Abstract
Background
SIVA is a transcriptional target of p53 that plays a potential role in the development and progression of cancer. In this study, we analyzed SIVA1 and SIVA2 expression, and its association with clinical features and TP53 and MDM2 expression in bone marrow cells from healthy donors and myelodysplastic syndrome (MDS) patients.
Methods
Fifty-five untreated patients with MDS and 22 healthy donors were included. Gene expression was evaluated by quantitative PCR. For statistical analysis, Mann–Whitney test, Spearman correlation analysis and Log-rank (Mantel-Cox) were used, as appropriate. A p value <0.05 was considered statistically significant.
Results
SIVA1 and SIVA2 transcripts were significantly decreased in bone marrow samples from MDS patients compared to healthy donors, and positively correlated with MDM2 and TP53 expression in MDS patients (all p < 0.05). MDM2 expression was also downregulated in bone marrow samples from MDS patients compared to healthy donors (p < 0.05). However, SIVA1, SIVA2, MDM2 and TP53 expressions did not impact on MDS outcomes.
Conclusions
SIVA1 and SIVA2 transcripts are downregulated in bone marrow samples from MDS patients.
Similar content being viewed by others
Background
Apoptosis resistance and genomic instability are hallmarks of cancer cells [1, 2]. The p53 tumor suppressor protein is a transcription factor that regulates several signaling pathways involved in the cell response to stress, suppressing malignant transformation by cell cycle arrest, DNA repair, induction of apoptosis and initiation of senescence [3]. Deregulation of p53 is a common event in hematological malignancies. In acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS), strong p53 protein expression has been associated with TP53 mutations [4,5,6,7] and TP53 mutations have been associated with poor prognosis [8,9,10,11]. In low-risk MDS patients, high p53 protein expression is an independent predictor of transformation into AML [4].
SIVA is a transcriptional target of p53 that was initially described as a proapoptotic protein and acts on both extrinsic and intrinsic apoptotic pathways [12, 13]. Two alternatively-spliced transcript variants encoding distinct proteins have been described, SIVA1 and SIVA2. MDM2 is a negative regulator of p53 and may modulate the expression of SIVA through regulation of the stability and activation of p73 and E2F1 transcription factors, which represent a p53-independent mechanism of SIVA regulation [13, 14].
SIVA1 binds to BCL2 and BCL-XL, and abrogates their anti-apoptotic activity [15, 16]. SIVA modulates BAX oligomerization, binds to XIAP, and balances NFκB and JNK signaling, promoting apoptosis [17, 18]. In acute lymphoblast leukemia cell lines, both SIVA isoforms play an important role in the apoptotic pathway, induced through CD27 antigen by activation of BID, with a consequent release of cytochrome C and activation of caspases 9 and 3 [19]. In leukemia cell lines, SIVA1 also binds to and inhibits Stathmin 1 activity, preventing tumor growth [20]. In contrast to the tumor suppression functions initially described for SIVA, recent studies indicate that the conditional knockout of SIVA1 reduced tumorigenesis in KRAS-driven lung cancer murine model [21] and high SIVA1 expression was associated with worse survival rates in AML patients [22]. In the present study, we characterized SIVA1 and SIVA2 expressions in healthy controls and MDS patients, and their correlation with clinical features, as well as the expression of SIVA-related genes: TP53 and MDM2.
Methods
Bone marrow samples
Bone marrow samples collected from 55 untreated patients with MDS and 22 healthy donors from related bone marrow transplantation (median age 33 years [range 18–56]) were analyzed. Patient’s characteristics are described in Table 1. The present study was approved by the Ethics Committee of the University of Campinas in accordance with the Helsinki Declaration. Written informed consent was obtained from all healthy donors and MDS patients who participated in this study. Patients who attended the clinic between 2005 and 2013 and signed the informed consent for the study were included. Diagnosis was made by clinical data, peripheral blood counts, bone marrow (BM) cytology and histology and cytogenetics. Deficiency anemias, autoimmune diseases and viral infections were excluded [23]. The cases were classified by the WHO 2008 criteria and risk stratification was made according to IPSS-R [24].
Quantitative polymerase chain reaction (qPCR)
Total RNA was obtained from total bone marrow cells, after removal of erythrocytes by hemolysis, using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Genomic DNA was eliminated using DNAse I treatment (Invitrogen). cDNA was obtained from 1 μg of RNA using RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas, St. Leon-Rot, Germany). A total of 120 ng of cDNA was used for gene expression analysis by quantitative PCR (qPCR) in the ABI 7500 Sequence Detection System (Applied Biosystem, Foster City, CA, USA) using specific primers for SIVA1, SIVA2, TP53, MDM2 and HPRT1. Primer sequences and concentrations are described in Table 2. HPRT1 was used as the reference gene. The relative gene expression was calculated using the equation 2-ΔΔCT [25]. A negative ‘No Template Control’ was included for each primer pair. The dissociation protocol was performed at the end of each run to check for non-specific amplification. Three replicas were run on the same plate for each sample.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, Inc., San. Diego, CA, USA) or SAS System for windows 9.2 (SAS Institute, Inc., Cary, NC, USA). Mann–Whitney test was used for measured factors; Spearman correlation analysis was used for ranking correlation tests and Log-rank (Mantel-Cox) was used to estimate overall survival (OS) and event free survival (EFS). OS was defined from time of sampling to date of death or last seen. For MDS patients, EFS was defined as time of sampling to date of progression to high-risk MDS or AML with myelodysplasia-related changes, or date of death. A p value <0.05 was considered as statistically significant.
Results
SIVA1 and SIVA2 transcripts are downregulated in bone marrow cells from MDS patients
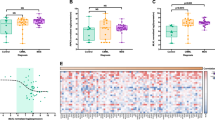
SIVA1 and SIVA2 transcripts were significantly decreased in bone marrow samples from MDS patients compared to cells from healthy donors (SIVA1: median 0.71 [range 0.00–10.28] versus (vs.) 2.18 [0.23–25.88], p < 0.0001, Fig. 1a; SIVA2: 0.85 [0.04–18.14] vs. 4.69 [0.81–35.53], respectively, p < 0.0001, Fig. 1b). No difference was observed on SIVA1 and SIVA2 expression when MDS patients were stratified by IPSS-R into very low/low vs. intermediate/high/very high (p > 0.05, Additional file 1: Figure S1). Similar results were observed when MDS patients were stratified by WHO 2008 classification (refractory anemia (RA)/refractory anemia with ringed sideroblasts (RARS)/refractory cytopenia with multilineage dysplasia (RCMD) vs. refractory anemia with excess blast-1 (RAEB-1)/refractory anemia with excess blast-2 (RAEB-2) group; p > 0.05, Additional file 2: Figure S2. Spearman correlation analysis showed a significant positive correlation between SIVA1 and SIVA2 expression in normal (r = 0.74, p < 0.0001, Fig. 1c) and MDS (r = 0.75, p < 0.0001, Fig. 1d) bone marrow samples, indicating a similar regulation for both isoforms of SIVA in hematopoietic cells.
SIVA1 and SIVA2 expression in bone marrow cells from healthy donors and patients with myelodysplastic syndromes (MDS). SIVA1 (a) and SIVA2 (b) mRNA expression in total bone marrow cells from healthy donors and MDS patients. Horizontal lines indicate medians. The numbers of subjects studied and p values are indicated; Mann–Whitney test. Correlation analysis between SIVA1 expression with SIVA2 expression in total bone marrow cells from healthy donors (c) and MDS patients (d). The p and r values are indicated; Spearman correlation test
SIVA1 and SIVA2 expression correlates with MDM2 and TP53 expression in MDS bone marrow cells
MDM2 expression was downregulated in bone marrow cells from MDS patients compared to healthy donors (1.08 [0.18–10.07] vs. 1.59 [0.24–5.52], p = 0.03, Figure 2a). TP53 expression was similar between MDS patients and healthy donors (TP53: 0.95 [0.00–33.41] vs. 1.10 [0.04–7.32], Fig. 2b). TP53 expression was significantly increased in the IPSS-R intermediate/high/very-high risk MDS compared to the IPSS-R very low/low risk groups (1.25 [0.16–33.41] vs. 0.72 [0.00–4.65], p = 0.03; Additional file 1: Figure S1), and no differences were observed for MDM2 expression (p > 0.05). No difference was observed in MDM2 and TP53 expression when MDS patients were stratified by WHO 2008 classification into RA/RARS/RCMD group vs. RAEB-1/RAEB-2 group (p > 0.05, Additional file 2: Figure S2). MDM2 and TP53 expressions were positively correlated with SIVA1 and SIVA2 in bone marrow samples from MDS patients (MDM2/SIVA1: r = 0.39, p = 0.003; MDM2/SIVA2: r = 0.44, p = 0.0007; TP53/SIVA1: r = 0.48, p < 0.002; TP53/SIVA2: r = 0.32, p = 0.02; Figure 2c). In healthy donors, SIVA1 and SIVA2 expression correlated only with MDM2 expression, but not with TP53 expression (Additional file 3: Figure S3). In our cohort of MDS patients, the factors that were significantly associated with EFS and OS were gender, WHO 2008 classification and IPSS-R by univariate analysis. Male gender and RAEB1/2 classification negatively impact on EFS and OS by multivariate analysis (Table 3).
TP53 and MDM2 expression and their correlation with SIVA1 and SIVA2 levels in bone marrow cells from patients with myelodysplastic syndromes (MDS). MDM2 (a) and TP53 (b) mRNA expression in total bone marrow cells from healthy donors and MDS patients. Horizontal lines indicate medians. The numbers of subjects studied and p values are indicated; Mann–Whitney test. c Correlation analysis between SIVA1 or SIVA2 with MDM2 and TP53 expression in total bone marrow cells from MDS patients. The p and r values are indicated; Spearman correlation test
Discussion
Herein, we analyzed the expression of SIVA1 and SIVA2 in normal and MDS bone marrow samples, and their correlation with MDM2 and TP53 expression. Regarding SIVA expression in MDS, our results are in agreement with a previous microarray study that showed a downregulation of SIVA in bone marrow mononuclear cells from MDS patients, when compared to healthy donors [26], and provide further evidence of the participation of SIVA in hematological malignancies.
We also observed a downregulation of MDM2 in MDS patients. Pellagatti and colleagues [27], using microarray analysis, reported that the ATM signaling pathway is deregulated in high-risk MDS, which included downregulation of MDM2. The positive correlation between SIVA transcripts and MDM2 may be related to the fact that both genes are transcription targets of p53 [13, 28], suggesting a defective transcriptional activity of p53 protein. SIVA1 binds to and regulates p53 stability by acting as an adapter protein between p53 and MDM2 [29, 30], and SIVA1 acts as an ubiquitin ligase for ARF and indirectly regulates p53 stability [31]. Given that there is a reduced expression of SIVA1 in bone marrow samples from MDS, herein identified, further studies are necessary to verify whether SIVA1 downregulation may be involved in aberrant p53 signaling pathway reported in MDS cells [32].
Conclusion
In conclusion, we demonstrated that SIVA expression is impaired in MDS. The downregulation of SIVA and its correlation with MDM2 may be due to defective p53 transcriptional machinery in this disease. Future studies are necessary to verify the effects of SIVA in hematopoietic cells and their participation in the malignant phenotype.
Abbreviations
- AML:
-
Acute myeloid leukemia
- BAX:
-
BCL2-associated X protein
- BCL2:
-
B cell leukemia/lymphoma 2
- BCL-XL:
-
BCL2-like 1
- BID:
-
BH3 interacting domain death agonist
- BM:
-
Bone marrow
- EFS:
-
Event free survival
- HPRT1:
-
Hypoxanthine phosphoribosyltransferase 1
- IPSS-R:
-
Revised International Prognostic Scoring System
- JNK:
-
c-Jun N-terminal kinase
- MDM2:
-
MDM2proto-oncogene
- MDS:
-
Myelodysplastic syndromes
- NFκB:
-
Nuclear factor kappa B
- OS:
-
Overall survival
- p53:
-
Protein 53
- q-PCR:
-
Quantitative polymerase chain reaction
- RA:
-
Refractory anemia
- RAEB-1:
-
Refractory anemia with excess blast-1
- RAEB-2:
-
Refractory anemia with excess blast-2
- RARS:
-
Refractory anemia with ringed sideroblasts
- RCMD:
-
Refractory cytopenia with multilineage dysplasia
- SIVA:
-
SIVA1 apoptosis inducing factor
- TP53:
-
Tumor protein 53
- WHO:
-
World Health Organization
- XIAP:
-
X-linked inhibitor of apoptosis.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–37.
Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22.
Saft L, Karimi M, Ghaderi M, Matolcsy A, Mufti GJ, Kulasekararaj A, et al. p53 protein expression independently predicts outcome in patients with lower-risk myelodysplastic syndromes with del(5q). Haematologica. 2014;99:1041–9.
Loghavi S, Al-Ibraheemi A, Zuo Z, Garcia-Manero G, Yabe M, Wang SA, et al. TP53 overexpression is an independent adverse prognostic factor in de novo myelodysplastic syndromes with fibrosis. Br J Haematol. 2015;171:91–9.
Duarte FB, Goncalves RP, Barbosa MC, Rocha Filho FD, de Jesus Dos Santos TE, Dos Santos TN, et al. Tumor suppressor p53 protein expression: prognostic significance in patients with low-risk myelodysplastic syndrome. Rev Bras Hematol Hemoter. 2014;36:196–201.
Bektas O, Uner A, Buyukasik Y, Uz B, Bozkurt S, Eliacik E, et al. Clinical and pathological correlations of marrow PUMA and P53 expressions in myelodysplastic syndromes. APMIS. 2015;123:445–51.
Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–506.
Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19:1405–13.
Padua RA, Guinn BA, Al-Sabah AI, Smith M, Taylor C, Pettersson T, et al. RAS, FMS and p53 mutations and poor clinical outcome in myelodysplasias: a 10-year follow-up. Leukemia. 1998;12:887–92.
Quintas-Cardama A, Hu C, Qutub A, Qiu YH, Zhang X, Post SM, et al. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 2017;31:1296–305.
Prasad KV, Ao Z, Yoon Y, Wu MX, Rizk M, Jacquot S, et al. CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc Natl Acad Sci U S A. 1997;94:6346–51.
Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, Callaghan SM, et al. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 2004;279:28706–14.
Ray RM, Bhattacharya S, Johnson LR. Mdm2 inhibition induces apoptosis in p53 deficient human colon cancer cells by activating p73- and E2F1-mediated expression of PUMA and Siva-1. Apoptosis. 2011;16:35–44.
Chu F, Barkinge J, Hawkins S, Gudi R, Salgia R, Kanteti PV. Expression of Siva-1 protein or its putative amphipathic helical region enhances cisplatin-induced apoptosis in breast cancer cells: effect of elevated levels of BCL-2. Cancer Res. 2005;65:5301–9.
Xue L, Chu F, Cheng Y, Sun X, Borthakur A, Ramarao M, et al. Siva-1 binds to and inhibits BCL-X(L)-mediated protection against UV radiation-induced apoptosis. Proc Natl Acad Sci U S A. 2002;99:6925–30.
Resch U, Schichl YM, Winsauer G, Gudi R, Prasad K, de Martin R. Siva1 is a XIAP-interacting protein that balances NFkappaB and JNK signalling to promote apoptosis. J Cell Sci. 2009;122:2651–61.
Barkinge JL, Gudi R, Sarah H, Chu F, Borthakur A, Prabhakar BS, et al. The p53-induced Siva-1 plays a significant role in cisplatin-mediated apoptosis. J Carcinog. 2009;8:2.
Py B, Slomianny C, Auberger P, Petit PX, Benichou S. Siva-1 and an alternative splice form lacking the death domain, Siva-2, similarly induce apoptosis in T lymphocytes via a caspase-dependent mitochondrial pathway. J Immunol. 2004;172:4008–17.
Machado-Neto JA, Lazarini M, Favaro P, de Melo CP, Scopim-Ribeiro R, Franchi Junior GC, et al. ANKHD1 silencing inhibits Stathmin 1 activity, cell proliferation and migration of leukemia cells. Biochim Biophys Acta. 1853;2015:583–93.
Van Nostrand JL, Brisac A, Mello SS, Jacobs SB, Luong R, Attardi LD. The p53 target Gene SIVA enables non-small cell lung cancer development. Cancer Discov. 2015;5:622–35.
Park IK, Blum W, Baker SD, Caligiuri MA. E3 ubiquitin ligase Cbl-b activates the p53 pathway by targeting Siva1, a negative regulator of ARF, in FLT3 inhibitor-resistant acute myeloid leukemia. Leukemia. 2017;31:502–5.
Reis-Alves SC, Traina F, Harada G, Campos PM, Saad ST, Metze K, et al. Immunophenotyping in myelodysplastic syndromes can add prognostic information to well-established and new clinical scores. PLoS One. 2013;8:e81048.
Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–65.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8.
Bar M, Stirewalt D, Pogosova-Agadjanyan E, Wagner V, Gooley T, Abbasi N, et al. Gene expression patterns in Myelodyplasia underline the role of apoptosis and differentiation in disease initiation and progression. Transl Oncogenomics. 2008;3:137–49.
Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Della Porta MG, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24:756–64.
Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–9.
Du W, Jiang P, Li N, Mei Y, Wang X, Wen L, et al. Suppression of p53 activity by Siva1. Cell Death Differ. 2009;16:1493–504.
Mei Y, Wu M. Multifaceted functions of Siva-1: more than an Indian god of destruction. Protein Cell. 2012;3:117–22.
Wang X, Zha M, Zhao X, Jiang P, Du W, Tam AY, et al. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun. 2013;4:1551.
Zhang L, McGraw KL, Sallman DA, List AF. The role of p53 in myelodysplastic syndromes and acute myeloid leukemia: molecular aspects and clinical implications. Leuk Lymphoma. 2017;58:1777–90.
Acknowledgments
The authors would like to thank Raquel S. Foglio and Andy Cumming for English review, and Tereza Salles for her valuable technical assistance.
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Availability of data and materials
Please contact author for data requests.
Authors’ contributions
JAM-N performed all the experiments, statistical analyses, patient database, manuscript preparation, completion and final approval. PMC, PF, ML and RS-R participated in the interpretation of manuscript data, clinical data collection, manuscript editing, and final approval. IL-M, FFC and STOS participated in revised the diagnoses, patient follow up, manuscript editing and final approval. FT participated in the overall design of the study and experiments, statistical analyses, patient follow up, manuscript preparation, editing, completion and final approval. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the University of Campinas in accordance with the Helsinki Declaration. Written informed consent was obtained from all healthy donors and MDS patients who participated in this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1: Figure S1.
SIVA1, SIVA2, MDM2, TP53 and BCL2 expression in bone marrow cells from myelodysplastic syndromes (MDS) stratified by Revised International Prognostic Scoring System (IPSS-R). SIVA1 (A), SIVA2 (B), MDM2 (C) and TP53 (D) mRNA expression in total bone marrow cells from MDS patients stratified by IPSS-R into very low/low risk group and intermediate/high/very high risk group. Horizontal lines indicate medians. The numbers of subjects studied and p values are indicated; Mann–Whitney test. (PDF 32 kb)
Additional file 2: Figure S2.
SIVA1, SIVA2, MDM2 and TP53 expression in bone marrow cells from myelodysplastic syndromes (MDS) stratified by World Healthy Organization (WHO) 2008 classification. SIVA1 (A), SIVA2 (B), MDM2 (C) and TP53 (D) mRNA expression in total bone marrow cells from MDS patients stratified by WHO classification into refractory anemia (RA)/refractory anemia with ringed sideroblasts (RARS)/refractory cytopenia with multilineage dysplasia (RCMD) group and refractory anemia with excess blast-1 (RAEB-1)/refractory anemia with excess blast-2 (RAEB-2) group. Horizontal lines indicate medians. The numbers of subjects studied are indicated. (PDF 31 kb)
Additional file 3: Figure S3.
TP53 and MDM2 expressions and their correlation with SIVA1 and SIVA2 levels in bone marrow cells from healthy donors. Correlation analysis between SIVA1 or SIVA2 with MDM2 or TP53 expression in total bone marrow cells from healthy donors. The p and r values are indicated; Spearman correlation test. (PDF 25 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Machado-Neto, J.A., de Melo Campos, P., Favaro, P. et al. SIVA, a target of p53, is downregulated in myelodysplastic syndromes. Appl Cancer Res 37, 25 (2017). https://doi.org/10.1186/s41241-017-0033-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41241-017-0033-9