T. diversus is known to have insecticidal activity against the armyworm Spodoptera litura (Herlinda et al. 2020). This result proved that Talaromyces sp. has the potential as entomopathogen of S. frugiperda.
Sarocladium sp. is associated with attacks on plants. Sarocladium strictum is often isolated from dead larvae and pupae. Sarocladium sp. is known as the most common endophytic fungi, grows on the leaf surfaces of vascular plants, is widely distributed in the atmosphere, and often seen to assist other primary fungi such as powdery mildew or rust in colonizing their hosts. The present results showed the real influence of S. strictum isolates as seen from its ability to cause up to 70% death of the tested larvae. These results have also been reported in other studies that S. strictum can show activity as a natural insecticide through interference with the larval, pupal, and developmental stages of Spodoptera littoralis (El Sayed et al. 2020).
Aspergillus terreus is widely distributed fungus in nature and has a wide range of habitats such as soil, plants, compost, and stored grain (Mahata et al. 2022). The results of the study revealed that the efficacy of A. terreus caused death in the tested larvae. This high mortality rate is thought to be caused by the activity of secondary metabolite compounds produced by EPF. This result is in line with other study which reveals that A. terreus can control insect pests (Singh et al. 2023). Moreover, A. terreus was found as an effective and friendly mycoinsecticide in control Artemia nauplii which is a model organism for acute and eco-toxicity tests in marine ecosystems (Ragavendran et al. 2018). The reduction in the severity of the disease and the recovery from eggplant blight disease due to the application of A. terreus indicated the ability of this agent to suppress the spread of the disease (Attia et al. 2022).
The highest mortality for S. frugiferda larvae was observed in MLN8 treatment with a mortality rate of over 80% is Aspergillus terreus based on the DN sequencing. This finding corresponds with previous research about the effectiveness of Aspergillus sp. According to Rosa et al. (2020) Aspergillus sp is a fungal entomopathogen that can defeat Helopeltis sp. by infected the insects by introducing hyphae from the spores into the insect's body by the enzymes. Moreover, Aspergillus sp. controlled the melon fly Bactrocera cucurbitae (Yang et al. 2015) and Rhizoctonia solani, a spot disease on rice plants (Qin et al. 2023).
Conclusions
Sarocladium strictum, Talaromyces purpureogenus, and Aspergillus terreus are projected as entomopathogenic agents with the ability to cause death to FAW pest within a period of 2 to 4.33 days after application. Further study is needed to determine the potential of these entomopathogens as mortality agents and to develop efficacious formulations of the pathogens for use in the field. Exploration of biological agents is one appropriate way to enrich the diversity of entomopathogens controlling insect pests. Through exploration in certain areas, various types of beneficial and location-specific microorganisms can be collected and identified for various control purposes.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alexopoulos CJ, Mims CW, Blackwell M (1996) Introductory mycology. John Wiley and Sons, Hoboken
Attia MS, Hashem AH, Badawy AA, Abdelaziz AM (2022) Biocontrol of early blight disease of eggplant using endophytic Aspergillus terreus: improving plant immunological, physiological and antifungal activities. Bot Stud 63:26
Day R, Abrahams P, Bateman M et al (2017) Fall armyworm: impacts and implications for Africa. Outlooks Pest Manag. https://doi.org/10.1564/v28_oct_02
Djouhri Y, Saidi-Touati M, Benelmouffok A, Bouchtout MN (2022) Susceptibility of nymphs and adults of Blattella germanica (Dictyoptera: Blattellidae) to Aspergillus terreus (Eurotiales: Trichocomaceae) infested via oral ingestion, contact, and bait methods: a comparative study. Appl Entomol Zool 57:137–145
El Sayed AM, Basam SM, Bellah A El-Naggar E-M et al (2020) LC–MS/MS and GC–MS profiling as well as the antimicrobial effect of leaves of selected Yucca species introduced to Egypt. Sci Rep 10:17778
HadjTaieb K, Gharsallah H, Ksentini I et al (2020) Screening of biological activities of fungi associated with pistachio bark beetle, Chaetoptelius vestitus (Coleoptera, Curculionidae), infesting pistachio cultivations in Tunisia. J Appl Microbiol 128:1472–1485
Herlinda S, Efendi RA, Suharjo R et al (2020) New emerging entomopathogenic fungi isolated from soil in South Sumatra (Indonesia) and their filtrate and conidial insecticidal activity against Spodoptera litura. Biodiversitas 21:5102–5113
Hernández MC, Pildain MB, Novas MV et al (2007) Mycobiota associated with larval mines of Thrypticus truncatus and T. sagittatus (Diptera, Dolichopodidae) on waterhyacinth, Eichhornia crassipes, in Argentina. Biol Control 41:321–326
Humber RA (1997) Fungi: identification. In: Manual of techniques in insect pathology. Elsevier, pp 153–185.
Islam W, Adnan M, Shabbir A et al (2021) Insect-fungal-interactions: a detailed review on entomopathogenic fungi pathogenicity to combat insect pests. Microb Pathog 159:105122
Jing W, Huang C, Li C et al (2021) Biology, invasion and management of the agricultural invader: Fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). J Integr Agric 20:646–663
Kasoma C, Shimelis H, Laing MD et al (2021) Outbreaks of the fall armyworm (Spodoptera frugiperda), and maize production constraints in Zambia with special emphasis on coping strategies. Sustainability 13:10771
Mahata PK, Dass RS, Pan A, Muthusamy B (2022) Substantive morphological descriptions, phylogenetic analysis and single nucleotide polymorphisms of aspergillus species from Foeniculum vulgare. Front Microbiol 13:832320
Mirsam H, Suriani AM et al (2022) Molecular characterization of indigenous microbes and its potential as a biological control agent of Fusarium stem rot disease (Fusarium verticillioides) on maize. Heliyon 8:e11960. https://doi.org/10.1016/j.heliyon.2022.e11960
Montezano DG, Sosa-Gómez DR, Specht A et al (2018) Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr Entomol 26:286–300
Nicoletti R, Becchimanzi A (2021) Talaromyces–insect relationships. Microorganisms 10:45
Oliveira I, Pereira JA, Lino-Neto T et al (2012) Fungal diversity associated to the olive moth, Prays oleae Bernard: a survey for potential entomopathogenic fungi. Microb Ecol 63:964–974
Prasanna BM, Huesing JE, Eddy R, Peschke VM (2018) Fall armyworm in Africa: a guide for integrated pest management. USAID, CIMMYT, CGIAR.
Qin X, Chang Y, Wang Y et al (2023) Aspergillus sp. R3, a new producer for cyclopyazonic acid, inhibits rice sheath blight fungus Rhizoctonia solani Kühn. Physiol Mol Plant Pathol 125:102007
Ragavendran C, Natarajan D (2015) Insecticidal potency of Aspergillus terreus against larvae and pupae of three mosquito species Anopheles stephensi, Culex quinquefasciatus, and Aedes aegypti. Environ Sci Pollut Res 22:17224–17237
Ragavendran C, Srinivasan R, Kim M, Natarajan D (2018) Aspergillus terreus (Trichocomaceae): a natural, eco-friendly mycoinsecticide for control of malaria, filariasis, dengue vectors and its toxicity assessment against an aquatic model organism Artemia nauplii. Front Pharmacol 9:1355
Rosa E, Ekowati CN, Handayani TT et al (2020) Characterization of entomopathogenic fungi as a natural biological control of American cockroaches (Periplaneta americana). Biodiversitas 21:11
Singh S, Kaur S, Kaur R, Kaur A (2023) Impact of plant symbiotic endophytic fungus, Aspergillus terreus on insect herbivore Spodoptera litura (Fabricius)(Lepidoptera: Noctuidae). Neotrop Entomol 52:932–944
Suliman EA, Mohammed YO (2012) The activity of Aspergillus terreus as entomopathogenic fungi on different stages of Hyalomma anatolicum anatolicum under experimental conditions. J Entomol 9:343–351
Sun B-D, Liu X-Z (2008) Occurrence and diversity of insect-associated fungi in natural soils in China. Appl Soil Ecol 39:100–108
Thiyam G, Dufossé L, Sharma AK (2020) Characterization of Talaromyces purpureogenus strain F extrolites and development of production medium for extracellular pigments enriched with antioxidant properties. Food Bioprod Process 124:143–158
Tudi M, Daniel Ruan H, Wang L et al (2021) Agriculture development, pesticide application and its impact on the environment. Int J Environ Res Public Health 18:1112
Vidal S (1996) Changes in suitability of tomato for whiteflies mediated by a non-pathogenic endophytic fungus. In: Proceedings of the 9th international symposium on insect-plant relationships. Springer, pp 272–274.
Yang Y, Zhang Y, Wang M et al (2015) Bioefficacy of entomopathogenic Aspergillus strains against the melon fly, Bactrocera cucurbitae (Diptera: Tephritidae). Appl Entomol Zool 50:443–449
Yilmaz N, Visagie CM, Houbraken J et al (2014) Polyphasic taxonomy of the genus Talaromyces. Stud Mycol 78:175–341
Yuan G, Czajka JJ, Dai Z et al (2023) Rapid and robust squashed spore/colony PCR of industrially important fungi. Fungal Biol Biotechnol 10:15
Zhang D, Xiao Y, Xu P et al (2021) Insecticide resistance monitoring for the invasive populations of fall armyworm, Spodoptera frugiperda in China. J Integr Agric 20:783–791
Ziganshina EE, Mohammed WS, Shagimardanova EI et al (2018) Fungal, bacterial, and archaeal diversity in the digestive tract of several beetle larvae (Coleoptera). Biomed Res Int 2018:15
Acknowledgements
We acknowledge Balai Pengujian Standar Instrumen Tanaman Serealia, Ministry of Agriculture, for providing opportunities to carry out research in the laboratory of plant pests and diseases and genetic laboratory.
Funding
The funding source for his research was internal funding of Research Organization for Agriculture and Food, National Research and Innovation Agency of Indonesia. The funding covers the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
(Grant number - IDR 25000000)
Author information
Authors and Affiliations
Contributions
EN involved in investigation, formal analysis, writing original draft, data curation; AS took part in investigation, methodology, writing original draft, data curation, editing; AT involved in conceptualization, project investigator, research supervision, writing review, and editing; MY involved in project investigator, validation, manuscript review, and editing; SW took part in investigation, writing original draft; ED involved in investigation, writing original draft; AA involved in manuscript original draft, visualization, software; S took part in investigation, writing original draft; R involved in visualization, data curation, manuscript original draft, review, and editing; MSS took part in supervision, writing original draft, review, and editing; AF participated in supervision, writing original draft, review, and editing; TK involved in research consultant; B took part in research consultant; A involved in investigation; H participated in investigation.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable in this section.
Consent for publication
Not applicable in this section.
Competing interests
The authors declare that they have no competing interests in all financial and non-financial sections.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Najamuddin, E., Sebayang, A., Tenrirawe, A. et al. Selection and molecular identification of specific entomopathogens in South Sulawesi and the pathogenicity to fall armyworm (Spodoptera frugiperda JE. Smith) (Lepidoptera: Noctuidae). Egypt J Biol Pest Control 34, 22 (2024). https://doi.org/10.1186/s41938-024-00786-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-024-00786-4