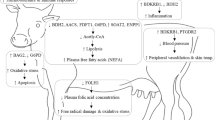
Abstract
Environmental heat stress in dairy cattle leads to poor health, reduced milk production and decreased reproductive efficiency. Multiple genes interact and coordinate the response to overcome the impact of heat stress. The present study identified heat shock regulated genes in the peripheral blood mononuclear cells (PBMC). Genome-wide expression patterns for cellular stress response were compared between two genetically distinct groups of cattle viz., Hariana (B. indicus) and Vrindavani (B. indicus X B. taurus). In addition to major heat shock response genes, oxidative stress and immune response genes were also found to be affected by heat stress. Heat shock proteins such as HSPH1, HSPB8, FKB4, DNAJ4 and SERPINH1 were up-regulated at higher fold change in Vrindavani compared to Hariana cattle. The oxidative stress response genes (HMOX1, BNIP3, RHOB and VEGFA) and immune response genes (FSOB, GADD45B and JUN) were up-regulated in Vrindavani whereas the same were down-regulated in Hariana cattle. The enrichment analysis of dysregulated genes revealed the biological functions and signaling pathways that were affected by heat stress. Overall, these results show distinct cellular responses to heat stress in two different genetic groups of cattle. This also highlight the long-term adaptation of B. indicus (Hariana) to tropical climate as compared to the crossbred (Vrindavani) with mixed genetic makeup (B. indicus X B. taurus).
Similar content being viewed by others
Introduction
Dairy farming significantly contributes to the socio-economic development of communities, countries and regions [1, 2]. Dairy milk has nutritional value. Consequently, there is an increasing demand for milk and milk products in the human diet [3, 4]. Bovine milk is a primary source (up to 83%) of all the total diary milk production throughout the world [5]. Among several challenges faced by the cattle farming, environmental stress is considered as one of the major threats for sustainability of higher milk production [6].
Impending climate change scenario has remained as an unanimously accepted reality [7]. The Intergovernmental Panel on Climate Change (IPCC) has predicted temperature rise of 1.5 °C in the next two decades [8]. The climate change has a complex effect on the livestock production. Temperature stress would be one major environmental factor to influence the health and productivity of dairy cattle [9]. For example, when the outside temperature reaches to 35 °C, quantity of milk is decreased by 33% and at the temperature of 40oC by 50% [10]. Heat stressed cows at higher temperature humidity index (THI = 83.4) have had significantly lesser milk yield (31.2 vs. 38.6 kg/d) than the control cows at thermo-neutral conditions (THI = 75.7) [11]. Similar effects on the milk yield were observed during the season with high temperature and humidity [12, 13]. These deleterious effects on milk production and health of animals stem from the heat stress induced molecular changes that alter normal physiological mechanisms [14, 15].
Gene networks within the cells and tissues co-ordinate metabolism and milk production in dairy cattle [16]. The most recognized pathways of thermal stress responses include the central role of heat shock proteins (HSP) [17]. Heat shock proteins are associated with heat stress response in cattle [18]. Apart from heat shock proteins, recent genome-wide expression studies identified differences in the other transcripts between heat stressed and their respective control cells (bovine mammary epithelial cells and peripheral blood mononuclear cells) [19]. An iTRAQ proteomic study revealed heat stress induced changes in mammary tissues of dairy cows, independent of feed intake by the animals [20]. Relatively heat tolerant and susceptible cattle respond differently to heat stress. The comparative study on relatively heat tolerant and susceptible dairy cattle identified potential regulatory genes responsible for differences in heat stress response mechanisms [21]. Similar approach with limited data found gene expression differences between the zebu and crossbred cattle [22, 23].
The tropical Zebu cattle (B. indicus) are low producers compared to their European counterparts (B. taurus). However, B. indicus are relatively tolerant to environmental heat stress owing to their long-term adaptation in the tropical climate conditions [24]. Cross-breeding programs were initiated between B. indicus and exotic breeds (Holstein Friesian, Jersey and Brown-Swiss) with the aim to enhance the milk production [25,26,27]. In the Indian subcontinent, this has resulted in new breeds of dairy cattle (Jersind, Jerthar, Karan-Swiss, Karan fries, Sunandini, Frieswal, Phule Triveni and Vrindavani) and these synthetic animals with exotic genetic background are capable of producing more milk than the native Indian breeds [28]. However, these animals are relatively susceptible to environmental stress as they are yet to be adapted to tropical climatic conditions [29]. The susceptibility of the crossbred dairy cattle to environmental stress is considered as a major drawback of the cross breeding program. In the recent years, environmental extremes caused by global warming puts them at higher risk of being affected by heat stress. Enhancement of thermo-tolerance (with no adverse effect on milk yield) in crossbred cattle would make them the most desirable population for the tropical dairy.
Any intervention to enhance the thermo-tolerance in crossbred animals would require thorough understanding of molecular mechanism regulating thermo-tolerance. Towards this end, differences in heat stress responses and underlying molecular changes between the zebu and crossbred cattle need to be delineated. In the present study, we compared heat stress response between Hariana breed (B. indicus) and its crossbred counterpart, Vrindavani (Holstein–Friesian/ Brown Swiss /Jersey [50–75%] X Hariana [25–50%]). Our hypothesis included: 1) heat stress dysregulates multiple genes as a part of intricate network of cellular stress response; 2) genetic background of cattle (B. indicus versus B. indicus X B. taurus) contributes to differences in cellular stress responses. Our genome-wide expression analysis revealed activation of distinct genes during heat stress with notable differences between Hariana and Vrindavani cattle. The functional analysis revealed potential molecular mechanisms involved in cellular heat stress responses in these animals.
Materials and methods
Experimental animals and design
Animals from Indian native Zebu (Hariana) and its corresponding crossbred, Vrindavani (Hariana [25–50%] X Holstein–Friesian/ Brown Swiss /Jersey [50–75%]) were included in the present study. Six healthy male animals (12–18 months and N = 3 per breed) housed at ambient conditions with adequate feed and water. The experiments were approved by the Institute Animal Ethics Committee (IAEC) and conducted at Indian Veterinary Research Institute (IVRI), Bareilly, UP, India. The experiments were carried out during winter months (December to February) when environmental temperature ranges between 15–18 °C with 40–50% average humidity.
PBMC isolation and in vitro heat shock treatment
Isolation of PBMC from blood samples of experimental animals and heat shock treatment were done as per the procedures explained in the earlier studies [30]. Briefly, the blood samples were collected from experimental animals in the morning hours (10.00 am) using sodium heparin coated vacutainers. Blood samples were diluted with PBS and layered on Histopaque-1077 (Sigma-Aldrich). Density gradient centrifugation was used for separation of peripheral blood mononuclear cells (PBMC). The isolated PBMCs were suspended in RPMI 1640 medium (Gibco, Thermo Scientific Inc.) supplemented with 10%FBS (Gibco) and antibiotic antimycotic solution (Gibco). PBMC were plated at 1 × 106cells/well in 12-well plates and incubated at 37ºC in a humidified CO2 incubator (New Brunswick, Eppendorf SE). After 24 h of incubation, the control group cultures (CN) were exposed to 37 °C continuously, whereas the heat shock group (HS) cells were subjected to 42 °C for 3 h. After heat stress treatment, both CN and HS treated cells were processed for total RNA extraction.
RNA isolation, cDNA library construction and sequencing
Total RNA were isolated from samples using Trizol (Ambion, Austin, TX, USA) as per the manufacturer’s protocol. RNA concentration, purity and integrity were first measured with NanoDrop ND-1000 UV–vis Spectrophotometer (NanoDrop Technologies) followed by RNA Nano 6000 Assay kit on Bioanalyzer 2100 system (Agilent Technologies). cDNA libraries were generated from total RNA with Illumina TruSeq RNA Sample Preparation v2 Kit (Illumina, San Diego, CA, USA) following manufacturer's instructions. Quality of sequencing libraries was assessed with Bioanalyzer 2100 system. Sequencing was performed for total 12 sequencing libraries using Illumina HiSeq 2500 and 100 bp paired sequencing reads were produced.
Quality control and alignment to reference genome
Quality of the raw reads was assessed using FastQC. Adapters and low quality bases were trimmed with fastX clipper. The quality trimmed reads were separately aligned to the B. taurus genome (ARS-UCD1.2, GCA_002263795.2) using DRAGEN RNA splice aware aligner. Using annotation files (GTF), gene level and transcript level read count files were generated for all the samples for further analysis.
Identification of differentially expressed genes (DEG)
Differentially expressed genes (DEG) were identified using the DESeq2 protocol. Transcripts per million (TPM) counts were subjected to shrinkage estimators for dispersions followed by fold change analysis with generalized linear model (GLM). Benjamini–Hochberg method was applied for calculation of false discovery rate (FDR) (Padj) values. The criteria of log2 fold change ≥ 1 with padj < 0.05 were used for selecting the DEG.
Functional enrichment analysis and gene annotation
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used for functional enrichment analysis. The analysis were performed based on DEGs identified between control (CN) and heat stress (HS) conditions for each breed of cattle. GO terms significantly associated with the gene lists were determined based on their FDR. Functional evidence of the relationship between the significant GO terms (FDR < 0.05) and the DEGs were identified. The Protein Analysis Through Evolutionary Relationships (PANTHER) [31] and Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 were used to perform functional annotation for the genes.
Quantitative real-time PCR (qRT-PCR) validation
The total RNA was isolated from PBMC samples using TRIzol reagent (15,596,026, Thermo Scientific Inc.) following manufacturer's protocol. The purity was checked by nanodrop spectrophotometer (NanoDrop 3300, Thermo Scientific Inc). The total RNA was used for synthesis of cDNA using First strand cDNA synthesis kit (K1622 Revert Aid, Thermo Scientific Inc.). The gene expression changes were analyzed by using Brilliant III SYBR green kit (Agilent Technologies Inc.). Beta-Actin (ACTB) was used as internal control gene. Initial fold changes were calculated using 2^(–ΔΔCT) method later transformed into log2 fold change values. The list of primers designed and used in the gene expression studies is included as supplementary table (Table S1).
Results
Quality filtered RNA sequencing reads aligned to the bovine reference genome
A little more than 2.7 billion, 100-base paired-end total reads were generated in the present study. On an average there were 220 million reads per sample. More than 98% reads were mapped to the B. taurus reference genome (ARS-UCD1.2, GCA_002263795.2). The samples had an average of 18,493 genes detected that accounted for about 84.5% transcripts of all the 21,880 annotated cow genes (ARS-UCD1.2). The count data for each samples were filtered by removing the genes that had TPM smaller than 1 to eliminate genes with low counts. This resulted in final count table of 11526 and 11,174 genes respectively for Hariana (control and HS treated) and Vrindavani (control and HS treated) samples for differential expression of genes analysis. A summary of sequencing data, quality analysis and alignment results are included in Table 1. Exploratory analysis of the sample data revealed hierarchical clustering of control (CN) and heat stress (HS) samples in both Hariana and Vrindavani (Supplementary Fig. S1A and B). The principal component analysis (PCA) results show that the samples are grouped based on the heat stress treatment in both breeds indicating the effect of heat stress treatment on the overall gene expression patterns (Supplementary Fig. S1C and D).
Heat shock alters the gene expression patterns in bovine PBMC
Differential expression analysis between the control (CN) and heat stress (HS) treatment group (FDR < 0.05 and log2 fold change > ± 1.0) revealed multiple DEG (differentially expressed genes) in both Hariana and its crossbred, Vrindavani. These results are represented through volcano plot (Fig. 1A and B). In Hariana breed, there were 368 DEG, of which 130 were up-regulated and 238 were down-regulated. Similarly, in Vrindavani breed, there were 337 DEG, of which 139 were up-regulated and 198 were down-regulated. The major DEG depicted in the heat map showing their fold change of expression based on the normalized counts of all individual samples (n = 3) of each control and heat stress group (Fig. 2A and B). Based on the lowest FDR and highest log2 fold change the top 10 up-regulated and top 10 down-regulated genes are presented in the Table 2.
Representation of heat shock induced differential expression of genes in Hariana (A) and Vrindavani (B). The up-regulated genes were represented as red dots whereas the down-regulated genes were represented as green dots. The criteria of significance considered log2 fold change values higher than 2 with FDR value smaller than 0.05
The pattern of heat shock induced gene expression in Hariana (A) and Vrindavani (B). The differentially expressed genes were represented based on the normalised gene counts across all the three (N = 3) replicates for each breed of cattle. The hierarchical clustering of the replicate samples clusters together based on the heat stress treatment
Zebu and crossbred cattle show differences in heat induced gene expression
After identifying the heat stress induced gene expression changes separately in each breed of cattle, we set out to find the differences between these breeds. Out of 368 and 337 DEG identified in response to heat stress treatment in Hariana and Vrindavani respectively, 94 genes were common between both the breeds (Fig. 3). Among these 52 common DEG were up-regulated, while 40 common DEG were down-regulated. However, there were two genes (PPP1R15A and HMOX1) that were up-regulated in Vrindavani but down-regulated in Hariana. Apart from these common DEG, 276 and 245 distinct DEGs were unique to for Hariana and Vrindavani cattle, respectively.
Differentially expressed genes are related to significant cellular functions
To determine biological functions of the heat shock induced genes at molecular level, the DEG were subjected to the gene set enrichment analysis with GO and KEGG databases. The enriched GO terms were categorized as activated or suppressed. In case of Hariana breed heat stress treatment activated mainly chaperone mediated protein folding and refolding. The suppressed biological processes included cell motility, localization of cells and cellular response to stimulus (Fig. 4A). In Vrindavani, the heat stress treatment activated cellular response to stress, positive regulation of metabolic process and responses to inorganic substance or chemicals. The suppressed biological processes include possible defense response to Gram-positive bacterium, cellular amino acid catabolic process, regulation of GTPase activity and microtubule binding (Fig. 4B). The majority of the genes involved in activation of protein folding, refolding and response to temperature stimulus had higher expression whereas the majority of the genes involved in response to hypoxia had lower expression in both Hariana and Vrindavani cattle (Fig. 5A and B).
Functional annotation of DEG under heat stress. Gene enrichment analysis for Gene ontology (GO) terms and top 5 activated and top 5 suppressed terms were represented for Hariana (A) and Vrindavani (B). The size of the dot indicates number of genes enriched and color indicates the statistical significance (FDR value)
Further, the differentially expressed genes (DEG) were used to know the enrichment of different cellular signaling pathways. In Hariana cattle, the 5 major pathways enriched by upregulated DEG included cellular response to heat stress, regulation of HSF-1 mediated heat shock response, mRNA processing, protein processing in endoplasmic reticulum and estrogen signaling pathway (Fig. 6A). The activated pathways in Vrindavani cattle are similar to those of Hariana cattle, except that the lipid and atherosclerosis pathway was enriched instead of the mRNA processing pathway (Fig. 6B). The potentially suppressed pathways in Hariana included TNF signaling pathway, HIF-1 signaling pathway, IL-17 signaling pathway, cytokine-cytokine receptor interaction and fluid shear stress and atherosclerosis (Fig. 6C). However, differences were observed in Vrindavani in terms of potentially suppressed pathways such as hematopoietic cell lineage, graft-versus-host disease, antigen processing presentation, MHC class II antigen presentation and Th1 and Th2 cell differentiations (Fig. 6D).
Major categories of genes are affected by heat stress in zebu and crossbred cattle
To narrow down the differences between breeds, DEG obtained after analysis were categorized based on their biological functions. Three major categories such as stress response genes, oxidative response genes and immune response genes were compared for their expression patterns between the breeds (Fig. 7). The heat stress response genes in the two cattle breeds displayed different patterns of gene expression, but these differences were merely in the degree of gene expression, as all genes in this category were upregulated in both breeds. However, crossbred cattle (Vrindavani) showed higher degree of up-regulation for most of the genes compared to the native zebu cattle (Hariana) (Fig. 7A). In case of the oxidative response category, the genes such as BNIP3, HMOX1, JUND, RELA and RHOB were found up-regulated in the Vrindavani cattle, whereas the same were down-regulated in the Hariana cattle (Fig. 7B and D). Similarly, in the immune response category, the major genes such as FSOB, GADD45B, JUN, VEGFA and PPP1R15A were up-regulated in Vrindavani, whereas the same were down-regulated in the Hariana cattle (Fig. 7C and E). These results indicated that Vrindavani and Hariana cattle show differences in the expression patterns of certain immune response and oxidative response genes when challenged with the heat stress.
Comparison of expression patterns of different categories of genes between Hariana and Vrindavani. The heat map shows expression differences of heat stress response genes (A), oxidative stress response genes (B) and immune response genes (C). The bar diagram shows significantly different expression patterns of genes involved in oxidative stress response (D) and immune response (E)
Protein–Protein interaction networks indicated heat shock response
The protein–protein interaction networks were constructed by the involvement of differentially expressed proteins. The BioGRID protein interaction network repository was used for extracting the interactions. Further, differentially expressed highly connected genes (DEHC) (fold change > ± 2 and padj < 0.05 with degree of connections > 20) were used to determine potential and significant protein–protein interaction networks (Fig. 8A and B). There were 23 and 49 DEHC genes in Hariana and Vrindavani cattle, respectively. Out of DEHC genes, 17 were found common between Hariana and Vrindavani cattle. Most of these proteins represented major hub proteins such as HSP90AA1, HSPB1, HSPD1, CALCOCO2, BAG2, HSP90AB1, HSPA4, MAPK3, ACOT7 and HSPE1.These genes were related to upregulated genes of heat shock, oxidative stress and immune related genes. Among these genes, only MAPK3 was found down-regulated and the remaining genes were up-regulated. The change in expression of major hub proteins indicates their potential importance in coordinating the cellular heat stress response.
Protein–protein interaction (PPI) network of hub genes for Hariana (A) and Vrindavani (B). The size of edges indicate the degree of connections of the respective genes and width of node lines indicate the strength of interactions. Up-regulated genes are marked red and down-regulated genes are marked green
Validation of RNA sequencing results using qRT-PCR
Five major up-regulated genes (HSPB1, FKBP4, HSP90AA1, ACOT7, HSPA1A) and four down-regulated genes (RBM3, SDS, BNIP3 and IL6) as observed in the RNA sequencing results were analyzed with qRT-PCR. The expression of these genes obtained through qRT-PCR is presented in comparison with the results of RNA sequencing (Fig. 9). The direction and the level of expression (Log2fold change) of these genes indicated that qRT-PCR and RNA sequencing results are similar and comparable.
Validation of major differentially expressed genes using qRT-PCR. The up-regulated genes (HSPB1, FKB4, HSP90AA1, ACOT7, HSPA1A) and down-regulated genes (RBM3, SDS, BNIP3, IL6) as identified in RNA sequencing data were evaluated using qRT-PCR. The results of qRT-PCR were compared with the expression levels (log2 fold change) obtained in the RNA sequencing
Discussion
Heat stress adversely impacts health, production and reproduction parameters in dairy cattle [32,33,34]. Curtailing the effects of heat stress and its consequent losses has been an important objective of farmers and animal researchers. One of the strategies is to understand the heat stress responses and utilize the better adaptability of animals to reduce the impact of heat stress [35, 36]. The present study delineates the molecular mechanisms of heat stress response in cattle of different genetic background. We compared genome-wide expression patterns between relatively thermo-tolerant breed of B. indicus (Hariana) and its crossbred, B. indicus X B. taurus (Vrindavani).
Different types of heat shock proteins (HSP90, HSP70) and other related candidate genes have been individually characterized in zebu and crossbred cattle [37, 38]. However, stress response is a complex biological process regulated by multiple genes [39]. Transcriptomics using high-throughput sequencing and other techniques allow capture of genome-wide expression patterns [40,41,42]. Using this approach, the present study identifies the involvement of multiple genes that act through interacting networks during cellular stress response. There were 368 and 337 DEG in the heat stress exposed Hariana and Vrindavani cattle respectively. The up-regulated genes mainly belong to the molecular chaperones (HSPB1, HSPA1A, HSP90AA1, DNAJB1), co-chaperone gene (FKBP4) and solute carrier family (Sodium/Myo-Inositol Cotransporter) protein (SLC5A3). The major down-regulated genes were immune response genes such as interluekin-6 (IL6), interleukin 1 receptor antagonist (IL1RN), Nitric oxide synthase 2 (NOS2) and bovine major histocompatibility complex (BoLA) genes that play important role in antigen presentation. Earlier, similar transcriptomics approach identified DEG in response to heat stress in cattle [43]. Heat stress induced up-regulation of heat shock genes (DNAJB1, HSPA1A, HSP90AA1, HSPB8, DNAJB4) was similarly found in PBMC of Holstein dairy cattle [39]. Apart from up-regulated heat shock genes, interestingly, we found expression of immune response related genes down-regulated. These findings were also corroborated by earlier studies that showed down-regulated expression of BoLA (BoLA-DRB3) in bovine mammary cells [44]. The down-regulation of major immune response genes indicates that animals become more susceptible to infection during the heat stress period.
Many of the genes identified in the present study were found related the heat tolerance traits of cattle in the previous studies [45,46,47]. Polymorphisms in different heat shock proteins (HSP90, HSPA6, HSPB7) and heat shock factor 1 (HSF1) were associated with the heat tolerance traits in cattle [45, 48, 49]). In addition, mutations in other related genes such as prolactin (PRL) and prolactin receptor (PRLR) were associated with thermoregulation and hair morphology in cattle [50].
The present study recorded GO enrichment for DEGs as activation of cellular response to stress and chaperone mediated protein folding. In contrast to Hariana (zebu) cattle, heat stress in Vrindavani (crossbred) cattle resulted in a suppressed response to bacteria and defense response to Gram-positive bacteria, suggesting that the Vrindavani cattle is more susceptible to bacterial infection during heat stress period. Experiments in Holstein calves revealed that heat stress induced genes were mainly categorized for protein folding functions [51]. PBMCs of Holstein dairy cattle subjected to heat stress showed enrichment of similar biological process such as protein folding, response to endoplasmic reticulum stress, negative regulation of cell proliferation, inclusion body assembly and apoptotic process [39]. We found major enriched signaling pathways by up-regulated DEGs such as HSF1mediated heat shock response, estrogen signaling pathway, MAPK singling pathway. Whereas the down-regulated genes enriched mainly immune responses such as TNF signaling, IL-17 signaling, Chemokine signaling and MHC class II antigen presentation. These results indicate that heat stress activates protective mechanisms to prevent damage or misfolding of cellular proteins. However, major immune response pathways were dampened indicating the vulnerability of animals to possible challenge of infectious agents. In earlier studies, heat stress to Jersey and Holstein steers was found to alter the proportion of immune cells and expression of cytokines (IL-10, IL-17A and IL6) indicating enhanced susceptibility of heat-stressed steers to diseases [52]. The effect of heat stress on immune cells and gene expression pattern was also found breed specific for Holstein and Jersey cattle [52, 53]. Conversely, different immune categories of Holstein cows (ranked as high, average and low immune responders) were found to exhibit differences in the heat stress response [54]. One of the highlight of our study is that there are differences in the expression of immune system related genes between Hariana and Vrindavani in responses to heat stress. The immune pathways of Vrindavani cattle were found to be more dysregulated in comparison to Hariana cattle when subjected to heat stress. These results may suggest that Vrindavani cattle are more vulnerable to infections during heat stress compared to Hariana cattle.
Our analysis focused on comparison of DEG and it revealed up-regulation of certain oxidative stress related genes (BNIP3, HMOX1, VEGFA, RHOB) in Vrindavani cattle. However, those genes were down-regulated in Hariana cattle. Bcl2 interacting protein 3 (BINP3) was found induced in hypoxic stress [55]. BNIP3 was found to promote apoptosis in response to energy stress by inhibiting mTORC1 function [56]. Heme oxygenase-1 (HMOX1) was reported to be upregulated during different types of stressors mainly during cellular oxidative stress response [57, 58]). Role of HMOX1 was highlighted in protecting the cells [59, 60]. The distribution of HMOX1, HSP70 and glutathione together depict cellular stress tolerance [61]. Vascular endothelial growth factor-A (VEGFA) is a major hypoxia response protein induced by hypoxia inducible factor alpha (HIF1α). However, it is also upregulated without the help of HIF1α during unfolded protein response (UPR) [62]. Heat stress treatment in rats significantly increased expression of VEGF along with HSP70 [63]. VEGF expression was up-regulated in heat stressed Jersey cattle. VEGF signaling pathway plays an important role in immunosuppression and oxidative stress response in the affected animals [43]. Small GTP binding protein RhoB (RHOB) was found rapidly up-regulated by genotoxic and heat stress [64]. Upregulation of RhoB significantly inhibited heat stress induced apoptosis and played cytoprotective role [65].
Apart from differences in the oxidative stress response gene, certain immune response related (FSOB, JUN, GADD45B) genes were found up-regulated in the Vrindavani, whereas those genes were found down-regulated in the Hariana cattle. FSOB and JUN proteins are produced in response to any cellular stimuli and contributes to the formation of transcription factor activator protein-1 (AP-1). Activation of AP-1 in turn regulates genes involved in proliferation, migration and immune response [66,67,68]. Studies on rats indicated that FOSB proteins potentially have a role in long-lasting adaptation changes in the cellular stress responses [69]. In the present study, we found significant up-regulation of FSOB in Vrindavani indicating its possible role in differential heat stress response compared to the Hariana cattle. Also, Vrindavani cattle are likely to actively undergoing adaption process to tropical climate. Under environmental stressors, there was an up-regulation of GADD45B (growth arrest DNA damage inducible beta protein) gene expression and this causes arrests of cells growth or proliferation. GADD45B activates p38/JNK pathway via MTK1/MEKK4 kinase [70]. Earlier it was shown that GADD45B levels were increased in stress susceptible but not resilient mice [71]. This finding corroborates with our results as relatively heat stress susceptible crossbred Vrindavani cattle showed higher expression of GADD45B compared to Hariana cattle. Further, interestingly its important paralogue GADD45A showed significantly differential methylation in Angus cattle during heat stress challenge [72]. Hence based on the long-term adaptation of animals to certain environmental conditions, epigenetic mechanisms may play a role in producing differences in stress responses.
Conclusions
The present study revealed genome-wide expression patterns in the PBMC of cattle subjected to heat stress. Apart from heat stress response, we found mainly altered oxidative stress and immune response mechanisms as the genes related to these processes were found dysregulated. There were differences in the number and types of dysregulated genes categorized as common and specific for each of these breeds. Even though with same expression patterns, heat stress response genes (HSPH1, HSPB8, FKB4, DNAJ4 and SERPINH1) were up-regulated at higher fold change in Vrindavani compared to the Hariana cattle. Further, certain oxidative stress response genes (HMOX1, BNIP3, RHOB and VEGFA) and immune response genes (FSOB, GADD45B and JUN) were up-regulated in Vrindavani whereas the same were down- regulated in Hariana cattle. These differences in cellular stress responses highlight the long-term adaptation of B. indicus (Hariana) to tropical climate compared to the mixed breed (Vrindavani) with contribution from B. taurus.
Availability of data and materials
Data and materials presented in the manuscript will be made available upon request to the corresponding author.
References
Chaudhary B, Upadhyaya M. Socio-Economic Impacts of Dairy Cooperative. Econ J Dev Issues. 2015;15(1–2):15–23. https://doi.org/10.3126/ejdi.v15i1-2.11859.
Arvidsson Segerkvist K, Hansson H, Sonesson U, Gunnarsson S. Research on Environmental, Economic, and Social Sustainability in Dairy Farming: A Systematic Mapping of Current Literature. Sustainability. 2020;12(14):5502. https://doi.org/10.3390/su12145502.
Visioli F, Strata A. Milk, dairy products, and their functional effects in humans: a narrative review of recent evidence. Adv Nutr. 2014;5(2):131–43. https://doi.org/10.3945/an.113.005025.
Haug A, Høstmark AT, Harstad OM. Bovine milk in human nutrition–a review. Lipids Health Dis. 2007;6:25. https://doi.org/10.1186/1476-511X-6-25.
OECD/FAO (2020), OECD-FAO Agricultural Outlook 2020-2029, OECD Publishing, Paris/FAO, Rome. https://doi.org/10.1787/1112c23b-en.
Gauly M, Ammer S. Review: Challenges for dairy cow production systems arising from climate changes. Animal. 2020;14(S1):s196–203. https://doi.org/10.1017/S1751731119003239.
National Academy of Sciences. Climate Change: Evidence and CausesWashington. DC: The National Academies Press; 2020. https://doi.org/10.17226/25733.
Ara Begum R, Lempert E, Ali TA, Benjaminsen T, Bernauer W, Cramer X, et al. Point of Departure and Key Concepts. In: Climate Change 2022: Impacts, Adaptation, and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change [Pörtner HO, Roberts DC, Tignor ES, Poloczanska K, Mintenbeck A, Alegría M, Craig S, Langsdorf S, Löschke V, Möller A, Okem B, Rama (eds.)]. Cambridge University Press, Cambridge, UK and New York, NY, USA, 121-196. https://doi.org/10.1017/9781009325844.003.
Thornton P, Nelson G, Mayberry D, Herrero M (2022). Impacts of heat stress on global cattle production during the 21st century: a modelling study. Lancet Planet Health:e192-e201 https://doi.org/10.1016/S2542-5196(22)00002-X
West JW (2003) Effects of heat-stress on production in dairy cattle. J Dairy Sci2131–2144:https://doi.org/10.3168/jds.S0022-0302(03)73803-X
Safa S, Kargar S, Moghaddam GA, Ciliberti MG, Caroprese M. Heat stress abatement during the postpartum period: effects on whole lactation milk yield, indicators of metabolic status, inflammatory cytokines, and biomarkers of the oxidative stress. J Anim Sci. 2019;97(1):122–32. https://doi.org/10.1093/jas/sky408.
Niyonzima YB, Strandberg E, Hirwa CD, Manzi M, Ntawubizi M, Rydhmer L. The effect of high temperature and humidity on milk yield in Ankole and crossbred cows. Trop Anim Health Prod. 2022;54(2):85. https://doi.org/10.1007/s11250-022-03092-z.
Chen X, Dong JN, Rong JY, Xiao J, Zhao W, Aschalew ND, Zhang XF, Wang T, Qin GX, Sun Z, Zhen YG. Impact of heat stress on milk yield, antioxidative levels, and serum metabolites in primiparous and multiparous Holstein cows. Trop Anim Health Prod. 2022;54(3):159. https://doi.org/10.1007/s11250-022-03159-x.
Li H, Zhang Y, Li R, Wu Y, Zhang D, Xu H, Zhang Y, Qi Z (2021) Effect of seasonal thermal stress on oxidative status, immune response and stress hormones of lactating dairy cows. Anim Nutr:216–223.: https://doi.org/10.1016/j.aninu.2020.07.006
Mishra SR. Behavioural, physiological, neuro-endocrine and molecular responses of cattle against heat stress: an updated review. Trop Anim Health Prod. 2021;53(3):400. https://doi.org/10.1007/s11250-021-02790-4.
Collier RJ, Collier JL, Rhoads RP, Baumgard LH. Invited review: genes involved in the bovine heat stress response. J Dairy Sci. 2008;91(2):445–54. https://doi.org/10.3168/jds.2007-0540.
Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40(2):253–66. https://doi.org/10.1016/j.molcel.2010.10.006.
Pires BV, Stafuzza NB, Lima SGPNP, Negrão JA, Paz CCP. Differential expression of heat shock protein genes associated with heat stress in Nelore and Caracu beef cattle. Livest Sci. 2019;230:1871–1413. https://doi.org/10.1016/j.livsci.2019.103839.
Garner JB, Chamberlain AJ, Vander Jagt C, Nguyen TTT, Mason BA, Marett LC, Leury BJ, Wales WJ, Hayes BJ. Gene expression of the heat stress response in bovine peripheral white blood cells and milk somatic cells in vivo. Sci Rep. 2020;10(1):19181. https://doi.org/10.1038/s41598-020-75438-2.
Ma L, Yang Y, Zhao X, Wang F, Gao S, Bu D (2019) Heat stress induces proteomic changes in the liver and mammary tissue of dairy cows independent of feed intake: An iTRAQ study. PLoS One14(1):e0209182: https://doi.org/10.1371/journal.pone.0209182.
Liu S, Yue T, Ahmad MJ, Hu X, Zhang X, Deng T, Hu Y, He C, Zhou Y, Yang L (2020) Transcriptome Analysis Reveals Potential Regulatory Genes Related to Heat Tolerance in Holstein Dairy Cattle. Genes (Basel)11(1):68:https://doi.org/10.3390/genes11010068.
Singh AK, Upadhyay RC, Chandra G, Kumar S, Malakar D, Singh SV, Singh MK. Genome-wide expression analysis of the heat stress response in dermal fibroblasts of Tharparkar (zebu) and Karan-Fries (zebu × taurine) cattle. Cell Stress Chaperones. 2020;25(2):327–44. https://doi.org/10.1007/s12192-020-01076-2.
Khan RIN, Sahu AR, Malla WA, Praharaj MR, Hosamani N, Kumar S, Gupta S, Sharma S, Saxena A, Varshney A, Singh P, Verma V, Kumar P, Singh G, Pandey A, Saxena S, Gandham RK, Tiwari AK (2022) Systems biology under heat stress in Indian cattle. Gene 30, 805:145908.: https://doi.org/10.1016/j.gene.2021.145908
Cooke RF, Cardoso RC, Cerri RLA, Lamb GC, Pohler KG, Riley DG, Vasconcelos JLM (2020) Cattle adapted to tropical and subtropical environments: genetic and reproductive considerations. J Anim Sci98(2) https://doi.org/10.1093/jas/skaa015
Rajapurohit AR. Cross-Breeding of Indian Cattle: An Evaluation. Economic and Political Weekly. 1979;14:9–24. http://www.jstor.org/stable/4367454.
Galukande E, Mulindwa H, Wurzinger M, Roschinsky R, Mwai A, Sölkner J. Cross-breeding cattle for milk production in the tropics: Achievements, challenges and opportunities. Animal Genetic Resources. 2013;52:111–25. https://doi.org/10.1017/S2078633612000471.
Osei-Amponsah R, Asem KE and Yeboah Obese F (2020) Cattle crossbreeding for sustainable milk production in the tropics. International Journal of Livestock Production :108–113. https://doi.org/10.5897/IJLP2020.0717
Singh CV. Cross-breeding in Cattle for Milk Production: Achievements, Challenges and Opportunities in India-A Review. Adv Dairy Res. 2016;4:3. https://doi.org/10.4172/2329-888X.1000158.
Berian S, Gupta SK, Sharma S, Ganai I, Dua S, Sharma N. Effect of Heat Stress on Physiological and Hemato-biochemical Profile of Cross Bred Dairy Cattle. Journal of Animal Research. 2019;9(1):95–101. https://doi.org/10.30954/2277-940X.01.2019.13.
Masroor S, Aalam MT, Khan O, Tanuj GN, Gandham RK, Dhara SK, Gupta PK, Mishra BP, Dutt T, Singh G, Sajjanar BK. Effect of acute heat shock on stress gene expression and DNA methylation in zebu (Bos indicus) and crossbred (Bos indicus × Bos taurus) dairy cattle. Int J Biometeorol. 2022;66(9):1797–809. https://doi.org/10.1007/s00484-022-02320-3.
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 8, 47(D1):D419-D426.: https://doi.org/10.1093/nar/gky1038
Dahl GE, Tao S, Laporta J. Heat Stress Impacts Immune Status in Cows Across theLife Cycle. Front Vet Sci. 2020;7:116. https://doi.org/10.3389/fvets.2020.00116.
Monteiro APA, Tao S, Thompson IMT, Dahl GE. In utero heat stress decreases calf survival and performance through the first lactation. J Dairy Sci. 2016;99(10):8443–50. https://doi.org/10.3168/jds.2016-11072.
Tao S, Orellana Rivas RM, Marins TN, Chen YC, Gao J, Bernard JK. Impact of heat stress on lactational performance of dairy cows. Theriogenology. 2020;150:437–44. https://doi.org/10.1016/j.theriogenology.2020.02.048.
König S, May K (2019) Invited review: Phenotyping strategies and quantitative-geneticbackground of resistance, tolerance and resilience associated traits in dairycattle. Animal13(5):897–908.: https://doi.org/10.1017/S1751731118003208. Epub 2018Dec 7. Review. PubMed [citation] PMID: 30523776
Pryce JE, Haile-Mariam M. Symposium review: Genomic selection for reducingenvironmental impact and adapting to climate change. J Dairy Sci. 2020;103(6):5366–75. https://doi.org/10.3168/jds.2019-17732.
Deb R, Sajjanar B, Singh U, Kumar S, Singh R, Sengar G, Sharma A. Effect of heat stress on the expression profile of Hsp90 among Sahiwal (Bos indicus) and Frieswal (Bos indicus × Bos taurus) breed of cattle: a comparative study. Gene. 2014;536(2):435–40. https://doi.org/10.1016/j.gene.2013.11.086.
Bhanuprakash V, Singh U, Sengar G, Sajjanar B, Bhusan B, Raja TV, Alex R, Kumar S, Singh R, Kumar A, Alyethodi RR, Kumar S, Deb R. Differential effect of thermal stress on HSP70 expression, nitric oxide production and cell proliferation among native and crossbred dairy cattle. J Therm Biol. 2016;59:18–25. https://doi.org/10.1016/j.jtherbio.2016.04.012.
Fang H, Kang L, Abbas Z, Hu L, Chen Y, Tan X, Wang Y, Xu Q (2021) Identification of key genes and pathways associated with thermal stress in peripheral blood mononuclear cells of Holstein dairy cattle. Front Genet 10(12):662080:https://doi.org/10.3389/fgene.2021.662080
O’Brien MA, Costin BN, Miles MF. Using genome-wide expression profiling to define gene networks relevant to the study of complex traits: from RNA integrity to network topology. Int Rev Neurobiol. 2012;104:91–133. https://doi.org/10.1016/B978-0-12-398323-7.00005-7.
Kadam US, Lossie AC, Schulz B, Irudayaraj J (2013) Gene Expression Analysis Using Conventional and Imaging Methods. In: Erdmann V, Barciszewski J (eds) DNA and RNA Nanobiotechnologies in Medicine: Diagnosis and Treatment of Diseases. RNA Technologies. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-36853-0_6
Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T (2017) Transcriptomics technologies. PLoS Comput Biol 13(5):e1005457.: https://doi.org/10.1371/journal.pcbi.1005457
Wang J, Xiang Y, Jiang S, Li H, Caviezel F, Katawatin S, Duangjinda M. Involvement of the VEGF signaling pathway in immunosuppression and hypoxia stress: analysis of mRNA expression in lymphocytes mediating panting in Jersey cattle under heat stress. BMC Vet Res. 2021;17(1):209. https://doi.org/10.1186/s12917-021-02912-y.
Yue S, Wang Z, Wang L, Peng Q, Xue B. Transcriptome Functional Analysis of Mammary Gland of Cows in Heat Stress and Thermoneutral Condition. Animals (Basel). 2020;10(6):1015. https://doi.org/10.3390/ani10061015.
Sajjanar B, Deb R, Singh U, et al. Identification of SNP in HSP90AB1 and its association with the relative thermotolerance and milk production traits in Indian dairy cattle. Anim Biotechnol. 2015;26:45–50. https://doi.org/10.1080/10495398.2014.882846.
Zeng L, Qu K, Zhang J, Huang B, Lei C. Genes related to heat tolerance in cattle-a review. Anim Biotechnol. 2022;15:1–9. https://doi.org/10.1080/10495398.2022.2047995.
Hariyono DNH, Prihandini PW. Association of selected gene polymorphisms with thermotolerance traits in cattle - A review. Anim Biosci. 2022;35(11):1635–48. https://doi.org/10.5713/ab.22.0055.
Zeng L, Cao Y, Wu Z, et al. A missense mutation of the HSPB7 gene associated with heat tolerance in Chinese indicine cattle. Animals. 2019;9:554. https://doi.org/10.3390/ani9080554.
Baena MM, Tizioto PC, Meirelles SLC, Regitano LC, de A. HSF1 and HSPA6 as functional candidate genes associated with heat tolerance in Angus cattle. Rev Bras Zootec. 2018;47:e20160390.: https://doi.org/10.1590/rbz472016039
Littlejohn MD, Henty KM, Tiplady K, Johnson T, Harland C, Lopdell T, Sherlock RG, Li W, Lukefahr SD, Shanks BC, Garrick DJ, Snell RG, Spelman RJ, Davis SR. Functionally reciprocal mutations of the prolactin signalling pathway define hairy and slick cattle. Nat Commun. 2014;18(5):5861. https://doi.org/10.1038/ncomms6861.
Srikanth K, Kwon A, Lee E, Chung H. Characterization of genes and pathways that respond to heat stress in Holstein calves through transcriptome analysis. Cell Stress Chaperones. 2017;22(1):29–42. https://doi.org/10.1007/s12192-016-0739-8.
Park DS, Gu BH, Park YJ, Joo SS, Lee SS, Kim SH, Kim ET, Kim DH, Lee SS, Lee SJ, Kim BW, Kim M. Dynamic changes in blood immune cell composition and function in Holstein and Jersey steers in response to heat stress. Cell Stress Chaperones. 2021;26(4):705–20. https://doi.org/10.1007/s12192-021-01216-2.
Joo SS, Lee SJ, Park DS, Kim DH, Gu BH, Park YJ, Rim CY, Kim M, Kim ET (2021) Changes in Blood Metabolites and Immune Cells in Holstein and Jersey Dairy Cows by Heat Stress. Animals 11(4) pii: 974.: https://doi.org/10.3390/ani11040974
Cartwright SL, McKechnie M, Schmied J, Livernois AM, Mallard BA. Effect of in-vitro heat stress challenge on the function of blood mononuclear cells from dairy cattle ranked as high, average and low immune responders. BMC Vet Res. 2021;17(1):233. https://doi.org/10.1186/s12917-021-02940-8.
Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27(17):6229–42.
Lin A, Yao J, Zhuang L, Wang D, Han J, Lam EW, Gan B. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene. 2013;33(24):3183–94. https://doi.org/10.1038/onc.2013.273.
Poss KD, Tonegawa S. Reduced stress defense in heme oxygenase 1-deficient cells. Proc Natl Acad Sci USA. 1997;94(20):10925–30.
Ryter SW, Choi AM. Heme oxygenase-1: redox regulation of a stress protein in lung and cell culture models. Antioxid Redox Signal. 2005;7(1–2):80–91.
Gozzelino R, Jeney V, Soares MP. Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol. 2010;50:323–54. https://doi.org/10.1146/annurev.pharmtox.010909.105600.
Yachie A (2021) Heme Oxygenase-1 Deficiency and Oxidative Stress: A Review of 9 Independent Human Cases and Animal Models. Int J Mol Sci 22(4) pii:1514: https://doi.org/10.3390/ijms22041514
Calabrese V, Scapagnini G, Ravagna A, Fariello RG, Giuffrida Stella AM, Abraham NG. Regional distribution of heme oxygenase, HSP70, and glutathione in brain: relevance for endogenous oxidant/antioxidant balance and stress tolerance. J Neurosci Res. 2002;68(1):65–75. https://doi.org/10.1002/jnr.10177.
Ghosh R, Lipson KL, Sargent KE, Mercurio AM, Hunt JS, Ron D, Urano F (2010) Transcriptional regulation of VEGF-A by the unfolded protein response pathway. PLoS One5(3):e9575.: https://doi.org/10.1371/journal.pone.0009575
Gong B, Asimakis GK, Chen Z, Albrecht TB, Boor PJ, Pappas TC, Bell B, Motamedi M. Whole-body hyperthermia induces up-regulation of vascular endothelial growth factor accompanied by neovascularization in cardiac tissue. Life Sci. 2006;79(19):1781–8.
Canguilhem B, Pradines A, Baudouin C, Boby C, Lajoie-Mazenc I, Charveron M, Favre G. RhoB protects human keratinocytes from UVB-induced apoptosis through epidermal growth factor receptor signaling. J Biol Chem. 2005;280(52):43257–63.
Li YD, Liu YP, Cao DM, Yan YM, Hou YN, Zhao JY, Yang R, Xia ZF, Lu J. Induction of small G protein RhoB by non-genotoxic stress inhibits apoptosis and activates NF-κB. J Cell Physiol. 2011;226(3):729–38. https://doi.org/10.1002/jcp.22394.
Gavala ML, Hill LM, Lenertz LY, Karta MR, Bertics PJ. Activation of the transcription factor FosB/activating protein-1 (AP-1) is a prominent downstream signal of the extracellular nucleotide receptor P2RX7 in monocytic and osteoblastic cells. J Biol Chem. 2010;285(44):34288–98. https://doi.org/10.1074/jbc.M110.142091.
Moon YM, Lee SY, Kwok SK, Lee SH, Kim D, Kim WK, Her YM, Son HJ, Kim EK, Ryu JG, Seo HB, Kwon JE, Hwang SY, Youn J, Seong RH, Jue DM, Park SH, Kim HY, AhnChoML SM. The Fos-Related Antigen 1-JUNB/Activator Protein 1 Transcription Complex, a Downstream Target of Signal Transducer and Activator of Transcription 3, Induces T Helper 17 Differentiation and Promotes Experimental Autoimmune Arthritis. Front Immunol. 2017;18(8):1793. https://doi.org/10.3389/fimmu.2017.01793.
Atsaves V, Leventaki V, Rassidakis GZ, Claret FX (2019) AP-1 Transcription Factors as Regulators of Immune Responses in Cancer. Cancers 11(7). pii: E1037: https://doi.org/10.3390/cancers11071037
Kovács LÁ, Füredi N, Ujvári B, Golgol A, Gaszner B (2022) Age-Dependent FOSB/ΔFOSB Response to Acute and Chronic Stress in the Extended Amygdala, Hypothalamic Paraventricular, Habenular, Centrally-Projecting Edinger-Westphal, and Dorsal Raphe Nuclei in Male Rats. Front Aging Neurosci14:862098.: https://doi.org/10.3389/fnagi.2022.862098
Miyake Z, Takekawa M, Ge Q, Saito H. Activation of MTK1/MEKK4 by GADD45 through induced N-C dissociation and dimerization-mediated trans autophosphorylation of the MTK1 kinase domain. Mol Cell Biol. 2007;27(7):2765–76. https://doi.org/10.1128/MCB.01435-06.
Labonté B, Jeong YH, Parise E, Issler O, Fatma M, Engmann O, Cho KA, Neve R, Nestler EJ, Koo JW. Gadd45b mediates depressive-like role through DNA demethylation. Sci Rep. 2019;9(1):4615. https://doi.org/10.1038/s41598-019-40844-8.
Del Corvo M, Lazzari B, Capra E, Zavarez L, Milanesi M, Utsunomiya YT, Utsunomiya ATH, Stella A, de Paula Nogueira G, Garcia JF, Ajmone-Marsan P (2021) Methylome Patterns of Cattle Adaptation to Heat Stress. Front Genet 28(12):633132: https://doi.org/10.3389/fgene.2021.633132
Acknowledgements
Authors acknowledge the Director, ICAR-IVRI for extending the facilities to carry over the present work.
Funding
This work was supported by the SERB-DST, Gov’t of India, CRG Grant no: CRG/2019/005515.
DST-SERB,Govt of India,CRG/2019/005515,Basavaraj Sajjanar
Author information
Authors and Affiliations
Contributions
BS, GS, TD and BPM designed the work. BS, MTA, OK, GNT performed the experiments. APS, GBM, RKG, SD and PKG involved in analysing the data. SM & MTA wrote the manuscript and BS, GS editing the final draft. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All the experiments were approved by Institutional Animal Ethics Committee (IAEC) of Indian Veterinary Research Institute (IVRI).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file1: Fig. S1.
Exploratory analysis for genome-wide expression patterns of samples. Table S1. Primer sequences used in qRT-PCR for gene expression analysis. Table S2. Major heat shock induced differentially expressed genes and their functions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sajjanar, B., Aalam, M.T., Khan, O. et al. Genome-wide expression analysis reveals different heat shock responses in indigenous (Bos indicus) and crossbred (Bos indicus X Bos taurus) cattle. Genes and Environ 45, 17 (2023). https://doi.org/10.1186/s41021-023-00271-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41021-023-00271-8