Abstract
The entomopathogenic fungus, Beauveria bassiana causes a disease known as the white muscadine disease in insects. Most of the entomopathogenic fungi are soil borne nature an attempt was made to collect the soil samples from various ecosystems. Therefore, the present study aimed to isolate native strains of B. bassiana and study their genetic diversity at Taif region, Saudi Arabia, using ITS of nuclear DNA and COI gene analysis by comparing of different isolates from occasional localities through the world. Ninety-four soil samples were collected from different regions at Taif. Only 11 samples had B. bassiana fungus with a ratio of 11.7%. Sequences of ITS (593 bp), and partial COI (437 bp) of these isolates were carried out and revealed that four differed in their genetics. These sequences were deposited in the DDBJ GenBank database with eight accession numbers. Consequently, the three analytical methods (MP, NJ, and ML) executed a single tree with identical topology. The tree identified the studied isolates into two main clusters; the first contained Uzbekistan and USA isolates. The Chinese isolate clustered with this group in the MP tree and was diverged as a single cluster in the NJ tree. The second clade included the newly studied four isolates from Taif and the Italian isolate. The pairwise genetic distances among the four studied isolates (D ranged between 0.002 and 0.008) showed that they were genetically closely related. Further studies are needed to indicate more differentiation among these isolates and to evaluate their efficacy against the important insect pests in Saudi Arabia.
Similar content being viewed by others
Background
Soil is the main reservoir of entomopathogenic fungi (EPF) which have an essential influence on the occurrence and expansion of insect mycoses (Ignoffo et al. 1978). There is an increasing interest in the exploitation of EPF for the control of insect pests (Inglis et al. 2001). The entomopathogenic fungus, Beauveria bassiana causes a disease known as the white muscadine disease in insects. Taking into account of the substantial information available on the effectiveness and potential use of B. bassiana in control of many insect species and also most of EPF are soil borne nature, an attempt was made to collect the soil samples from various ecosystems (Zimmermann 1986). EPF are already available commercially for the control of various pest species of thrips, aphids, and other pests (Goettel et al. 1990; Upadhyay 2003). Indigenous isolates or strains of mycopathogens from different hosts or localities provide control programs with available and specific tool of controlling certain indigenous pests because they are more adapted with the environment anywhere (Zayed 2003).
Isolation of these fungi have been based on insect cadavers or soil (Zimmermann 1986; Abdo et al. 2008; Glare et al. 2008). Both two methods with a susceptible insect host and selective media have been utilized to the isolation of EPF from soil (Zimmermann 1998). Using of insect bait is a very sensitive detection method and entomopathogenic fungi can be selectively isolated. However, some insect species may be selected for specific fungal pathogens and difficult to quantify inoculums levels. By contrast, selective media have some advantages for the mass collection of positive EPF and quantitative data. Therefore, various selective media have been developed for the mass collection of EPF from soil (Meyling 2007).
B. bassiana represents the first example describing a micro-organism as the agent of a contagious disease. It contains a diverse assemblage of genotypes and probably comprises species complexes. Therefore, it is conceivable to have individual isolates or pathotypes which exhibit a substantially restricted host range (Inglis et al. 2001).
The advent of the molecular taxonomy of microorganisms was important to distinguish species more accurately (Fierer 2008). The internal transcribed spacer (ITS1 and ITS2) and 5.8 S regions of the nuclear ribosomal repeat unit are the most widely used for identification of EPF (White et al. 1990; Arnold et al. 2007; Sabbahi et al. 2009; Shin et al. 2010). Other studies such as Mondal and Baksi (2018) used 28S ribosomal DNA for identification of isolates of B. bassiana and to differentiate them from other isolates. Moreover, a set of five mitochondrial probes derived from a strain of B. bassiana was used to evaluate the similarity of mtDNAs from 15 B. bassiana isolates and five genera of other EPF (Hegedus and Khachatourians 1993). The present study aimed to isolate native strains of B. bassiana and study their genetic diversity at Taif, Saudi Arabia, throughout an analysis of ITS of nuclear DNA and COI gene.
Methods
Samples collection
Ninety-four cultivated soil accumulated samples from various locations at Taif region, Saudi Arabia were collected by collecting the topsoil down to 15 cm depth, with a help of a scoop like tool made up of stainless steel. Three samples were collected from each site and mixed to represent a single sample of ½ kg. Samples were collected in sterile zipper polyethylene bags, brought to the laboratory, and stored at 4 °C before processing.
Fungi isolation by selective medium
The selective DOC2-PDA medium for EPF was prepared [0.2 g CuCl2, 2 mg crystal violet, 39 g PDA (Potato Dextrose Agar), 1000 ml distilled water, pH 4.0 with HCl] according to Shin et al. (2010). A 0.2 g of soil sample was placed in a 1.5 ml micro tube with 1.3 ml of 0.02% Tween-80 solution and was vortexed for 15 min. The resulting suspension was serially diluted (10− 1) and plated on DOC2-PDA medium. After incubation for 6 days at 25 °C, the putative EPF were selected by morphological characteristics (aspects of the colonies, such as color, diameter, and mycelia texture). The fungus, B. bassiana was identified mainly based on the morphological characteristics of reproductive structures according to the taxonomical key of Samson et al. (1988).
Genomic DNA isolation
Fungal genomic DNA was extracted from the hyphae using a partially modified chemical lysis method (St. Leger and Wang 2009). Each fungal isolate was inoculated into 1.5 ml micro tube with PDA (Potato Dextrose Agar) and was incubated for 4 days at 25 °C on a shaker at 250 rpm. The mycelia was pelleted by centrifuging at 10,000 rpm for 10 min and was suspended in 400 μl of fungal DNA extraction buffer (0.2 M Tris-Cl, 0.5 M NaCl, 10 mM EDTA (pH 8.0), and 1% (w/v) SDS). Then, 400 μl of phenol-chloroform-isoamylalcohol (25:24:1) was added, and the mixture was vortexed for 5 min. and centrifuged at 10,000 rpm for 8 min. Then, the aqueous upper layer was transferred to a new micro centrifuge tube. After adding 1 μl of RNase solution (20 mg/ml) into the sample, it was incubated at 37 °C for 30 min and purified again with a phenol-chloroform-isoamylalcohol (25:24:1). The DNA in the aqueous phase was precipitated with 2.5 volume of 100% ethanol. The sample was centrifuged at 4 °C/12,000 rpm for 10 min, and the pellet was washed with 70% ethanol, dried, and suspended in 25 μl of distilled water. The extracted DNA solutions were used as a template for PCR.
PCR for ITS and COI and sequencing
The ITS regions of the ribosomal DNA were amplified by PCR with specific primers for ITS1 and ITS2 in nuclear DNA and for Cytochrome Oxidase I (COI) in mitochondrial DNA. These specific primers were designed according to the accession numbers of KC753394 and KR733105 for ITS region and COI gene, respectively. These primers are presented in Table 1. PCR amplifications were carried out in a final volume of 50 μl by mixing 2 μl of DNA with 0.5 mM of each primer, 150 mM of dNTPs, 1 U of Taq DNA polymerase (Promega), and PCR reaction buffer. Amplification was carried out in a thermal cycler with an initial denaturation of 3 min at 94 °C followed by 35 cycles of 1 min at 94 °C, 1 min at suitable annealing temperature for both pair primer (52 °C), 1 min at 72 °C, and a final extension of 10 min at 72 °C. Aliquots of PCR products were checked by electrophoresis on a 1% agarose gel with ethidium bromide and were visualized by UV trans illumination. The PCR products were excised after electrophoresis and purified using BioFlux BioSpin Gel Extraction kit (Bioer technology Co., Ltd). The purified products were sequenced using the same primers used in the PCR by an automated DNA sequencer (Macrogen, Korea). Obtained sequences in this study were compared with the GenBank database, using the BLAST software on the NCBI website. Sequences were submitted to GenBank on the DDBJ website.
ITS analysis
The segment spanning ITS-1 to ITS-2 was sequenced for all collected isolates. The same fragment of other related isolates was collected from the database by Blast program and was used for phylogenetic analysis. The analysis was primarily done by maximum-likelihood (ML) (Swofford 2002). Heuristic searches with the nearest-neighbor interchange branch swapping and 10 random taxon additions were adjusted and bootstrapping was set to 10,000 replications. The best model test to estimate the data was TrN + I (Posada and Crandall 1998). Maximum-parsimony (MP) and neighbor joining (NJ) with 10,000 bootstrap replications for MP and 500 for NJ were used to confirm the robustness of ML tree topology. NJ was executed with Tamura-Nei distance option.
Results and discussion
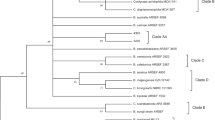
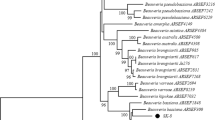
Out of the 94 soil samples collected from different regions at Taif region, only 11 samples contained B. bassiana fungus with a ratio of 11.7%. The sequences of the amplified ITS region and the mitochondrial COI gene were identical for 8 of the 11 samples while the other 3 samples were different. Therefore, four different isolates of B. bassiana in their genomics were obtained. The PCR products of ITS and COI for the different four isolates Sp1 to Sp4) are indicated in Fig. 1. The sequences of the three samples and a representative of the eight identical isolates were used together with other four different GenBank isolates in the phylogenetic analysis (Fig. 2).
ML tree constructed from 477 bp of ITS-I, 5.8 gene, ITS-II sequenced in this study. The values at some nodes refer to the bootstrapping of maximum-parsimony, neighbor-joining and maximum-likelihood, respectively which were shown when they were over 50%. The numbers between brackets are the accession numbers of the sequences collected from the GenBank database
Sequencing of partial COI (437 bp) indicated that all specimens were identical. Meanwhile, four differentiated specimens in ITS sequences with a total 593 bp were obtained. The 593 bp were 18S ribosomal RNA gene, partial sequence (53 bp); internal transcribed spacer 1, complete sequence (160 bp); 5.8S ribosomal RNA gene, complete sequence (158 bp); internal transcribed spacer 2, complete sequence (163 bp); and 28S ribosomal RNA gene, partial sequence (95 bp). These sequences were deposited in the DDBJ GenBank database with the accession numbers LC338054 to LC338057 for ITS and LC338058 to LC338061 for COI. All single nucleotide polymorphism (SNP) in these sequences were found in ITS1, 5.8S, and ITS2. Therefore, gabs were deleted in the analysis. The isolates were compared with five isolates in the GenBank from China, Uzbekistan, Italy, Spain, and USA (Table 2 and Fig. 2). Nucleotides of 477 sites of ITS-I, 5.8 gene ITS-II were analyzed for the 4 samples which acquired base composition of A = 23.6%, C = 31.5%, G = 25.4%, and T = 19.4%. Of these sites, 462 were constant and 15 were variables. Nine of the variable sites were parsimony uninformative and six were informative. MP tree exhibited consistency index (CI = 0.875), homology index (HI = 0.125), retention index (RI = 0.769), and rescaled consistency index (RC = 0.673).
The score of the ML tree was -lnL = 758.24 log likelihood. Chinese isolate exhibited different position in the MP and NJ trees; otherwise the three analytical methods executed similar tree topology (Fig. 1). Chinese isolate clustered with Uzbekistan and USA isolates in MP tree and it was out of both isolates in NJ tree. GTR + G was the best-fit model to explore the dataset (substitution rate matrix R a = 2.58, b = 5.727, c = 4.22, d = 0.000, e = 5.727, and f = 1.00). The proportion of invariable sites (I) was 0.00 and gamma distribution shape parameter = 0.134.
The tree identified the studied isolates into two main clusters. The first contained Uzbekistan and USA isolates which grouped with reasonable bootstrap support (63, 75, and 63% for MP, NJ, and ML, respectively). The Chinese isolate clustered with this group in the MP tree and was diverged as a single cluster in the NJ tree; however, this divergence was not statistically supported. The second clade included the newly studied four isolates from Taif and the Italian isolate (bootstrapping = 69, 76, and 61 for MP, NJ, and ML, respectively).
In this sense, several studies on the analysis of ITS genes have demonstrated the association between B. bassiana genetic groups and world-wide climatic zones (Ghikas et al. 2010). Moreover, phylogenetic studies based on nuclear ITS sequences have demonstrated the monophyly of Beauveria and the existence of at least two lineages within B. bassiana (Rehner and Buckley 2005).
The pairwise genetic distances among the studied isolates are shown in Table 2. The isolates (Sp1 to Sp4) were genetically closely related (D ranged between 0.002 and 0.008). Similarly, previous studies have reported genetic homogeneity among B. bassiana isolates originating from a particular (usually small) geographical region (Glare and Inwood 1998; Muro et al. 2005; Fernandes et al. 2009). However, these isolates and their cluster showed similar genetic distances to the other isolates of Uzbekistan and Chinese isolates. Meanwhile, both USA and Uzbekistan isolates were closely related to Chinese isolate. In this sense, the ITS-RFLP and ITS sequences did not detect significant genetic variation among 104 isolates of B. bassiana obtained from seven countries in the Middle East and West Asia (Muro et al. 2005). On the other hand, Berretta et al. (1998), using RAPDs with fluorescent labels, did not detect correlations with geographical origin or host as they found that some isolates of B. bassiana from distinct geographical origins and hosts were closely related. Generally, geographical distance between populations apparently is an important factor influencing genotypic variability among B. bassiana populations (Wang et al. 2003; Fernandes et al. 2009). Accordingly, greater geographical distances were associated with higher genetic distances. The isolation by distance of B. bassiana played an important role in its phylogenetic diversity (Rehner et al. 2006). Valero-Jiménez et al. (2016) indicated that the genome sequences of five isolates of B. bassiana provide a better understanding of the natural variation in virulence and will offer a major resource for future researches on this important biological control agent. Generally, the molecular genetic markers were useful for the identification of genetic types of B. bassiana and related species (Coates et al. 2002; Bhattacharya et al. 2005). The ITS1-5.8S-ITS2 region sequence analysis provided more information on polymorphism among 10 isolates of B. bassiana, allowing them to be clustered by relative similarity into three large groups (Costa et al. 2011).
Conclusions
The ITS sequences did detect slightly genetic variation among 11 (eight of them are identical) isolates of B. bassiana obtained from Taif, Saudi Arabia. These data recommended that the analyses yielding the tree topology are not enough to resolute the confident relationship among the studied isolates. It could be concluded that the four current isolates from Taif were closely related to each other and to the Italian isolate other than any of the studied isolates. Further studies are needed to indicate more differentiations among these isolates and to evaluate their efficacy against the important insect pests in Saudi Arabia in order to use these indigenous isolates because they are matched with the environmental conditions more than other commercial isolates.
References
Abdo C, Nemer N, Nemer G, Abou Jawdah Y, Kawar N (2008) Isolation of Beauveria species from Lebanon and evaluation of its efficacy against the cedar web-spinning sawfly, Cephalcia tannourinensis. Biocontrol 53:341–352
Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R (2007) Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99:185–206
Berretta MF, Lecuona RE, Zandomeni RO, Grau O (1998) Genotyping isolates of the entomopathogenic fungus Beauveria bassiana by RAPD with fluorescent labels. J Invertebr Pathol 71:145–150
Bhattacharya D, Reeb V, Simon DM, Lutzoni F (2005) Phylogenetic analyses suggest reverse splicing spread of group I introns in fungal ribosomal DNA. BMC Evol Biol 5:68–79
Coates BS, Hellimich RL, Lewis LC (2002) Allelic variation of a Beauveria bassiana (Ascomycota: Hypocreales) minisatellite is independent of host range and geographic origin. Genome 45:125–132
Costa MB, Pimenta FC, Luz C, Oliveira V, Oliveira M, Bueno E, Petrofeza S (2011) Beauveria bassiana: Quercetinase production and genetic diversity. Brazil J Microbiol 42:12–21
Fernandes ÉKK, Moraes ÁML, Pacheco RS, Rangel DEN, Miller MP, Bittencourt VREP, Roberts DW (2009) Genetic diversity among Brazilian isolates of Beauveria bassiana: comparisons with non-Brazilian isolates and other Beauveria species. J Appl Microbiol 107:760–774
Fierer N (2008) Microbial biogeography: patterns in microbial diversity across space and time. In: Zengler K (ed) Accessing uncultivated microorganisms: from the environment to organisms and genomes and back. ASM Press, Washington, DC, pp 95–115
Ghikas DV, Kouvelis VN, Typas MA (2010) Phylogenetic and biogeographic implications inferred by mitochondrial intergenic region analyses and ITS1-5.8S-ITS2 of the entomopathogenic fungi Beauveria bassiana and B. brongniartii. BMC Microbiol 10:174
Glare TR, Inwood AJ (1998) Morphological and genetic characterization of Beauveria spp. from New Zealand. Mycol Res 102:250–256
Glare TR, Reay SD, Nelson TL, Moore R (2008) Beauveria caledonica is a naturally occurring pathogen of forest beetles. Mycol Res 112(3):352–360
Goettel MS, Poprawski TJ, Vandenberg JD, Li Z, Roberts DW (1990) Safety to nontarget invertebrates of fungal biocontrol agents. In: Lard M, Lacey LA, Davidson EW (eds). Safety of microbial insecticides. CRC Press, Florida, pp 209–231
Hegedus DD, Khachatourians GG (1993) Identification of molecular variants in mitochondrial DNAs of members of the genera Beauveria, Verticillium, Paecilomyces, Tolypocladium, and Metarhizium. Appl Environ Microbiol 59:4283–4288
Ignoffo CM, Garcia C, Hostetter DL, Pinnel RE (1978) Stability of conidia of endomopathogenic fungus Nomuraea rileyi in and on soil. J Invertebr Pathol 28:256–268
Inglis DG, Goettel MS, Butt TM, Strasser H (2001) Use of hyphomycetous fungi for managing insect pests. In: Butt TM, Jackson C, Magan N (eds) Fungi as biocontrol agents: progress, problems, and potential. CAB international, Wallingford, pp 23–69
Meyling NV (2007) Methods for isolation of entomopathogenic fungi from the soil environment. Lab Manual :1–18
Mondal S, Baksi S (2018) Strain-specific identification of Beauveria bassiana isolated from a novel habitat, using rDNA-based sequence analogy. Egypt J Biol Pest Control 28:29 8Pp
Muro MA, Elliott S, Moore D, Parker BL, Skinner M, Reid W, El Bouhssini M (2005) Molecular characterization of Beauveria bassiana isolates obtained from overwintering sites of Sunn pests (Eurygaster and Aelia species). Mycol Res 109:294–306
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98
Rehner SA, Posada F, Buckley EP, Infante F, Castrillo A, Vega FE (2006) Phylogenetic origins of African and Neotropical Beauveria bassiana s.l. pathogens of the coffee berry borer, Hypothenemus hampei. J Invertebr Pathol 93:11–21
Sabbahi R, Lavallee R, Merzouki A, Guertin C (2009) Differentiation of entomopathogenic fungus Beauveria bassiana (Ascomycetes: Hypocreales) isolates by PCR-RFLP. Phytoprotection 90(2):49–56
Samson RA, Evans HC, Latge JP (1988) Atlas of entomopathogenic fungi. Springer - Verlag, New York, p 187
Shin TY, Choi J, Bae S, Koo H, Woo S (2010) Industrial Entomology. Study on Selective Media for Isolation of Entomopathogenic Fungi. Int J Indust Entomol 20:7–12
St. Leger RJ, Wang C (2009) Entomopathonic fungi and the genomics era. In: Patricia SP, Vanderberg J, Boemare N, Glazer I (eds) Insect pathogens: molecular approaches and techniques. CABI, Wallingford, pp 365–400
Swofford DL (2002) PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Upadhyay RK (2003) Advances in microbial control of insect pests. Kluwer Academic/Plenum Publishers, Dordrecht
Valero-Jiménez CA, Faino L, Veld DS, Smit S, Zwaan BJ, van Kan JAL (2016) Comparative genomics of Beauveria bassiana: uncovering signatures of virulence against mosquitoes. BMC Genomics 17:986
Wang C, Shah FA, Patel N, Li Z, Butt TM (2003) Molecular investigation on strain genetic relatedness and population structure of Beauveria bassiana. Environ Microbiol 5:908–915
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: a guide to methods and applications. Academic Press Inc, New York, pp 315–322
Zayed A (2003) Pathogenecity of two Beauveria bassiana indigenous isolates towards the greater wax moth Galleria mellonella L. larvae in Egypt. Efflatounia 3:10–14
Zimmermann G (1986) The galleria bait method for detection of entomopathogenic fungi in soil. J Appl Entomol 102:213–215
Zimmermann G (1998) Suggestion for a standardized method for reisolation of entomopathogenic fungi from soil using the bait method, “insect pathogens and insect parasitic nematodes”. IOBC Bull 21(4):289
Acknowledgements
The authors would like to express their deepest gratitude to KACST, Saudi Arabia, for the financial support for this study through a research grant number: SG-36-95.
Funding
This work was financially supported by KACST, Saudi Arabia,
Availability of data and materials
The datasets generated and analyzed during the current study are available in DNA Data Bank of Japan (DDBJ) (https://www.ddbj.nig.ac.jp/).
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sayed, S.M., Ali, E.F., El-Arnaouty, S.A. et al. Isolation, identification, and molecular diversity of indigenous isolates of Beauveria bassiana from Taif region, Saudi Arabia. Egypt J Biol Pest Control 28, 47 (2018). https://doi.org/10.1186/s41938-018-0054-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-018-0054-z