Abstract
Sclerotium rolfsii Sacc. (the sclerotial state of Athelia rolfsii (Cruzi) Tu and Kimbrough), the soil-borne pathogen on several plants all over the world, has been previously reported from Turkey on certain plants. In this study, turfgrass areas in 9 provinces of Turkey were firstly surveyed for S. rolfsii, and samples showing chlorotic, reddish-brown, and frog-eye shaped circular patches were collected. Totally, 32 Sclerotium rolfsii isolates were obtained from these areas. One mycelial compatibility group (MCG) was identified among S. rolfsii isolates. Disease severity in pathogenicity tests carried out in the greenhouse ranged from 83.74 to 92.87%. Identification of fungal and bacterial isolates used in the study was performed by DNA sequencing analysis. Five antagonistic bacterial strains, previously found as effective on controlling some fungal pathogens, were tested to determine their antifungal effects against the southern blight by using seed coating method in greenhouse conditions. In consequence of the biological control studies, Bacillus cereus 44bac and Stenotrophomonas rhizophila 88bfp were found more effective than the other strains with the ratio of 91.00 and 90.11%, respectively.
Similar content being viewed by others
Background
Southern blight, caused by the soil-borne fungus Sclerotium rolfsii Sacc. (Atheliaceae: Athelia rolfsii (Cruzi) Tu and Kimbrough), is a serious disease for a wide range of plants, including vegetables, fruits, ornamental plants, and field crops (Mullen 2001). The fungus, also, attacks primarily bentgrass, bluegrass, fescues, and ryegrass (Smiley 1992).
The first symptoms of outstanding in turfgrass areas are round crescent-shaped yellow areas about 20 cm in diameter. Grasses grow yellowish over time and become sparse. As long as the disease continues to progress, diseased areas in the form of rings or patches die. But the grass in the center remains green. The color of the dead areas turns reddish brown over time. These rings, formed in dead turfgrass in summer and humid weather, expand quickly (about 20 cm per week). Sometimes, symptoms observed in these areas similar to the “frog eye.” In Agrostis and Poa species, the diseased areas caused by this disease are usually seen in the autumn. Under moist conditions, white mycelial growth which develops on the dead grass and later on sclerotia ranging from white or light to dark brown on the mycelium are observed (Smiley 1992).
S. rolfsii isolates can be diverged into different mycelial compatibility groups (MCGs) based on mycelial interactions among isolates. The role of MCGs is important in defining field populations of fungi and facilitating genetic variation in fungal species, where the sexual reproductive stage (teleomorph stage) of the life cycle has a minimal impact on the disease cycle (Kohn et al. 1991).
Nowadays, the plantation and conservation of turfgrass areas has become a huge industrial sector in the world. Cultural practices are not efficient for controlling the disease, that is why fungicide usage is very common and widely used in turfgrass areas all around the world. Chemical fungicides are extensively used in turfgrass areas in Turkey, and excessive use of chemical fungicides has led to deteriorating human health, environmental pollution, and development of pathogen resistance to fungicides (Balcı and Gedikli, 2012). Due to the harmful effects in controlling fungal diseases, new studies are needed to use alternative methods for plant protection, which are less dependent on chemicals and are more environmentally friendly. In this regard, biological control can be an alternative or supplement to current management practices for S. rolfsii (Sai et al. 2010).
The most commonly beneficial microorganisms used in the control of plant pathogens are the following bacterial strains: Bacillus, Pseudomonas, and Trichoderma spp. (Raaijmakers et al. 2010). For the biocontrol of S. rolfsii, some bacterial genera have been tested for their ability to control. Pseudomonas spp. and Bacillus spp. have been commonly studied to control S. rolfsii on various plants. It was detected that Pseudomonas and Bacillus strains restricted in vitro hyphal growth or reduced germination of sclerotia of S. rolfsii (Rakh et al. 2011 and Tonelli et al. 2011). Several commercial preparations, containing these bacterial and fungal agents, are also recommended on turfgrass diseases in the world. Bio-Trek 22G (Trichoderma harzianum ) is the first registered biopesticide for dollar spot, brown patch, and Pythium root rot on turfgrass (Harman and Lo 1996). Eco Guard TM (Bacillus licheniformis), Rhapsody (B. subtilis), Actinovate SP (Streptomyces lydicus WYEC 108), and Botrycid (Pseudomonas aureofaciens) are the other microbial biocides used against turfgrass diseases (Corwin et al. 2007). Among these, only Rapsody is recommended against the southern blight disease caused by S. rolfsii in turfgrass areas. But there is no registered microbial biocide against turfgrass diseases in Turkey so far.
The objective of this study was to molecularly identify S. rolfsii isolates in Turkey, and to detect their virulence and mycelial compatibility groups, using some domestic bacterial and fungal isolates under greenhouse conditions.
Materials and methods
Survey and isolation of the pathogens
The survey was performed and samples were collected from the turfgrass areas in İstanbul, Antalya, Ankara, İzmir, Kayseri, Bursa, Aydın, and Muğla Provinces in 2015. Segments of leaves and roots were sterilized for 1 min, in 1% sodium hypochlorite (NaOCl) solution, then washed with sterile water and air dried in a laminar flow cabinet before culturing on potato dextrose agar (PDA, Difco, USA) containing 50 mg/l streptomycin sulfate. Isolates were incubated under the light and dark regimes, respectively on 28 ± 1 °C for 7 days.
Bacterial isolates
Five antagonistic domestic bacterial strains (215b, 44bac, 88cfp, 166fp, and 88bfp) used in this study were isolated from the tomato and cucumber rhizospheres in a previous study, where isolates 166fp and 88cfp managed with Pythium deliense, Sclerotinia minor, and Alternaria solani on tomato were detected (Aşkın 2008). Also, isolate 44bac managed downy mildew on cucumber (Aşkın and Ozan 2013) under field conditions. Molecular identification of the 5 antagonistic bacterial isolates, used in this study, was first determined in this study.
Molecular identifications of fungal and bacterial isolates
Isolation of fungal DNA was carried out by Blood and Tissue Kit (QIAGEN Inc. Valencia, CA), as specified by the manufacturer. The PCR reaction mixture and conditions were made by modifying according to Mahadevakumar et al. (2016). DNA amplification was performed, using the optimized cycles optimized with Techne TC-5000 thermal cycler. Primers ITS-1 (5 ′TCC GTA GGT GAA CCT GCGG 3′) and ITS-4 (5 ′TCC TCC GCT TAT TGA TATGC 3′) were used for amplification of ITS regions (White et al. 1990). The polymerase chain reaction (PCR) was performed in a 50-μl reaction mixture containing 1 μl template DNA, 1 μl forward primer (10 mM), 1 μl reverse primer (10 mM), 5 μl reaction buffer (10×), 4 μl dNTP (each 2.5 mM), 0.5 μl Taq DNA Polymerase (5 U/μl), and 37.5 μl sterile double-distilled water. The PCR cycling protocol consisted of initial denaturation at 94 °C for 4 min, followed by 30 cycles of 94 °C for 45 s, 55 °C for 45 s, 72 °C for 2 min, and a final elongation step of 72 °C for 10 min. As a negative control, the template DNA was replaced by sterile double-distilled water.
Molecular definition of bacteria was made according to the protocol of DNA isolation from Blood and Tissue Kit (QIAGEN Inc. Valencia, CA). The 16S rDNA gene fragments were amplified by PCR using the universal primers 27F 5′AGAGTTTGATCMTGGCTCAG3′ and 1492R 5′TACGGYTACCTTGTTACGACTT3′ (Lane, 1991). The PCR reaction mixture and conditions were modified to carry out the PCR reaction. DNA replications were performed in the ABI Veriti (Applied Biosystem) thermal cycler using the following cycles:
-
1.
The initial denaturation consist of 5 min at 94 °C, 35 cycles of amplification step consisting of denaturation of 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 120 s, and final extension of 10 min at 72 °C (Lane 1991).
-
2.
The PCR product was directly subjected to Sanger sequence treatment in a special Arge Laboratory (BM Gene Research and Biotechnology Company, Ankara, Turkey).
-
3.
Bipartite raw sequence electropherograms were compared to the isolate sequences in Gen Bank after BLAST screening in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Determination of mycelial compatibility groups (MCGs) of S. rolfsii isolates
In order to determine the mycelial compatibility among 32 isolates, obtained from different areas grown with turfgrass areas, mating each isolate with themselves and with all other isolates was carried out (Punja and Grogan 1983). Mating studies were performed on PDA medium with 0.25% food coloring (Ponceau 4R, Turkey). Mycelial discs of two isolates were reciprocatively plated on PDA medium with a distance of 3–4 cm. Cultures were incubated at 25 ± 1 °C and colony growth was observed after 7–14 days (Kohn et al. 1991). Hyphal interaction among the isolates mate was observed at the end of 7 days after culturing. Compatibility between each of the two groups was evaluated according to a red line with separation in the region, where the hyphae collided. When the red line was seen, it was accepted as incompatible, otherwise not (Punja and Grogan 1983).
Fungal inoculums
S. rolfsii was grown on wheat bran medium in bottles of 500 ml, sterilized in an autoclave for 20 min. at 121 °C, for 15 days at 28 ± 1 °C (Aşkın 2008).
Pathogenicity tests
Pathogenicity tests of S. rolfsii isolates were conducted under greenhouse conditions. The fungus inoculums grown on wheat bran (4 g inoculums/kg soil) was added to the sterilized garden soil, fine sand, and burnt fertilizer mixed (2:1:1) and then distributed in the pots (10 cm in diameter). Control pots contained the sterilized garden soil, fine sand, and burnt fertilizer mixed (2:1:1) free from the inoculum. Three pots were replicated for each treatment. All pots were covered by a sanitized polyethylene nylon and incubated for 3 days. At the end of the duration, 30 seeds of turfgrass (cv. Festuca arundinacea) were placed on the soil surface, covered with 1 cm of sterile natural soil, and watered with 9–10 ml of water. The infected plants were counted 3 weeks later (Zhang et al. 2014) and recorded. Evaluation was made according to a scale of 0 to 5: 0 = no disease symptoms, 1 = 1–10% hypocotyl infecting and/or shortening, 2 = 11–30% hypocotyl infecting and/or shortening, 3 = 31–50% hypocotyl infecting and/or shortening, 4 = 51–80% hypocotyl infecting and/or shortening, and 5 = entire hypocotyl infecting and/or shortening (Ichielevich Auster et al. 1985). Disease severity was calculated according to the Townsend–Heuberger formula (Townsend and Heuberger 1943):
(N is the number of samples in the scale with different disease grades, V is the scale value, Z is the highest scale value, and N is the total number of samples observed)
Bacterial inoculums
Bacterial isolates were cultured in potato dextrose broth. After 24 h, the bacterial concentration was verified through spectrophotometry at a wavelength (λ) of 600 nm seeking for an absorbance between 0.9 and 1 equivalent to a concentration of 1 × 108 c/ml and by counting the colony forming units (cfu) per milliliter through the total viable count. Surface-disinfected seeds of turfgrass were inoculated with bacterial solutions by soaking with agitation for 12 h. Rhizobacterial stock cultures were maintained in nutrient agar medium amended with 15% glycerol and stored at − 80 °C. Before being used in the bioassays, stock cultures were streaked onto nutrient agar plates and incubated at 28 ± 1 °C for 48 h.
Biocontrol assays
This study was carried out using turfgrass seeds mixture containing 4 cvs: Festuca rubra, Lolium perenne, Poa pratensis, Festuca arundinacea, and the most virulent S. rolfsii isolate (Sr34-10). The soil used in the experiment was prepared in the form of a mixture of 2:1:1 garden soil to stream sand to burnt fertilizer. Soil mix was sterilized in an autoclave at 121 °C for 45 min. The inoculum of S. rolfsii was developed on wheat bran. Antagonistic bacteria were applied by coating to the seeds. Experiments were carried out in both sterilized and non-sterilized soils, where three treatments were performed: (1) negative control by sowing uncoated turfgrass seeds in non-infested soils, (2) positive control by sowing uncoated turfgrass seeds in infested soils to evaluate the varietal sensitivity, and (3) sowing coated turfgrass seeds in infested soils to evaluate the biocontrol efficacy of each antagonistic isolate against S. rolfsii. The mixture of inoculum and soil (5 g to 1 kg of soil) were filled in sterile plastic pots (10 cm in diameter). After 4–5 days, 30 coated and uncoated turfgrass seeds were sown at a depth of 1 cm per pot. The plants were grown in a plant growth medium containing 12 h of light, 12 h of darkness, and 25 ± 1 °C temperature. The experiments were carried out in 3 replicates, according to randomized plot design. After the inoculation, observations were made at intervals of 10 days and a 0–5 scale was used 30 days after sowing, and disease severity was calculated as mentioned before.
Statistical analysis
Variance analyses were carried out, using the SPSS GLM statistical program, to determine the differences among virulence levels of isolates and disease rates in biocontrol assay.
Disease assessment
Disease ratios were estimated according to the devised scale by Townsend-Heuberger formula (Townsend and Heuberger 1943). The calculated disease severity and the activity of bacterial isolates using the Abbott formula was determined from the disease severity values. Disease severity was compared by Tukey’s multiple comparison test on these ratios.
Results and discussion
Survey of the disease and identification of Sclerotium rolfsii
The survey was carried out in wide parks, golf courses, stadiums, and recreation areas of 8 provinces among the largest turfgrass areas in different regions in Turkey, and totally 1400 samples were collected. In consequence of isolations, distinguishing symptoms, which were chlorotic or reddish-brown frog-eye or crescent-shaped circular patches (Figs. 1 a and b), of 32 S. rolfsii isolates were obtained, based on both colony morphology and rDNA internal transcribed spacer (ITS) region sequences. The isolates showed a rapid and a radial mycelial development. The colony color was white on the PDA medium, with a large amount of round, 1–3-mm-diameter brown sclerotia formed (Fig. 1c).“Clamp connection” event in fungus hyphae was observed (Fig. 1d).
On the other hand, molecular studies were also performed on isolates according to ITS-1 and 4 regions, and amplicons displayed by gel transilluminator were found to be between 650–680 bp, which is specific for S. rolfsii. The obtained results are parallel to that of Poornima et al. (2018) who studied genetic variations among S. rolfsii isolates of groundnut by using ITS rDNA sequence data and obtained an amplification fragment of about 650–700 bp. The sequences of the isolates showed a 99–100% similarity rate compared to those of S. rolfsii deposited in the NCBI database. It was concluded that S. rolfsii caused more damage in warmer climates than in semi-arid areas in Turkey. S. rolfsii was previously found in soybean, peanut, sugar beet, tomatoes, pepper, eggplant, bean, and artichoke in Turkey, causing it to have different names according to crops (Yaşar and Türk 2016 and Aydoğdu et al. 2016). This is the first confirmed study on southern blight on turfgrass in Turkey.
Identification of the bacterial bioagents
Unidentified bacterial bioagents, which used in this study to see possible effects for controlling southern blight on turfgrass, were previously isolated from different crops (Aşkın 2008 and Aşkın and Ozan 2013). The sequence data of bacterial isolates showed 99–100% similarity with isolates in GenBank. The results of BLAST analysis showed that Stenotrophomonas rhizophila (100% similarity), Pseudomonas putida 166fp (100% similarity), P. putida 88cfp (99.64% similarity), Paenibacillus sp. (99.79% similarity), and Bacillus cereus (100% similarity) were identified. As a result of the study, amplicons displayed by gel transilluminator were found to be between 1500 and 1550 bp. It was detected that these Pseudomonas spp. and Bacillus spp. protected the infection by Pythium deliense, Sclerotinia minör, and Alternaria solani on tomato (Aşkın 2008) and downy mildew on cucumber (Aşkın and Ozan 2013) under field conditions.
Mycelial compatibility groups
In all mates, a red line was not observed in the contact area of the 2 colonies, and complete fusion was observed; all isolates were compatible with each other. Sclerotia were formed at the junction (Fig. 2). As a result of mating type analysis of 32 isolates, all of them were compatible with each other. Thus, it can be concluded that only one mycelial compatibility group (MCG) was seen (Table 1) in S. rolfsii in turfgrass, in this study.
To our knowledge, there is no MCG study on S. rolfsii on turfgrass in the world. However, in some studies performed in different crops, MCG of S. rolfsii isolates obtained from different hosts and geographical regions and even from the same regions and hosts were tested, and it has been reported that there is no relationship between geographic region and host specificization of the formation of MCG of S. rolfsii. But genetic variations among different MCGs have been reported in different crops (Adandonon et al. 2005).
Pathogenicity
Disease severity of 32 S. rolfsii isolates ranged between 83.74 and 92.87% in different turfgrass composition (Table 1). The fungus is a destructive pathogen that can infect more than 500 plant species commonly seen in tropical and subtropical regions (Smiley et al.1992). S. rolfsii has been found to cause damage to tomatoes, peppers, lettuce, bean, peanut, and sugarbeet in Turkey (Yaşar and Mert-Türk 2016). But, it has not been previously identified in turfgrass areas in Turkey.
Biocontrol assays
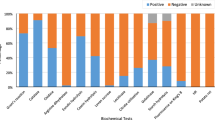
In greenhouse experiments, all identified bacterial isolates were found to be effective in controlling the disease comparing to the severity of the disease in control treatment. The lowest disease severities were found at the treatments of 44bac (B. cereus) and 88bpf (S. rhizophila), being 8.00 ± 3.138 and 8.80 ± 3.138 %, respectively (P < 0.0001) (Table 2 and Fig. 2). The highest disease severity was obtained in the treatment of 88cpf (P. putida), being 40.27 ± 3.138 % compared to that in the control treatment (Table 2). The highest protection effect was obtained by the isolate 44bac (B. cereus) being 91.00%, followed by the isolates 88bpf (Stenotrophomonas rhizophila), being 90.11%, and 215b (Paenibacillus sp.), being 64.60% (Table 2 and Fig. 3).
For the biocontrol of S. rolfsii, several bacterial strains have been studied in the world, and effective results have been obtained in these studies, most of them belonging to the biocontrol of S. rolfsii on vegetables with the genera Pseudomonas and Bacillus (Rakh et al. 2011 and Tonelli et al. 2011). But, there are no adequate studies related to S. rolfsii on turfgrass.
Conclusion
The presence of S. rolfsii as a causal of southern blight disease on the turfgrass areas in Turkey has firstly revealed with this study. Both Bacillus cereus 44bac and Stenotrophomonas rhizophila 88bpf were found to be effective in controlling S. rolfsii under greenhouse conditions. These bacterial strains could be recommended in the management programs of the southern blight disease in turfgrass areas. However, further studies are needed to be experimented under field conditions.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Adandonon A, Aveling TAS, van derMerwe NA, Sanders G (2005) Genetic variation among Sclerotium isolates from Benin and South Africa, determined using mycelial compatibility and ITS rDNA sequence data. Aust Plant Pathol 34:19–25
Aşkın A (2008) The effects of non-pathogenic Pseudomonas on damping-off of tomato seedlings caused by some fungal pathogens in Ankara province. Ph D. Thesis, Ankara Univ. Grad. School of Nat. and App. Scis. Dept. of Plant Protec. No. of pages?
Aşkın A, Ozan S (2013) Orta Anadolu Bölgesinde örtü altı hıyar yetiştiriciliğinde mildiyö (Pseudoperonospora cubensis Berk. And Curt.) mücadelesinde Bacillus spp. izolatlarının kullanım olanaklarının araştırılması. Bitki Koruma Ürünleri Ve Makineleri Kongresi, 2-4 Nisan; Antalya, 57-68.
Aydoğdu M, Kurbetli İ, Ozan S (2016) First report of Sclerotium rolfsii causing crown rot on globe artichoke in Turkey. Plant Dis 100(10):2161
Balcı V, Gedikli N (2012) Golf Alanlarında Kullanılan Kimyasal İlaçların ve Gübrelerin Çevre ve Uygulayıcılar Üzerine Etkileri-Organik Yaklaşımlar. Spormetre Beden Eğitimi ve Spor Bilimleri Dergisi, 2011 4(4):141–148
Corwin B, Tisserat N, Fresenburg B (2007) Integrated Pest Management. Identification and Management Of Turfgrass Diseases. Plant Protection Programs College of Agriculture, Food and Natural Resources, Columbia, p 55
Harman GE, Lo CT (1996) The First Registered Biological Control Product for Turf Disease: Bio-Trek 22G by G. E. Turfgrass Trends, Cornell University, USA, p 8–14
Ichielevich-Auster M, Sneh B, Koltin Y, Barash I (1985) Suppression of damping-off caused by Rhizoctonia species by a nonpathogenic isolate of R. solani. Phytopathology 75:1080–1084
Kohn LM, Stavoski E, Carbone I, Royer J, Anderson JB (1991) Mycelial incompatibility and molecular markers identify genetic variability in field populations of Sclerotinia sclerotiorum. Phytopathology 81:480–485
Lane DJ (1991) 16S/23S rRNA Sequencing In nucleic acid techniques in bacterial systematics. In: Stackebrandt E, Goodfellow M (eds) . John Wiley and Sons, New York, NY, USA, pp 115–175
Mahadevakumar S, Yadav V, Tejaswini GS, Janardhana GR (2016) Morphological and molecular characterization of Sclerotium rolfsii associated with fruit rot of Cucurbita maxima. Euro J Plant Pathol 145(1):215–219
Mullen J (2001) Southern blight, southern stem blight, white mold. The Plant Health Instructor. DOI: https://doi.org/10.1094/PHI-I-2001-0104-01. https://www.apsnet.org/edcenter/intropp/lessons/fungi/Basidiomycetes/Pages/SouthernBlight.asp.
Poornima Sunkad G, Sudini H (2018) Molecular variability among the isolates of Sclerotium rolfsii causing stem and pod rot of groundnut collected from Karnataka. India Inter J Curr Microbiol App Sci 7(5):2925–2934
Punja ZK, Grogan RG (1983) Hyphal interactions and antagonism among field isolates and single-basidiospore strains of Athelia (Sclerotium) rolfsii. Phytopathology 73:1279–1284
Raaijmakers JM, de Bruijn I, Nybroe O, Ongena M (2010) Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. Fems Microbiol Revs 34:1037–1062
Rakh RR, Raut LS, Dalvi SM, Manwar AV (2011) Biological control of Sclerotium rolfsii, causing stem rot of groundnut by Pseudomonas cf. monteilii 9. Recent Res Sci Technol 3:26–34
Sai LV, Anuradha P, Vijayalakshmi K, Reddy NPE (2010) Biocontrol of stem rot of groundnut incited by Sclerotium rolfsii and in vitro compatibility of potential native antagonists with fungicides. J Pure App Microbiol 4:565–570
Smiley R, Dernoeden P, Clarke B (1992) Compendium of Turfgrass Diseases, Second Edition, American Phytopathological Society. APS Press, St. Paul, p 96
Tonelli ML, Furlan A, Taurian T, Castro S, Fabra A (2011) Peanut priming induced by biocontrol agents. Physiol Molec Plant Pathol 75:100–105
Townsend GK, Heuberger JW (1943) Methods for estimating losses caused by diseases in fungicide experiments. Plant Dis Rep 27:340–343
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: A Guide to Methods and Applications. Academic Press, Inc, New York, pp 315–322
Yaşar İ, Mert-Türk F (2016) Mycelial compatible groups of the Sclerotium rolfsii isolates and comparison of virulence, VII International Scientific Agricultural Symposium "Agrosym 2016" Jahorina, 06-09 October 2016, Bosnia and Herzegovina.
Zhang XY, Yu XX, Yu Z, Xue YF, Qi LP (2014) A simple method based on laboratory inoculum and field inoculum for evaluating potato resistance to black scurf caused by Rhizoctonia solani. Breed Sci 64(2):156–163
Acknowledgements
A part of this study was supported by The Scientific and Technological Research Council of Turkey, project number: 114O400.
Funding
The part of the survey, isolation, and pathogenicity of S. rolfsii was funded by The Scientific and Technological Research Council of Turkey.
Author information
Authors and Affiliations
Contributions
FU contributed to the survey, isolation, identification, pathogenicity, mycelial compatibility groups, and biological control studies of S. rolfsii isolates. All authors designed the study and wrote the manuscript, AA contributed to the provision of bacterial isolates and biological control studies. EK contributed to the identification of bacterial isolates. MY analyzed the data (statistical analyses), mycelial compatibility groups studies, and read and approved the final manuscript. MÜB identified of turfgrass species. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ünal, F., Aşkın, A., Koca, E. et al. Mycelial compatibility groups, pathogenic diversity and biological control of Sclerotium rolfsii on turfgrass. Egypt J Biol Pest Control 29, 44 (2019). https://doi.org/10.1186/s41938-019-0144-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-019-0144-6