Abstract
The fungus Trichoderma reesei is a powerful host for secreted production of proteins. The promoter of cdna1 gene, which encodes a small basic protein of unknown function and high expression, is commonly used for constitutive protein production in T. reesei. Nevertheless, the production level of proteins driven by this promoter still needs to be improved. Here, we identified that the region 600- to 700-bp upstream of the start codon is critical for the efficiency of the cdna1 promoter. Increasing the copy number of this region to three improved the production of a heterologous β-mannanase by 37.5%. Screening of several stressful conditions revealed that the cdna1 promoter is heat inducible. Cultivation at 37 °C significantly enhanced the production of β-mannanase as well as a polygalacturonase with the cdna1 promoter compared with those at 30 °C. Combing the strategies of promoter engineering, multi-copy gene insertion, and control of cultivation temperature, β-mannanase of 199.85 U/mL and relatively high purity was produced in shake flask, which was 6.6 times higher than that before optimization. Taken together, the results advance the understanding of the widely used cdna1 promoter and provide effective strategies for enhancing the production of recombinant proteins in T. reesei.
Graphical Abstract

Similar content being viewed by others
Introduction
The filamentous fungus Trichoderma reesei is widely used for cellulase production in industry, and the production level of secreted proteins was reported to be higher than 80 g/L (Bischof et al. 2016; Fonseca et al. 2020). In addition, T. reesei has been engineered for the production of other kinds of enzymes and pharmaceutical proteins (e.g., glucoamylase, endo-1,4-beta-xylanase and interferon (Bodie et al. 2021; EFSA Panel on Food Contact Materials et al. 2018; Landowski et al. 2016)). Consequently, the development of a powerful genetic toolbox and efficient fermentation processes are important for using T. reesei as a host of protein production.
Strong promoters are essential to achieve high-level production of proteins. The cellulase genes in T. reesei are highly inducible by cellulose, sophorose, and lactose, and their promoters have been employed for driving the expression of heterologous proteins (Fitz et al. 2018; Sun et al. 2020; Zou et al. 2012). Because the cellulase inducers can trigger the expression of dozens of endogenous secreted proteins (Herpoël-Gimbert et al. 2008), cellulase gene promoters are unfavorable for downstream purification of target proteins (Chai et al. 2022). In contrast, glucose represses the expression of most extracellular proteins, providing a clean background for secreted protein production using constitutive promoters (Linger et al. 2015; Rantasalo et al. 2019). Li et al. (2012) screened the promoters of 13 carbon metabolic genes and found that pdc (encoding pyruvate decarboxylase) and eno (encoding enolase) promoters had higher efficiencies in glucose medium. Also, the cfe1 gene was found to be highly expressed under various conditions, and its promoter was used for gene overexpression in T. reesei (Beier et al. 2022). Nevertheless, the efficiencies of constitutive promoters in T. reesei are generally much lower than those of cellulase gene promoters (Nakari-Setäläas & Penttilä, 1995).
The gene cdna1 gene (Trire2_110879; NCBI Gene ID:18,482,212) was identified as one of the most highly expressed genes in glucose medium in T. reesei (Nakari et al. 1993). The promoter of cdna1 was reported to be 20 to 50 times more efficient than that of translation elongation factor 1α gene, tef1 (Nakari-Setäläas and Penttilä, 1995). This promoter has been used to produce relatively pure proteins in T. reesei in glucose-containing media (Sun et al. 2022; Uzbas et al. 2012; Zhang et al. 2022). However, the production level of proteins using the cdna1 promoter still needs to be improved. In addition, although the cdna1 promoter is functional in both glucose and cellulose media, whether it is regulated by environmental signals is unclear.
In this study, the characteristics of the cdna1 promoter was revisited. The key sequence important for its efficiency was identified, which allowed rational sequence engineering for higher gene expression. In addition, the promoter was found to be heat inducible. With the engineered cdna1 promoter and optimized cultivation condition, the production of a β-mannanase in T. reesei was significantly improved.
Materials and methods
Strain construction
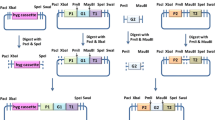
T. reesei QP4, a uracil auxotrophic strain generated from QM9414 by deleting the pyr4 gene (Zhong et al. 2016), was used as a parent for strain construction in this study. The plasmid pM5p containing the 2,028-bp upstream sequence of pyr4, followed by poman5A gene (GenBank accession of protein sequence: EPS31069.1) and its 500-bp downstream sequence from P. oxalicum, pyrG gene from Aspergillus nidulans as a selection marker, and the 2,183-bp downstream sequence of pyr4, was used for constructing gene expression cassettes (Additional file 1: Fig. S1). The ClonExpress II One Step Cloning Kit (Vazyme, China) was used to generate recombinant plasmids.
The cdna1 promoters of different lengths, as well as the key sequence fragments (for PcMC) or 5'-UTR of cel5A (for MCU5), were amplified from the genomic DNA of QP4 and then inserted into the restriction site Eco72I on pM5p. The MCU7 promoter was constructed using fusion and nested PCR, and cloned into the Eco72I site on pM5p. The obtained plasmids were digested with restriction enzyme PmeI, and the poman5A gene expression cassettes carrying homologous arms of pyr4 were recovered by purification. To construct poman5A randomly inserted strains, the PcMC-poman5A-pyrG fragment was cloned into the SacI and EcoRI sites of plasmid pUC19, and then the generated plasmid was linearized with restriction enzyme NdeI. The obtained cassettes were transformed into the protoplasts of QP4 as described by Penttilä et al. 1987. Transformants were screened and purified on minimal medium plates, and verified by diagnostic PCR. All the primers used for strain construction were listed in Additional file 1: Table S1.
Cultivation
All T. reesei strains in this study were maintained on Potato Dextrose Agar plates at 30 °C for 7 days for conidiation. Conidia were harvested by washing the plates with normal saline containing 0.01% (w/v) Tween 80. For protein production, biomass measurement, and RNA extraction, fresh conidia were inoculated into 50 mL seed medium at a final concentration of 106/mL, and the Erlenmeyer flasks were incubated in a rotary shaker at 200 rpm at 30 °C for 24 h. Then, 5 mL seed culture was inoculated to 50 mL fermentation medium for continued cultivation at 30 °C unless otherwise indicated.
The seed medium contained (g/L): glucose 20.0, (NH4)2SO4 5.0, KH2PO4 15.0, MgSO4·7H2O 0.6, CaCl2 0.6, peptone 2.0, FeSO4·7H2O 0.005, MnSO4·H2O 0.0016, ZnSO4·7H2O 0.0014, and CoCl2·6H2O 0.002. The fermentation medium contained (g/L): glucose 20.0, (NH4)2SO4 5.0, KH2PO4 5.0, MgSO4·7H2O 0.6, CaCl2 1.0, and yeast extract 20.0. For xylose fermentation medium, glucose was replaced by 20 g/L xylose. For pH 8.0 medium, 50 mM Tris-HCl buffer (pH 8.0) was used to prepare the medium. For low phosphate fermentation medium, the concentration of KH2PO4 was changed to 0.1 g/L and 50 mM sodium citrate buffer (pH 5.0) was used for medium preparation. Uracil with a final concentration of 1 g/L was added to the medium for the cultivation of QP4.
Enzyme assays and SDS-PAGE
For β-mannanase activity measurement, the crude enzyme solution was diluted to 500 μL using 0.2 M acetic acid–sodium acetate buffer (pH 4.8) and then mixed with 1.5 mL of 1% (w/v) locust bean gum (Sigma-Aldrich). The reaction system was incubated at 50 °C for 30 min and ended with the addition of 3 mL 3,5-dinitrosalicylic acid reagent. After boiling for 10 min and the addition of 20 mL distilled water, absorbance at 540 nm was determined. The concentration of released product was calculated according to the standard curve of mannose. One unit of β-mannanase activity was defined as the amount of enzyme required to release 1 μmol mannose from the substrate per minute under the above condition. Polygalacturonase activity was measured using the same method except that the substrate was replaced by 1% (w/v) polygalacturonic acid (Sigma-Aldrich). The culture supernatants were mixed with 5 × SDS-PAGE loading buffer (GenStar, China), boiled for 10 min, and centrifuged at 8,000 rpm for 1 min. Equal volumes (30 µl) of samples were loaded into 12.5% (w/v) SDS-PAGE gels for electrophoresis at 120 V for 1 h. Gels were stained with Coomassie Blue R-250 solution for 1 h and then washed with destaining solution (1:1:8 ethanol/acetic acid/water, v/v/v) until a clear background was obtained.
Biomass measurement
The weight of filter paper was recorded after drying to constant weight. Mycelia were harvested on filter paper by vacuum filtration and then placed in a 65 °C oven to dry to constant weight for weight measurement.
Nucleic acid extraction and quantitative PCR (qPCR)
The mycelia were collected on filter paper by vacuum filtration and then ground to powder in liquid nitrogen. DNA was extracted using the phenol/chloroform/isoamyl alcohol method as described by Ries et al. (Ries et al. 2014). Total RNA extraction and cDNA synthesis were performed using the RNAiso Reagent (TaKaRa, Japan) and HiScript III RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, China), respectively, according to the manufacturers’ instructions.
For qPCR, diluted genomic DNA or cDNA samples were mixed with primers and TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa, Japan) according to the manufacturer’s instructions. qPCR analysis was performed on LightCycler 480 II system (Roche) using the previously described conditions except setting the annealing temperature at 58 °C (Gao et al. 2017). The 2−ΔΔCT method was used for data analysis (Livak and Schmittgen 2001). To calculate the copy number of poman5A, the mixture of actin gene fragment and poman5A gene fragment with a molar ratio of 1:1 was used as a control. To determine relative changes in transcript abundances, the actin gene was used as a reference gene. The results were similar with those calculated with sar1 gene as a reference (Steiger et al. 2010). The primers used for qPCR are listed in Additional file 1: Table S2, Supporting Information.
Statistical analysis
Statistical significance tests of differences between samples were performed by calculating P values with one-tailed homoscedastic t test in the software Microsoft Office 2010 Excel (Microsoft, USA).
Results and discussion
Identification of a key sequence in cdna1 promoter
In the earliest report, the 1.2-kb sequence preceding the putative start codon of cdna1 gene was used as the promoter for gene expression (Nakari-Setäläas and Penttilä, 1995). However, the contribution of different regions within this sequence to downstream gene transcription was unknown. We constructed the expression cassettes of β-mannanase gene poman5A from P. oxalicum driven by the 1,159-bp cdna1 promoter and its 5′-truncated mutants. The PoMan5A enzyme recombinantly expressed in Pichia pastoris was found to be highly stable under high temperature and acidic conditions, making it suitable for many industrial applications (Liao et al. 2014). Each cassette was integrated into the pyr4 locus of the parent strain QP4 via homologous recombination, and the obtained strains were confirmed to contain only one copy of the poman5A gene (Additional file 1: Table S3). The production of β-mannanase using the 831- and 700-bp promoters were about 16% lower than that using the 1,159-bp promoter, while 71.4% and 85.4% declines were observed for the 600-bp and 505-bp promoters, respectively (Fig. 1A). In agree with this, SDS-PAGE analyses of the culture supernatants showed that the protein band of PoMan5A (about 70 kDa) was remarkably lighter in the strains containing 600-bp and 505-bp promoters (Fig. 1B). These results suggested that the sequence between 700- and 600-bp upstream of the start codon was critical for the efficiency of the cdna1 promoter.
Identification of the key sequence and engineering of cdna1 promoter. A Extracellular β-mannanase activities of strains expressing poman5A with different lengths of cdna1 promoter. The parent strain QP4 was used as a control. The samples were taken after 48 h of cultivation. B SDS-PAGE analysis of culture supernatants analyzed in panel A. C Schematic representation of the original (Pc1159) and engineered (PcMC) cdna1 promoter. D Extracellular β-mannanase activities of strains expressing poman5A with original and engineered cdna1 promoter. E SDS-PAGE analysis of the culture supernatants at 72 h analyzed in panel D. Data in panels A and D represent mean ± SD from triplicate cultivations
Increasing the copy number of key sequence improved the efficiency of cdna1 promoter
Increasing the copy number of cis-acting activating sequence is an effective strategy for enhancing the efficiency of promoters (Minetoki et al. 1998; Zhang et al. 2018). We constructed the strain expressing poman5A using the PcMC promoter containing three copies of the − 700 to − 600 bp sequence (Fig. 1C). Despite the lack of − 1,159 to − 700 bp sequence, the PcMC promoter resulted in 37.4% higher β-mannanase production than the 1,159 bp one at 48 h (Fig. 1D). The improved efficiency of the PcMC promoter was further confirmed by SDS-PAGE of the culture supernatants (Fig. 1E). In the future, detailed mapping of the key activating sequence within the − 700 to − 600 bp region and identification of the bound transcriptional activator(s) are expected to provide more efficient strategies to engineer the cdna1 promoter.
Replacing the 5′-untranslated region further improved β-mannanase production
Mapping the RNA-seq reads of T. reesei (NCBI SRA accession: SRR1057948) to the genomic sequence suggested an about 186-bp 5′-untranslated region (UTR) in the mRNA of cdna1. The highly expressed cellulase genes in T. reesei contain shorter 5′-UTR sequences than cdna1 (Saloheimo et al. 1988; Shoemaker et al. 1983), and might contribute to their high production levels. Therefore, we constructed chimeric promoters via replacing the 186-bp 3′ sequence of PcMC by the 5′-UTRs of two major cellulase genes (Fig. 2A). As shown in Fig. 2B and C, replacement of the 5′-UTR in PcMC promoter by those from cel7A/cbh1 and cel5A/eg2 improved the production of β-mannanase by 23 and 35%, respectively. The possible efficiency-determining factors in the three 5′-UTRs were compared (Dvir et al. 2013). All three sequences contain adenine at the -3 position, and only that of cel5A contains an upstream AUG. We supposed that the different efficiencies of the three 5′-UTRs might be related to their secondary structures.
Engineering of the 5′-untranslated region of cdna1 promoter. A Schematic representation of the 5′-UTR replaced promoters. Yellow and green boxes indicate 5′-UTRs from cel5A and cel7A promoters, respectively. The 123-bp intron in the UTR of cel5A is indicated by a thick line. B Extracellular β-mannanase activities of strains expressing poman5A with different promoters at 48 h. Data represent mean ± SD from triplicate cultivations. C SDS-PAGE analysis of the culture supernatants analyzed in panel B
Increased temperature induced the expression of genes driven by the cdna1 promoter
The cdna1 gene encodes an 88-amino acid protein of unknown function with a calculated isoelectric point of 11.51. Although cdna1 is highly transcribed in glucose medium, we speculated that such a small basic protein might be induced under some specific conditions. Therefore, we cultivated pre-grown mycelia of the Pc1159-poman5A strain under several different conditions. Cultivation with xylose as sole carbon source, or at pH 8.0, or with low phosphate, resulted in similar or lower β-mannanase production levels. In contrast, cultivation at 37 °C led to remarkably higher β-mannanase production than that at 30 °C (Fig. 3A). Further experiments showed that the increased temperature indeed improved the expression of PoMan5A while inhibiting cell growth (Fig. 3B–D). The abundances of both cdna1 and poman5A were significantly higher at 37 °C relative to 30 °C, indicating that the cdna1 promoter is heat inducible (Fig. 3E). In addition, the production of a heterologous endopolygalacturonase PoPga (GenBank accession of protein sequence: EPS32977.1, Gao et al. 2022) driven by cdna1 promoter was also enhanced at 37 °C (Fig. 3F, G), suggesting that elevating the temperature might be a universal strategy for improving protein production with this promoter.
The effect of temperature on protein expression driven by cdna1 promoter. A β-Mannanase production with Pc1159 promoter under different conditions. The samples were taken after 24 h of cultivation. Data represent mean ± SD from duplicate cultivations. Xyl, xylose. Low P, low phosphate. B The effect of temperature on β-mannanase production of the strain expressing poman5A with Pc1159. The parent strain was used as a control. C SDS-PAGE analysis of the culture supernatants analyzed in panel B. D The effect of temperature on cell growth. E The effect of temperature on the transcript abundances of cdna1 and poman5A in the strain expressing poman5A with Pc1159. RNA samples were taken at 24 h. F The effect of temperature on endopolygalacturonase production by the strain expressing popga with Pc1159. G SDS-PAGE analysis of the culture supernatants at 48 h analyzed in panel F. Data in panels B and D–F represent mean ± SD from triplicate cultivations. In panel A, the statistical significances of the difference between control and cultures under different conditions are shown. In panels B and F, the statistical significances of the difference between cultures at 30 °C and 37 °C are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Then, the responses of engineered cdna1 promoters to elevated temperature were investigated. Despite the decreased strength, the truncated promoter Pc600 maintained the characteristic of heat induction (Fig. 4A). However, the chimeric promoter MCU7 containing cel7A 5′-UTR showed similar efficiencies between 30 °C and 37 °C, suggesting that the replaced 186-bp sequence contains the putative heat-responsive element. Cultivation at 37 °C increased β-mannanase production with the PcMC promoter by 89%, which was confirmed by SDS-PAGE analysis of the culture supernatants (Fig. 4B).
β-Mannanase production by engineered cdna1 promoters at increased temperature. A Extracellular β-mannanase activities of strains expressing poman5A with different promoters at 48 h. Data represent mean ± SD from triplicate cultivations. B SDS-PAGE analysis of the culture supernatants analyzed in panel A
Combined genetic and process engineering enabled high-level production of β-mannanase
The above results showed that increasing the copy number of -700 to -600 bp sequence, replacement of 5′-UTR, and increasing the cultivation temperature could all enhance the production of β-mannanase. Considering that the 5′-UTR replaced promoters were not heat inducible any more, we constructed strains containing multiple copies of PcMC-poman5A gene expression cassettes via random genomic integration. One of the transformants, R-MC-22, was determined to contain two to three copies of the cassettes. The extracellular β-mannanase activity of this strain cultivated at 37 °C reached 199.85 U/ml at 72 h (Fig. 5), which was 6.6 times higher than that integrated with one Pc1159-poman5A cassette cultivated at 30 °C (Fig. 1D).
Conclusions
This study identified the key sequence for the transcription of downstream gene in the cdna1 promoter of T. reesei and found that increasing the cultivation temperature could further enhance the efficiency of this strong promoter. By combining the sequence modification and process improvement, the production level of β-mannanase was significantly enhanced. In the future, elucidation of the regulatory mechanism of the cdna1 promoter is expected to help the design of more efficient promoters for protein production in T. reesei.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Beier S, Stiegler M, Hitzenhammer E, Schmoll M (2022) Screening for genes involved in cellulase regulation by expression under the control of a novel constitutive promoter in Trichoderma reesei. Curr Res Biotech 4:238–246
Bischof RH, Ramoni J, Seiboth B (2016) Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Fact 15(1):106
Bodie E, Virag A, Pratt RJ, Leiva N, Ward M, Dodge T (2021) Reduced viscosity mutants of Trichoderma reesei with improved industrial fermentation characteristics. J Ind Microbiol Biotechnol 48(1–2):kuab014
Chai S, Zhu Z, Tian E, Xiao M, Wang Y, Zou G, Zhou Z (2022) Building a versatile protein production platform using engineered Trichoderma reesei. ACS Synth Biol 11(1):486–496
Dvir S, Velten L, Sharon E, Zeevi D, Carey LB, Weinberger A, Segal E (2013) Deciphering the rules by which 5’-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci USA 110(30):E2792-2801
EFSA Panel on Food Contact Materials, Enzymes and Processing Aids (CEP), Silano V, Barat Baviera JM, Bolognesi C, Brüschweiler BJ, Cocconcelli PS, Crebelli R, Gott DM, Grob K, Lampi E, Mortensen A, Rivière G, Steffensen IL, Tlustos C, Van Loveren H, Vernis L, Zorn H, Glandorf B, Marcon F, Penninks A, Aguilera J, Aguilera-Gomez M, Andryszkiewicz M, Arcella D, Gomes A, Kovalkovicova N, Liu Y, Chesson A (2018) Safety evaluation of the food enzyme endo-1,4-beta-xylanase from a genetically modified Trichoderma reesei (strain DP-Nzd22). EFSA J 16(11):e05479
Fitz E, Wanka F, Seiboth B (2018) The promoter toolbox for recombinant gene expression in Trichoderma reesei. Front Bioeng Biotechnol 6:135
Fonseca LM, Parreiras LS, Murakami MT (2020) Rational engineering of the Trichoderma reesei RUT-C30 strain into an industrially relevant platform for cellulase production. Biotechnol Biofuels 13:93
Gao L, Li Z, Xia C, Qu Y, Liu M, Yang P, Yu L, Song X (2017) Combining manipulation of transcription factors and overexpression of the target genes to enhance lignocellulolytic enzyme production in Penicillium oxalicum. Biotechnol Biofuels 10:100
Gao L, Liu G, Zhao Q, Xiao Z, Sun W, Hao X, Liu X, Zhang Z, Zhang P (2022) Customized optimization of lignocellulolytic enzyme cocktails for efficient conversion of pectin-rich biomass residues. Carbohydr Polym 297:120025
Herpoël-Gimbert I, Margeot A, Dolla A, Jan G, Mollé D, Lignon S, Mathis H, Sigoillot JC, Monot F, Asther M (2008) Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol Biofuels 1(1):18
Landowski CP, Mustalahti E, Wahl R, Croute L, Sivasiddarthan D, Westerholm-Parvinen A, Sommer B, Ostermeier C, Helk B, Saarinen J, Saloheimo M (2016) Enabling low cost biopharmaceuticals: high level interferon alpha-2b production in Trichoderma reesei. Microb Cell Fact 15(1):104
Li J, Wang J, Wang S, Xing M, Yu S, Liu G (2012) Achieving efficient protein expression in Trichoderma reesei by using strong constitutive promoters. Microb Cell Fact 11:84
Liao H, Li S, Zheng H, Wei Z, Liu D, Raza W, Shen Q, Xu Y (2014) A new acidophilic thermostable endo-1,4-beta-mannanase from Penicillium oxalicum GZ-2: cloning, characterization and functional expression in Pichia pastoris. BMC Biotechnol 14:90
Linger JG, Taylor LE 2nd, Baker JO, Vander Wall T, Hobdey SE, Podkaminer K, Himmel ME, Decker SR (2015) A constitutive expression system for glycosyl hydrolase family 7 cellobiohydrolases in Hypocrea jecorina. Biotechnol Biofuels 8:45
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Minetoki T, Kumagai C, Gomi K, Kitamoto K, Takahashi K (1998) Improvement of promoter activity by the introduction of multiple copies of the conserved region III sequence, involved in the efficient expression of Aspergillus oryzae amylase-encoding genes. Appl Microbiol Biotechnol 50(4):459–467
Nakari T, Alatalo E, Penttilä ME (1993) Isolation of Trichoderma reesei genes highly expressed on glucose-containing media: characterization of the tef1 gene encoding translation elongation factor 1 alpha. Gene 136(1–2):313–318
Nakari-Setäläas T, Penttilä M (1995) Production of Trichoderma reesei cellulases on glucose-containing media. Appl Environ Microbiol 61(10):3650–3655
Penttilä M, Nevalainen H, Rättö M, Salminen E, Knowles J (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61(2):155–164
Rantasalo A, Vitikainen M, Paasikallio T, Jäntti J, Landowski CP, Mojzita D (2019) Novel genetic tools that enable highly pure protein production in Trichoderma reesei. Sci Rep 9:5032
Ries L, Belshaw NJ, Ilmén M, Penttilä ME, Alapuranen M, Archer DB (2014) The role of CRE1 in nucleosome positioning within the cbh1 promoter and coding regions of Trichoderma reesei. Appl Microbiol Biotechnol 98(2):749–762
Saloheimo M, Lehtovaara P, Penttilä M, Teeri TT, Ståhlberg J, Johansson G, Pettersson G, Claeyssens M, Tomme P, Knowles JK (1988) EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene 63(1):11–22
Shoemaker S, Schweickart V, Ladner M, Gelfand D, Kwok S, Myambo K, Innis M (1983) Molecular cloning of exo-cellobiohydrolase I derived from Trichoderma reesei strain L27. Nat Biotechnol 1(8):691–696
Steiger MG, Mach RL, Mach-Aigner AR (2010) An accurate normalization strategy for RT-qPCR in Hypocrea jecorina (Trichoderma reesei). J Biotechnol 145(1):30–37
Sun X, Zhang X, Huang H, Wang Y, Tu T, Bai Y, Wang Y, Zhang J, Luo H, Yao B, Su X (2020) Engineering the cbh1 promoter of Trichoderma reesei for enhanced protein production by replacing the binding sites of a transcription repressor ACE1 to those of the activators. J Agric Food Chem 68(5):1337–1346
Sun Y, Qian Y, Zhang J, Yao C, Wang Y, Liu H, Zhong Y (2022) Development of a novel expression platform for heterologous protein production via deleting the p53-like regulator Vib1 in Trichoderma reesei. Enzyme Microb Technol 155:109993
Uzbas F, Sezerman U, Hartl L, Kubicek CP, Seiboth B (2012) A homologous production system for Trichoderma reesei secreted proteins in a cellulase-free background. Appl Microbiol Biotechnol 93(4):1601–1608
Zhang H, Yan JN, Zhang H, Liu TQ, Xu Y, Zhang YY, Li J (2018) Effect of gpd box copy numbers in the gpdA promoter of Aspergillus nidulans on its transcription efficiency in Aspergillus niger. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny154
Zhang J, Li J, Gao L, Waghmare PR, Qu J, Liu G (2022) Expression of a SARS-CoV-2 neutralizing nanobody in Trichoderma reesei. Chin J Biotech 38(6):2250–2258
Zhong L, Qian Y, Dai M, Zhong Y (2016) Improvement of uracil auxotrophic transformation system in Trichoderma reesei QM9414 and overexpression of β-glucosidase. CIESC Journal 67(6):2510–2518
Zou G, Shi S, Jiang Y, van den Brink J, de Vries RP, Chen L, Zhang J, Ma L, Wang C, Zhou Z (2012) Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering. Microb Cell Fact 11:21
Acknowledgements
The authors thank Zhifeng Li from State Key Laboratory of Microbial Technology of Shandong University for assistance in qPCR.
Funding
This work was supported by the National Key Research and Development Project of China (No. 2021YFC2101300 and 2018YFA0900500), National Natural Science Foundation of China (No. 32170037), the Young Scholars Program of Shandong University (YSPSDU to G.L.), and the Key research program of China National Tobacco Corporation (No. 110202102018).
Author information
Authors and Affiliations
Contributions
GL and LG conceived the study and designed the experiments. SJ conducted the major experiments. YW, QL, and QZ assisted with the experiments. XS, XL, and YQ analyzed data. SJ and GL wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
YQ is an editor of Bioresources and Bioprocessing.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Primers used for strain construction and verification in this study. Table S2. Primers used for qPCR in this study. Table S3. Copy numbers of poman5A gene in the constructed strains measured by qPCR. Figure S1. Map of plasmid pM5p.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, S., Wang, Y., Liu, Q. et al. Genetic engineering and raising temperature enhance recombinant protein production with the cdna1 promoter in Trichoderma reesei. Bioresour. Bioprocess. 9, 113 (2022). https://doi.org/10.1186/s40643-022-00607-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-022-00607-2