Abstract
Background
Two-spotted spider mite, Tetranychus urticae (Acari: Tetranychidae) incidence is a major constraint in vegetable cultivation. The indiscriminate use of acaricides is causing environmental threat and also residual effect in vegetables. To develop an eco-friendly management strategy, an investigation was made to access the natural occurrence of entomopathogenic fungi (EPF) infecting mites in bhendi, India ecosystem.
Results
Natural incidence of EPF was high during the month of December. Twelve EPF isolates were isolated from the mycosed T. urticae cadavers using Potato Dextrose Agar Medium. Morphological studies confirmed that the isolated fungi were Beauveria bassiana. PCR amplification of ITS region was carried out and the results showed, amplification at 560 bp. In NCBI database, the sequence of the virulent isolates had shown 99.2, 98.7, 99.1, 97.7 and 96.8% homology with other B. bassiana isolates, confirming the occurrence of B. bassiana mycosed mites in vegetable ecosystem.
Conclusion
The isolate MZ749636 (B.b-7) was found to be virulent against T. urticae causing 86% mortality of T. urticae at the conidial load of 1 × 108 conidia/ml at laboratory conditions and so it could be utilized for the eco-friendly management of T. urticae.
Similar content being viewed by others
Background
Bhendi [Abelmoschus esculentus (L.) Moench], a warm vegetable crop, grown over 509.0 million hectares in India. Among the biotic constraints in cultivation of bhendi, sucking pests like the mite (Tetranychus urticae Koch), aphid (Aphis gossypii Glover) and whitefly (Bemisia tabaci Gennadius) are considered important (Chinniah 2000). Prasad and Singh 2011, reported nearly six different mite species, including two-spotted spider mite (T. urticae Koch), Red spider mite (T. macfarlanei Baker and Pritchard), Red spider mite (T. ludeni Zacher), false spider mite (Brevipalpus phoenicis Geijskes), Broad mite (Polyphagotarsonemus latus Banks), and tomato russet mite (Aceria lycopersici Wolffenstein). Among these, T. urticae pose a major threat to bhendi cultivation, especially during the summer, spring and post-rainy seasons, leading in severe yield loss of 7 to 48% (Roseleen and Ramaraju 2010). T. urticae also infests the important vegetable crops in the region.
In India, T. urticae had been recorded in more than 900 hosts including bhendi, brinjal, cucumber, potato etc., (Migeon et al. 2010). Due to short life span of 9–10 days and high reproductive potential of more than 100 eggs/female, it had progressed to the position of a major pest of vegetables with a broader host range (Miyazaki et al. 2013).
Insecticides and acaricides are the preferred management tools in view of accessibility and quick control against phytophagous mites. However, acaricides are employed indiscriminately, leading in the development of acaricidal resistance (Simma et al. 2020). Therefore, non-chemical alternative methods have be explored for managing T. urticae.
Insect pathogens, particularly entomopathogenic fungi (EPF) have been employed to manage a variety of insect pests, particularly mites with great effectiveness. These EPF are known to infect the pests naturally leading to the death due to mycosis. Beauveria bassiana (Balsamo) Vuillemin (Jeyarani et al. 2011), Metarhizium anisopliae (Meetchnikoff) (McGuire and Northfield 2020) and similar EPF were known to cause natural infection on T. urticae. There lies a greater opportunity for integration of these EPF in applied biological control of two-spotted spider mites. Several authors have reported the variable levels of virulence of EPF against target pests in laboratory and field investigations. Hence, the present study aimed to find the diversity of various EPF causing infection on two-spotted spider mites, and exploiting them for eco-friendly management.
Methods
Survey and collection of samples
Survey was conducted in Coimbatore district of Tamil Nadu viz., TNAU Orchard (11.0122° N, 76.9354° E), TNAU insectary (11.02232° N, 76.9434° E) Thondamuthur (10.9899° N, 76.8409° E), Narasipuram (10.9880° N, 76.7740° E), Aandipatti (10.0015° N, 77.6164° E), Pichanur (10.8623° N, 76.8727° E), Theethipalayam (10.9694° N, 76.8956° E), Madampatti (10.9698° N, 76.8598° E), and Poosaripalayam (11.0062° N, 76.8668° E) during 2020–2021 to isolate the EPF infecting two-spotted spider mites. The mites were examined in the field using a 40-× magnifying hand lens and the mite cadavers showing natural external growths of fungi (Jeyarani et al. 2011) were collected in a 1.5 ml sterile eppendorf tube using a camel hair brush (000 size) and further investigations were carried out in the AICRIP- Acarology Laboratory, Department of Agricultural Entomology, TNAU, Coimbatore. The mites showing fungal infection were placed in a Petri plate layered with sterile moistened filter paper and incubated at 26 ± 2 °C for further mycelial development and sporulation (Goettel and Inglis 1997). Mites were also collected throughout the survey to confirm the species of two-spotted spider mite. The collected mites were mounted on the slide using the Hoyer's Medium. Morphological identification of the species was made using 40–100 × phase contrast microscope (Euromex iscope, USA).
Isolation of entomopathogenic fungi
Potato Dextrose Agar Medium (PDA) (Dinu et al. 2012) was used for the isolation of the EPF. Fungus with white powdery conidial mass on infected two-spotted spider mites were collected and kept at temperature of 25 ± 2 °C and relative humidity of 85–90%t under laboratory conditions for further sporulation. The sporulated two-spotted spider mites were surface sterilized, using 70% ethanol for a minute, followed by 0.1% sodium hypochlorite for a second and rinsed using distilled water for four to five times and the conidia from the infected specimen's surface were transferred to a 90 mm Petri plate containing PDA medium and were incubated at an ambient temperature for sporulation (Abdo et al. 2008). Sub-culturing was made by inoculating a single colony into 90 mm Petri plate containing PDA medium and incubated at a 25 ± 2 °C and RH 85–90% for two weeks in order to obtain the pure culture.
Morphological characterization
To study the morphological characters of the isolates, 15-day old culture was used (Malarvannan et al. 2010). The morphological characters, such as colony colour, mycelial character and spore structure were studied by phase contrast microscopy according to Samson et al. 2013.
Molecular characterization
The CTAB (cetyl-trimethyl ammonium bromide) method was used to extract genomic DNA as described by Zhang et al. (1925). Using a cork-borer, a single colony from the mother culture was inoculated into a 250 ml flask containing 100 ml Potato Dextrose Broth (PDB). For 1–2 weeks, the liquid culture was shaken at 250 RPM at 28 °C (Reddy et al. 2016). To separate mycelial mat and culture filtrate, the cultures in the flasks were aseptically filtered through Whatman No 1 filter paper (Viswanath et al. 2008). The mycelial mat was separated from the broth was dried on the Whatman no 1 filter paper for 2 h. One hundred mg of the mycelial mat was ground using 800 µl of 2% CTAB extraction buffer (2% CTAB, 1.4 M NaCl, 20 Mm EDTA, 100 Mm Tris–HCl, 1% PVP and 0.2% mercaptoethanol with the pH of 8) (Ashwathi et al. 2017). It was then transferred to a sterile Eppendorf tube and incubated for 1 h at 65ºC in a water bath. An equal volume of phenol:chloroform:iso amyl alcohol (25:24:1) was added and spun at 10,000 rpm in refrigerated centrifuge for 10 min at 4 °C. After centrifugation, the supernatant was transferred to another Eppendorf tube containing an equal volume of ice cold iso-propanol and incubated overnight at − 20 °C. Later, it was spun at 13,000 rpm for 10–15 min at 4 °C and supernatant was discarded after centrifugation and the pellet was washed with 70% ethanol and dried. The resultant pellet was then suspended in 30 µl of TE buffer (10 mM Tris HCl, 1 mM EDTA, pH 8.0) (Chaithra et al. 2020). The presence of DNA was confirmed using a Nano drop Spectrophotometer (Thermo Scientific Company, USA). 1 µl deionized water was placed on the bottom optical surface for blanking, and the lever arm was closed to guarantee the upper pedestal makes contact. After blanking, 1 µl of the sample was dispensed on the lowest pedestal and the lever arm was closed and the measurement was made as per the procedure by Desjardins and Conklin (2010).
PCR amplification- ITS region
The isolated DNA was amplified using a Polymerase Chain Reaction (PCR). The ITS-1(ITS1-F 5′-TCGGTAGGTAGGTGAACCTGCGG-3′) and ITS-4 (18s-ITS4 -R 5′-CAGGAGACTTGTACACGGTCCAG-3′) region were amplified using the method described. The PCR product consisted of 5 µl of reaction mixture (200 µM of each dNTP, 3.75 mM MgCl2 and 0.05 U of Taq DNA polymerase in 20 mM of pH 8.4 Tris HCl containing 50 mM KCl), 2 µl of nuclease free water, 1 µl of DNA and 1 µl each of forward and reverse primer. A negative control (without DNA) was included. The amplification temperature was set to 1 cycle of initial denaturation at 94 ºC for 3 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 ºC for 30 s followed by elongation at 72 ºC for 1 min each of 35 cycles and 1 cycle of final elongation at 72 ºC for 10 min. The PCR product was subjected to 1% agarose gel electrophoresis stained using ethidium bromide solution of 10 µg/ml and visualised under UV transilluminator. The PCR product was then sent for sequencing to Eurofins Genomics, Bengaluru. The nucleotide sequence obtained was aligned for its similarity using the BLAST software (Islam et al. 2017).
Rearing of two-spotted spider mites, T. urticae for pathogenicity testing
The mites collected from the field were reared in the Acarology Laboratory using freshly detached mulberry leaves. The leaves were placed in Petri plates with moistened sterile cotton. Collected mites were released in the Petri plate and allowed to multiplication. The cotton pads were moistened daily and the mulberry leaves were changed whenever it is necessary (Hassan et al. 2019). After multiplication of the mites, it was transferred to the caged potted bhendi (Bhendi hybrid COBh H 1) plants maintained at Insectary, Department of Agricultural Entomology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu for further multiplication.
Pathogenicity of the isolated entomopathogenic fungi isolates against T. urticae
For testing the pathogenicity of EPF, fully sporulated (15 days old) culture were used (Malarvannan et al. 2010). With a sterile surgical knife, the fungal surface was scraped with 10 ml of sterile distilled water containing 0.01% Tween 80 as a wetting agent (Rombach et al. 1986). The spore concentration was found using the Neubaur haemocytometer. The desired spore concentration (1 × 108 conidia/ml) was prepared by diluting the spore suspension with sterile distilled water.
Mulberry leaf discs were cut to the size of 5 cm diameter and they were dipped in the prepared EPF suspensions (1 × 108 conidia/ml) and dried over a filter paper at the temperature of 25 ± 1ºC and allowed to air dry. Mulberry leaves dipped in the sterile water containing 0.01% Tween 80 served as a control. The treated leaves were placed in Petri plates with moist sterile filter paper and 30 uniform aged adult mites were released to each Petri plate (Lee et al. 1997). The experimental setup was replicated thrice. The mortality of mites was calculated on 24 h interval and percentage mortality was calculated by the formula:
The percentage reduction over the control was calculated using the formula,
The dead mites were kept in a separate Petri plates with sterile moist filter paper to allow the fungus to grow on the surface of the cadavers and re-isolation of the pathogen was done to confirm the death due to EPF infection.
Statistical analysis
Mortality days after inoculation (DAI) were adjusted using Abbott's formula (Abbott 1925). Analysis of variance was used to examine the differences in mortality between the fungal isolates and the control group (ANOVA). The statistical software SPSS 16.0 was used to perform all of the analyses.
Results
Survey and morphological identification of two-spotted spider mites
The identity of live spider mite specimens that were collected during the survey was confirmed based on the morphological characters described by Seeman and Beard (2011). In this study, slides for both male and female mites were prepared and examined. The genital apertures, striations were the primary parameters considered. The present study revealed the occurrence of T. urticae in bhendi. The mycosed dead mites were collected from various locations (Fig. 1) and the per cent infection was calculated (Table 1) and it was maximum (8.42%) at TNAU Orchard. A total of 12 isolates were obtained and they were used for the morphological and molecular confirmation in the present studies.
Identification of EPF isolates
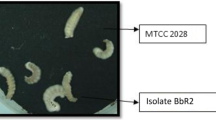
Identification of EPF isolates were done according to Humber (1997). Morphological aspects such as: colony color, colony structure, mycelial texture, conidial shape (Figs. 2, 3) and size were examined under the phase contrast microscope and tabulated (Table 2). The morphological identification revealed that all the isolates produced whitish colony or yellowish white colony with hyaline and septate mycelium and the conidia is either globose or sub-globose indicating that all the isolates were B. bassiana. The conidial size of all the isolates were in the range of 2.56–1.95 µm, which further confirms that all the isolates were B. bassiana.
Molecular characterization
PCR amplification revealed that the presence of the amplification of ITS region in 560 bp, which was used for further confirmation of B. bassiana. The PCR products were sent for sequencing to Eurofins Genomics, Bengaluru and the sequence was obtained. The sequence of the effective isolates such as B.b-7, B.b-2, B.b-1, B.b-12 and B.b-4 were submitted to NCBI which had 99.2, 98.7, 99.1, 97.7 and 96.8% homology with the other isolates of B. bassiana, respectively. The accession number obtained were MZ749636 (B.b-7), MZ674187 (B.b-2), MZ779015 (B.b-1), MZ749645 (B.b-12) and MZ674400 (B.b-4).
Pathogenicity of EPF and selection of the highly virulent isolates
With a spore concentration of 1 × 108 conidia/ml, the pathogenicity of the 12 B. bassiana isolates was tested against the two-spotted spider mite, T. urticae. The isolate MZ749636 (B.b-7), showed a rapid mycosis in T. urticae with maximum mortality of 86.10%, followed by the isolate MZ674187 (B.b-2), which caused 73.90% mortality. In case of the isolates MZ779015(B.b-1), MZ749645 (B.b-12) and MZ674400(B.b-4) which caused mortalities of 65.00, 55.00 and 47.23% mortality, respectively at 1 × 108 conidia/ml after 12 days of treatment (Table 3). The isolates B.b-10 and B.b-11 showed the least mortality of 39.43% (Fig. 4). The mortality of the mites started third day after EPF treatment and increased up to 12 days after post inoculation. The present study has clearly indicated that the period from 3rd day to 9th day was required for the maximum mortality (86.10%) of the two-spotted spider mite and the infected mites acts as the inoculum for two-spotted spider mite to become infected.
Discussion
Entomopathogenic fungal (EPF) spore adheres to the skin of the insects and later it forms the germination nail and penetrates the epicuticule and reaches hypodermis with the help of protein degrading enzymes. These hyphae after reaching the hypodermis multiply and spread in the body cavity and kill the insect (Altinok et al. 2019). The chlamydospores formed by the fungi germinates to form conidiophores on the skin of the dead insect under appropriate conditions and the spores are ready for new infections (Goettel et al. 2005). The present study was made to explore the diversity of EPF infecting two-spotted spider mite in Coimbatore district, Tamil Nadu. Previous studies were conducted by Jeyarani et al. (2011) in Coimbatore region of Tamil Nadu and recorded 5.25% infection of B. bassiana occurred on T. urticae in bhendi ecosystem which showed similarity to the present study results as the per cent incidence of B. bassiana in Bhendi ecosystem ranged from 1.53 to 8.42%.
Gebremariam et al. (2021) characterized the EPF morphologically indicating the presence of white or yellowish white colony, globose or sub-globose conidia were essential for the confirmation of B. bassiana. In the present study, the isolated EPF isolates produced whitish and yellowish white colony, globose and sub-globose conidia were in accordance with the findings of (Gebremariam et al. 2021).
DNA barcoding is very useful for identification of fungal species. The internal transcribed spacer (ITS) regions of the nuclear ribosomal RNA gene cluster are used as the fungal barcode (Xu 2016). The major advantage of using ITS is making it possible to amplify the gene from small amounts of biological specimens. The entire ITS region was about 600 bp long and contained two variable spacers, ITS-1 and ITS-2 which were separated by the highly conserved 5.8S rRNA gene in fungi (White et al. 1990), which was similar to the present findings. The molecular characterization of B. bassiana isolates by Dhar et al. (2019) observed the amplification of ITS region in 524 bp which is in line with the findings of the present study as the amplification of ITS region was observed at 560 bp.
Seiedy et al. (2010) found that the B. bassiana isolate DEB1008 caused 74.68% mortality at the spore concentration of 1 × 108 conidia/ml, which is in accordance with the present study as the isolate MZ674187 (B.b-2) caused a maximum mortality of 73.90% in laboratory condition. Yanar et al. (2018) reported that the two isolates of F-12 and F-56 caused the maximum mortality of 78.3 and 76.7%, respectively at the conidial concentration of 1 × 108 conidia/ml, which showed similarity to the results of the present investigation. The isolates BGF14 and BCA32 caused 25.88–61.92 and 32.36–62.03% mortality of T. urticae at the conidial concentration of 108 conidia/ml, which is analogous with the present results (Yucel 2021). At a spore concentration of 1 × 108 conidia/ml in laboratory circumstances, the B. bassiana isolates PPRC 19, PPRC 27, PPRC 29, and PPRC 66 were moderately virulent against T. urticae, producing 60–80% death, which corresponds to the findings of the present study and the isolates 9614 and 9609 were highly virulent with the mortality range of 86–90% (Negash et al. 2017), which is in further conformity to the present results as the isolate MZ749636 (B.b-7) resulted in 86.10% mortality of the two-spotted spider mite.
Conclusion
Under laboratory circumstances, Beauveria bassiana isolates MZ749636 (B.b-7), MZ674187 (B.b-2) and MZ779015 (B.b-1) were highly successful in controlling the two-spotted spider mite, T. urticae. These three isolates can be utilized in future for effective control in field conditions.
Availability of data and materials
Not applicable.
Abbreviations
- ANOVA:
-
Analysis of variance
- CTAB:
-
Cetyl-trimethyl ammonium bromide
- DAI:
-
Mortality days after inoculation
- EPF:
-
Entomopathogenic fungi
- PCR:
-
Polymerase chain reaction
- PDA:
-
Potato Dextrose Agar
- PDB:
-
Potato Dextrose Broth
References
Abbott WS (1925) A method of computing the effectiveness of an insecticide. J Econ Entomol 18(2):265–267. https://doi.org/10.1093/jee/18.2.265a
Abdo C, Nemer N, Nemer G, Abou JY, Atamian H, Kawar NS (2008) Isolation of Beauveria species from Lebanon and evaluation of its efficacy against the cedar web-spinning sawfly, Cephalcia tannourinensis. BioControl 53(2):341–352. https://doi.org/10.1007/s10526-006-9062-0
Altinok HH, Altinok MA, Koca AS (2019) Modes of action of entomopathogenic fungi. Curr Trends Nat Sci 8(16):117–124
Ashwathi S, Ushamalini C, Parthasarathy S, Nakkeeran S (2017) Morphological, pathogenic and molecular characterisation of Pythium aphanidermatum: a causal pathogen of coriander damping-off in India. J Pharm Innov 6(11):44–48
Chaithra M, Vanitha S, Ramanathan A, Jegadeeshwari V, Rajesh V, Hegde V, Apshara E (2020) Morphological and molecular characterization of endophytic fungi associated with Cocoa (Theobroma cacao L.) in India. Curr Appl Sci Technol 38:1–8. https://doi.org/10.9734/CJAST/2019/v38i630447
Chinniah C (2000) Evaluation of acaricides and insecticides against the spider mite and aphid on bhendi. Madras Agric J 87(7/9):483–486
Desjardins P, Conklin D (2010) NanoDrop microvolume quantitation of nucleic acids. JoVE 22(45):1–5. https://doi.org/10.3791/2565\
Dhar S, Jindal V, Jariyal M, Gupta VK (2019) Molecular characterization of new isolates of the entomopathogenic fungus Beauveria bassiana and their efficacy against the tobacco caterpillar, Spodoptera litura (Fabricius)(Lepidoptera: Noctuidae). Egypt J Biol Pest Control 29(1):1–9. https://doi.org/10.1186/s41938-019-0110-3
Dinu MM, Lupaştean D, Cardaş G, Andrei AM (2012) New Beauveria bassiana (Bals.) Vuill. isolate from Ips duplicatus (Sahlberg). Rom J Plant Prot 5:12–15
Gebremariam A, Chekol Y, Assefa F (2021) Phenotypic, molecular, and virulence characterization of entomopathogenic fungi, Beauveria bassiana (Balsam) Vuillemin, and Metarhizium anisopliae (Metschn.) Sorokin from soil samples of Ethiopia for the development of mycoinsecticide. Heliyon 7(5):70–91. https://doi.org/10.1016/j.heliyon.2021.e07091
Goettel GM, Inglis GD (1997) Fungi: hyphomycetes. In: Lacey L (ed) Manual of techniques in insect pathology. Academic Press, San Diego, pp 143–151
Goettel MS, Eilenberg J, Glare T (2005) Entomopathogenic fungi and their role in regulation of insect populations. In: Gilbert LI, Iatrou K, Gill SS (eds) Comprehensive molecular insect science. Elsevier, Amsterdam, pp 361–405
Hassan FR, Abdullah SK, Assaf LH (2019) Pathogenicity of the entomopathogenic fungus, Beauveria bassiana (Bals.) Vuill. endophytic and a soil isolate against the squash beetle, Epilachna chrysomelina (F.) (Coleoptera: Coccinellidae). Egypt J Biol Pest Control 29(1):1–7. https://doi.org/10.1186/s41938-019-0169-x
Humber RA (1997) Fungi: identification. In: Lacey LA (ed) Manual of techniques in insect pathology. Academic Press, Cambridge, pp 153–185
Islam T, Biswas MJ, Howlader MT, Ullah MS (2017) Laboratory evaluation of Beauveria bassiana, some plant oils and insect growth regulators against two-spotted spider mite, Tetranychus urticae Koch (Acari: Tetranychidae). Persian J Acarol. https://doi.org/10.22073/pja.v6i3.29338
Jeyarani S, Banu JG, Ramaraju K (2011) First record of natural occurrence of Cladosporium cladosporioides (Fresenius) de Vries and Beauveria bassiana (Bals.-Criv.) Vuill on two spotted spider mite, Tetranychus urticae Koch from India. J Entomol 8(3):274–279. https://doi.org/10.3923/je.2011.274.279
Lee S, Tsao R, Peterson C, Coats JR (1997) Insecticidal activity of monoterpenoids to western corn rootworm (Coleoptera: Chrysomelidae), two spotted spider mite (Acari: Tetranychidae), and house fly (Diptera: Muscidae). J Econ Entomol 90(4):883–892. https://doi.org/10.1093/jee/90.4.883
Malarvannan S, Murali PD, Shanthakumar SP, Prabavathy VR, Nair S (2010) Laboratory evaluation of the entomopathogenic fungi, Beauveria bassiana against the Tobacco caterpillar, Spodoptera litura Fabricius (Noctuidae: Lepidoptera). J Biopestic 3(1):126
McGuire AV, Northfield TD (2020) Tropical occurrence and agricultural importance of Beauveria bassiana and Metarhizium anisopliae. Front Sustain Food Syst 4:6. https://doi.org/10.3389/fsufs.2020.00006
Migeon A, Nouguier E, Dorkeld F (2010) Spider mites web: a comprehensive database for the Tetranychidae. Trends Acarol. https://doi.org/10.1007/978-90-481-9837-5_96
Miyazaki J, Wilson LJ, Stiller WN (2013) Fitness of twospotted spider mites is more affected by constitutive than induced resistance traits in cotton (Gossypium spp). Pest Manag Sci 69(10):1187–1197
Negash R, Dawd M, Azerefegne F (2017) Efficacy of Ethiopian Beauveria bassiana and Metarhizium anisopliae isolates on spotted spider mites, Tetranychus urticae (Acari: Tetranychidae) under laboratory conditions. Ethiop J Agric Sci 27(2):61–71
Prasad R, Singh J (2011) Status of mite pest fauna prevailing in brinjal agro-ecosystem. Uttar Pradesh J Zool 31(1):15–23
Reddy GB, Vijayavani S, Swarnabala G, Reddy KV (2016) Evaluation of locally available substrates for conidial biomass production of Beauveria bassiana MCC0044 employing solid substrate fermentation. IOSR J Agric Vet Sci 9(7):2319–2372
Rombach MC, Aguda RM, Shepard BM, Roberts DW (1986) Infection of rice brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae), by field application of entomopathogenic Hyphomycetes (Deuteromycotina). Environ Entomol 15(5):1070–1073. https://doi.org/10.1093/ee/15.5.1070
Roseleen SSJ, Ramaraju K (2010) Host-plant resistance in okra against the two-spotted spider mite, Tetranychus urticae Koch. Pest Manag Econ Zool 18(1/2):179–187
Samson RA, Evans HC, Latge JP (2013) Atlas of entomopathogenic fungi. Springer, Berlin, p 11
Seeman OD, Beard JJ (2011) Identification of exotic pest and Australian native and naturalised species of Tetranychus (Acari: Tetranychidae). Zootaxa 1:1–72. https://doi.org/10.11646/zootaxa.2961.1.1
Seiedy M, Saboori A, Allahyari H, Talaei-Hassanloui R, Tork M (2010) Laboratory investigation on the virulence of two isolates of the entomopathogenic fungus Beauveria bassiana against the two spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Int J Acarol 36(6):527–532. https://doi.org/10.1080/01647954.2010.519718
Simma EA, Hailu B, Jonckheere W, Rogiers C, Duchateau L, Dermauw W, Van Leeuwen T (2020) Acaricide resistance status and identification of resistance mutations in populations of the two-spotted spider mite Tetranychus urticae from Ethiopia. Exp Appl Acarol 82(4):475–491. https://doi.org/10.1007/s10493-020-00567-2
Viswanath B, Chandra MS, Pallavi H, Reddy BR (2008) Screening and assessment of laccase producing fungi isolated from different environmental samples. Afr J Biotechnol 7(8):1129–1133
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc, New York, pp 315–322
Xu J (2016) Fungal DNA barcoding. Genome 59(11):913–932
Yanar D, Yanar Y, Belgüzar S, Eser I, Unalan HK (2018) Efficacy of entomopathogenic fungus Beauveria bassiana isolates against the two-spotted spider mite, Tetranychus urticae Koch (Acari: Tetranychidae). Appl Ecol Environ Res 16(6):7903–7911
Yucel C (2021) Effects of local isolates of Beauveria bassiana (Balsamo) Vuillemin on the two-spotted spider mite, Tetranychus urticae (Koch)(Acari: Tetranychidae). Egypt J Biol Pest Control 31(1):1–7. https://doi.org/10.1186/s41938-021-00409-2
Zhang YP, Uyemoto JK, Kirkpatrick BC (1925) A small-scale procedure for extracting nucleic acids from woody plants infected with various phytopathogens for PCR assay. J Virolo Methods 71(1):45–50. https://doi.org/10.1016/S0166-0934(97)00190-0
Acknowledgements
The author thankfully acknowledges the Department of Agricultural Entomology, Tamil Nadu Agricultural University, Coimbatore for the facilities offered for the successful conduct of the experiment.
Funding
The author thankfully acknowledges the Department of Agricultural Entomology, Tamil Nadu Agricultural University, Coimbatore for the financial assistance provided for the successful conduct of the research.
Author information
Authors and Affiliations
Contributions
The author (VPA) carried out the experiment, recorded data, interpreted the results and wrote the manuscript. The second author (RV) corrected the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declares that there are no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abarna, V.P., Vishnupriya, R. DNA barcoding reveals the natural occurrence of Beauveria bassiana (Balsamo) Vuillemin in two-spotted spider mite, Tetranychus urticae Koch in Bhendi [Abelmoschus esculentus (L.) Moench] ecosystem in Coimbatore district of Tamil Nadu. Egypt J Biol Pest Control 32, 128 (2022). https://doi.org/10.1186/s41938-022-00623-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-022-00623-6