Abstract
Background
The threatening impact of conventional agriculture (CA) on soils could be due to the detrimental effects on soil microbial communities. Conversely, organic agriculture (OA) is envisaged as potentially enhancing helpful microbial communities and is proposed as environmentally sustainable. The soil microbiome influences soil health and quality, hence, it requires deeper investigation and understanding. In this study, applying 16S metabarcoding and qPCR techniques, we compared the microbial patterns of long-term organically and conventionally managed soils to explore their similarities and differences.
Results
Total DNA quantification showed an over 20-fold higher amount of DNA in OA soils (mean = 22.1 ± 3.92 μg g−1), compared to CA soils (mean = 0.95 ± 0.17 μg g−1). While 16S metabarcoding evidenced the absence of significant differences among communities of the two farming systems in terms of ecological indices, the qPCR analyses targeting functional genes reported a significantly higher abundance of all considered targets in OA sites spanning up to four-fold log increases. While OA and CA did not appear to affect overall bacterial diversity or evenness per se, qPCR-based functional analysis in OA showed a consistently higher abundance of all the salient microbial genes tested, when compared to CA, underlying a potentially beneficial impact on soil fertility and sustainability.
Conclusions
In essence, the sequencing-based analysis of absolute bacterial diversity could not differentiate the farming systems based on the amount of diversity but identified a unique set of taxa defining each. Hence, pairing this evaluation with the qPCR-based functional gene analyses can be a suitable approach to distinguish the exerted effects of CA or OA on soils.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
The global population, which is predicted to reach nine billion people by 2030 by the United Nations [1], is demanding the modern agricultural sector to achieve several goals that need careful long-term planning [2, 3]. Agriculture is expected to provide an increasing quantity of high-quality food to suppress hunger and malnutrition, but at the same time, prevent further impact on the surrounding environment [4]. The dilemma of choosing between nutritional versus environmental sustainability requires a thoughtful search for reliable solutions. Conventional agriculture has achieved several benefits for agricultural productivity, including increased yields and reduced crop losses, due to the utilization of external inputs [5, 6]. However, these practices have led to an intensification of the environmental pressure [7, 8] and a diffuse detrimental impact on ecosystems across the world [9, 10]. By reducing the use of external inputs and integrating several practices considered more environmentally friendly, organic agriculture strives to address the challenge of limiting the anthropogenic factors that might cause the degradation of ecosystems [11].
Sugar beet (Beta vulgaris L.), which serves as the primary source of approximately 20% to 30% of the annual global sugar production, constitutes an essential crop within the agricultural landscape of 52 countries situated in the temperate climate zone [12,13,14]. According to Eurostat [15], EU-27 countries grew ~ 1.5 million ha of sugar beet in 2019 and harvested ~ 111.6 million t (74.4 t ha−1 on average), which accounts for about half of the world’s total production. The largest sugar beet producers in the EU-27 are France, Germany, and Poland, which together produce more than 60.0% of the EU-27’s sugar beets (France 31.0%, Germany 22.8%, and Poland 12.1%). The demand for organic sugar is increasing in line with the growth of the organic market. In Europe, sugar production from organically grown beets is still embryonic with production reported only in Italy, Sweden, Denmark, and the Netherlands. One of the reasons for the low availability of organically produced sugar beet is the vulnerability to failure due to the management gap in organic farming. Moreover, organic sugar beet farming requires further improvements to achieve true multifunctionality that can assure a root yield comparable to the one identifying conventional farming along with the provision of several collateral ecological services that contribute to the preservation of the agroecosystem [16, 17].
The preservation of the agroecosystem revolves around the concepts of protection of soil health, quality and implementation of sustainable soil management practices. Soil, in this respect, has been defined as an indispensable complex natural resource that provides ecological functions, such as gene reservoir, and non-ecological functions, like archaeological artifact protection [18]. However, seeing soil as a granted commodity rather than the delicate result of the continuous delivery of plant organic compounds, and their microbially mediated breakdown and re-synthesis risks shifting its understanding from its actual nature. The live components of soil, encompassed within the soil microbiome pool, have a crucial role in building and maintaining its structure, which in turn regulates water availability, hosts the transformation and cycling of nutrients, the catabolism of toxins, mainly by microbes that account for a sizeable portion of the global genetic diversity [19]. Therefore, it is crucial to deepen our knowledge of soil microbes and the extent of their active services in agricultural contexts, to protect soil ecosystems and assure human welfare.
Improved molecular biology and high-throughput sequencing-based methods have been used in recent years to better understand microbial communities over traditional microbiology methods. By employing culture-independent techniques, the limitations associated with obtaining comprehensive information from the challenging cultivation of microorganisms collected from the environment have been effectively overcome [20]. DNA metabarcoding techniques using high-throughput sequencing, sequence alignment tools using bioinformatics methods, and databases of annotated microbes contribute to the determination of microbial species and knowledge of their genome functions in the soil. This is crucial in determining soil fertility and productivity with the goal of improving the agricultural system by relying more on microbial ecosystem services and less on the utilization of external input factors. Lately, multi-amplicon-based sequencing approaches provide a robust compositional structure of individual microbial community members and quantitative Real-Time PCR (qPCR) based approaches are used downstream for precise quantification of microbial targets [21]. Additionally, significant improvements in the algorithms and analysis methods of 16S analysis have been undertaken for community diversity estimation. Particularly, amplicon sequence variants (ASVs)-based clustering by predicting sequencing errors and denoising the reads [22] outperformed the operational taxonomical units (OTUs)-based approach [23].
The hypothesis underlying this study posits that soil bioindicators have the potential to display discernible variations when comparing conventional and organic farming practices. By investigating these bioindicators, the study is aimed to identify suitable markers from the soil microbiome in differentiating the effects of conventional versus organic farming practices by testing: (a) total soil DNA content, (b) 16S bacterial community using metabarcoding, and (c) specific functional gene quantitation via qPCR amplification.
Methods
Sites’ location and soil sampling
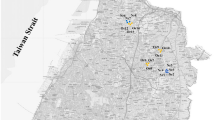
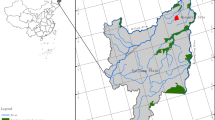
Soil samples were collected in six agricultural sites located in the Po River valley, Northeast Italy (Fig. 1). Among these sites, three were under organic agriculture (OA), and three were under conventional agriculture (CA). The three organically managed sites were located in Canal Dei Cuori farm (Loreo, RO, N 45° 05′ 02″, E 12° 10′ 55″), Le Barbarighe farm (San Martino di Venezze, RO, N 45° 06′ 10″, E 11° 53′ 03″) and Terre Emerse farm (Lozzo Atestino, PD, N 45° 17′ 25″, E 11° 35′ 00″), respectively. One of the three conventionally managed sites was located in Marsilio farm (Pozzonovo, PD, N 45° 10′ 51″, E 11° 47′ 48″), and the other two were located in the Cooperativa Produttori Bieticoli (CoProB) farm (Minerbio, BO, CoProB 1 N 44° 36′ 56″, E 11° 31′ 0.4″; CoProB 2 N 44° 38′ 12″, E 11° 33′ 49″). The soil classification, reported in Table 1, was based on the soil maps produced and published by the Regional Agency for Prevention and Environmental Protection of Veneto (ARPAV) [24] and by the Emilia-Romagna Region (RER) [25]. Regarding the OA sites, Canal Dei Cuori and Le Barbarighe soils were classified as Endoaquolls [26] and Gleyic Phaeozem [27], and Terre Emerse soil was classified as Haplustept [26] and Haplic Cambisol [27]. Concerning the CA sites, Marsilio soil was classified as Haplustept [26], and Fluvic Cambisol [27], and CoProB soils were classified as Haplustepts [26], and Fluvic Cambisol [27]. All sampling sites, which have been applying their respective soil management for over 20 years, were under sugar beet cultivation at the sampling time, which occurred in the month of August 2020. At the sampling time, all sugar beets within the sampled fields had attained the harvestable size (BBCH-49) [28]. Sugar beet yield traits such as root yield, sugar yield, and processing quality-related traits were evaluated in 2020 and 2021 (Additional file 1: Table S1). Soil sampling was performed by applying the grid sampling technique [29]. A manual auger was used to collect forty-eight sub-samples at a 15 cm depth from each sampling site. Sub-samples were, then, mixed to obtain two biological replicates from each farm, referred to as sample “a” and sample “b”. The composite samples were subjected to a 72-h air-drying process at room temperature [30], followed by sieving through a nested 0.5 mm wire mesh and subsequent immediate physicochemical analyses and total soil DNA extraction.
Soil physicochemical analyses
Soil samples were profiled by applying physical and chemical analysis techniques. Particle size distribution, compiled in Table 1, was investigated for each sample using laser diffraction analysis (Mastersizer 2000, Malvern Panalytical, Malvern, United Kingdom) [31]. Soil pH was measured potentiometrically by applying the soil/ultra-pure water ratio 1:2.5 w/v. A combustion analysis was performed to assess the concentration of total N, total C (Elementar Vario MACRO CNS, Elementar Analysensysteme GmbH, Hanau, Germany) and organic C (Skalar PrimacsSNC−100, Skalar Analytical BV, Breda, The Netherlands). The content of extractable phosphorus was determined using the Olsen P method [32]. Exchangeable cations assessment was performed using inductively coupled plasma optical emission spectrometry (ICP–OES) (Spectro Arcos MV, Spectro Ametek, Kleve, Germany).
Total soil DNA extraction, multi-amplicon 16S rDNA sequencing, quantification of functional genes for ecosystem services and bacterial plant-growth-promoting traits
Total soil DNA extraction from 250 mg of air-dried soil was performed using the DNeasy PowerSoil Pro Kit (Qiagen, Germany), in accordance with the manufacturer's instructions. Purified nucleic acid quantification was carried out using a Qubit Flex fluorometer (Thermo Fisher Scientific, Carlsbad, CA) with a Qubit 1 × dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific, Carlsbad, CA).
16S rDNA meta-barcoding library preparation was performed using the 16S Ion Metagenomics Kit (Thermo Fisher Scientific, Carlsbad, CA) that carries two sets of pooled primers targeting seven hypervariable regions of the 16S rRNA gene. The templating of 400 bp was carried out in an Ion One Touch 2 instrument using an Ion 520 chip and sequenced on the Ion GeneStudio S5 System (Thermo Fisher Scientific, Carlsbad, CA).
Raw reads processing was performed conforming to the pipeline published in Maretto et al. [33]. Demultiplexed uBAM files from the sequencer were converted into FASTQ format using samtools bamtofastq (v1.10) [34]. Raw reads were trimmed for 20 nucleotides on both ends to remove primers using cutadapt (v3.5) [35]. A Quantitative Insights Into Microbial Ecology 2 (QIIME2) (v2020.08) [36] microbiome pipeline was further used to analyze the raw reads. Imported reads were first denoised and dereplicated using the “qiime dada2” plugin followed by taxonomic classification of ASVs by a “classify-consensus-blast” plugin using SILVA SSU (v138.1) [37] as the reference database. Alpha diversity rarefaction analysis was done using the “qiime alpha-diversity” plugin and corresponding results were plotted using ggplot2 R-package [38]. The feature abundance table and taxonomic assignment table were exported and further processed using RStudio (version R-4.2.2) [39, 40] in conjunction with the tibble package [41] and TaxaPhyloseq [42]. Partially classified taxonomic entries or entries with counts below 10 were excluded from the analysis. MicrobiotaProcess packages [43] in R were used to calculate the diversity indices, perform PCoA analysis and, counts normalization. All plots were generated using ggplot2 R-package.
The abundance of the 16S gene, and that of genes involved in the N biogeochemical cycle such as amoA (eubacterial, AOB and archaeal, AOA ammonia oxidase/nitrification), nifH (nitrogenase/nitrogen fixation), nirK (nitrite reductase/intermediate denitrification), and nosZ (nitrous oxide reductase/terminal denitrification), and the abundance of the gene coding for the gh48 cellulase enzyme, of the PKs (polyketides) and NRPs (non-ribosomal peptide) cluster genes that are thought to play a crucial role in the adaptation of bacteria to soil, and in plants' health and development [44], were analyzed by qPCR using a QuantStudio 12K-Flex apparatus (Thermo Fisher Scientific, Carlsbad, CA). Moreover, the presence of specific 16S rDNA from Asticcacaulis, Mesorhizobium, Nocardioides, Sphingobium and Sphingomonas, which are known prominent sugar beet growth-promoting rhizobacteria (PGPR) [45], was assessed using the same qPCR approach. All primers sequences used are reported in Table 2. The assays targeting the five PGPR have been designed based on the sequences retrieved from the 16S metabarcoding data generated for this study. The reaction mix was composed of 2.5 µL PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Carlsbad, CA), 0.15 µL each of forward and reverse primer, 1.2 µL PCR-grade water, and 1 µL template DNA. The numbers of gene copies in soil for the targeted genes were calculated starting from the Ct values on the basis of calibration curves constructed using specific concentrations of Escherichia coli for the 16S gene or of the targeted functional genes cloned in plasmids of known length for the other amplicons [46].
Overall statistical analyses, including those performed on soil chemical properties, were performed using RStudio and the dplyr package [47]. The evaluation of significant differences between the mean values occurred with the non-parametric Wilcoxon Rank Sum test [48]. Data are expressed as mean ± standard error.
Results
The analysis of the texture of the collected topsoil (Table 1), based on the size particle limits and abundance established by the United States Department of Agriculture (USDA) in 1951 [58], showed that all CA samples are classified as “Silt Loam soils” since their silt percentage is higher than 50% and, those of sand and clay are in the range of 20–30% and 15–30% respectively. The samples from Canal Dei Cuori and Le Barbarighe farms are classified as “Silty Clay Loam soils” since they have less than the 20% of sand and clay content in the range of 27–40%. The samples collected in Terre Emerse farm are classified as “Clay Loam soils” since their percentages of clay and sand are higher than 27% and 20%, respectively. All the soil chemical characterization results are compiled in Table 3. No significant differences were observed in soil pH, ranging between 7.60 and 8.11 in CA soils and 7.38–7.91 in OA soils. Total N content was not significantly different in the two farming systems. Both carbon forms explored in this study were significantly higher (p < 0.01) in OA soils than in CA soils. In detail, the mean total C content in OA soils was 164% higher than in CA soils. Moreover, the organic C mean was 231% higher in OA soils when compared to CA soils. Olsen P did not show a significant difference between the two groups of soils. Among the exchangeable cations evaluated in this study, Ca++ and Mg++ were significantly (p < 0.05) more abundant in OA soils than in CA soils. At the same time, Na+ and K+ showed no significant differences between OA and CA soils. The Principal Component Analysis (PCA) performed on the soil chemical analyses results showed (Fig. 2) that the main parameters influencing dimension 1 are the exchangeable cations and the total N, and the main parameters influencing dimension 2 are total C and organic C.
Total soil DNA quantification results highlighted that OA soils (mean = 22.1 ± 3.92 µg g−1, individual farms: Canal Dei Cuori = 33.45 ± 0.75 µg g−1, Le Barbarighe = 20.61 ± 0.27 µg g−1, Terre Emerse = 12.17 ± 0.08 µg g−1) held a significantly higher (p < 0.01) amount of DNA than CA soils (mean = 0.95 ± 0.17 µg g−1, individual farms: Marsilio = 1.45 ± 0.01 µg g−1, CoProB 2 = 0.80 ± 0.11 µg g−1, CoProB 1 = 0.59 ± 0.06 µg g−1).
The metabarcoding analysis of the bacterial 16S gene on the soil samples provided 9,682,353 single-end reads with an average length of 217 nucleotides. The 32,725 identified ASVs were eventually classified into 980 database-featured taxa names. 91.1% of the annotated reads were classified at the phylum rank level, 90.3% at the class level, 88.7% at the order level, 87.0% at the family level, the 80.4% at the genus level.
The sequences rarefaction analysis conducted to assess whether the samples had been sequenced to a sufficient depth showed (Additional file 2: Fig. S1) that all the soils reached the plateau on the curve ranked from Canal Dei Cuori, the one with the highest number of detected sequences, to CoProB 1, the one with the lowest. The alpha-diversity was examined within each taxonomical level by calculating three ecological indices such as Chao1, Shannon, and Simpson 1-D to evaluate community richness, diversity, and evenness, respectively. The results of the analyses conducted at the genus level, which is representative of all the higher taxonomic ranks, are reported in Fig. 3. It is visible that, for all the considered indices, although some OA soils manifest the highest absolute values of richness, diversity and evenness, the means are even lower than those of the conventional soils and moreover, for none of the indices there was any significant difference between the two farming systems.
To determine whether there could be any possible association between the multiple independent variables characterizing microbial communities, multivariate analyses were conducted. Figure 4 displays the results of the PCA, the Principal Coordinates Analysis (PCoA), and the Permutational Multivariate Analysis of Dispersion (PERMDISP2). Although PCA and PCoA approaches showed a partial overlap of the two clusters, it is noticeable that the OA samples clustered tightly compared to the CA samples. The PERMDISP2 analysis, based on the Bray–Curtis dissimilarity matrix, indicated a significant (p = 0.01035) separation of the ellipse’s centroids. The Permutational Multivariate Analysis of Variance (PERMANOVA) returned a p-value = 0.051 for 999 permutations, a p-value = 0.0463 for 9999 permutations, a p-value = 0.04703 for 99,999 permutations, and the Analysis of Similarities (ANOSIM) reported an R value = 0.2481 and a p-value = 0.0177. The results of the analysis of the set of microbial taxa that are characteristic of the two farming systems and the analysis of the taxa shared among OA sites and CA sites are reported in Fig. 5. The overall core microbiome analysis showed a higher abundance of detected taxa in CA soils compared to OA soils, however, for every considered taxonomical level, the analysis of the shared taxa reported that OA sites shared a higher number of taxa compared to CA sites. In detail, OA sites, from the phylum to the genus taxonomic level, shared 103%, 108%, 109%, 111% and 113% more taxa than CA sites.
Multivariate analyses for the bacterial communities sequencing data at the genus taxonomic level to evaluate the beta diversity in OA sites (green) and CA sites (purple). a Principal Component Analysis (PCA), b Principal Coordinate Analysis (PCoA) based on the Bray–Curtis dissimilarity matrix, c Permutational Multivariate Analysis of Dispersion (PERMDISP2) showing the distance of each sample from the group’s centroid
The qPCR analysis results, reported in Table 4, depict that OA sites when compared to CA sites, have a significantly higher abundance of bacterial 16S gene (p < 0.01) and a significantly higher number of copies of functionally relevant genes such as those involved in the N biogeochemical cycle (Archaeal-amoA p < 0.05, Eubacterial-amoA p < 0.001, nifH p < 0.001, nosZ p < 0.05, nirK p < 0.01), the one coding for the gh48 enzyme (p < 0.01) and those genes clusters coding for polyketides (PKs I p < 0.01, PKs II p < 0.001) and non-ribosomal peptides (p < 0.05). The relative abundance of the sugar beet-growth-promoting rhizobacteria in OA and CA samples is portrayed in Fig. 6. In this figure, the threshold cycle values (Ct) are shown (i.e., the lower the number the higher the abundance of the amplicon). All five bacterial genera were detected in all samples irrespective of the soil management but were significantly (p < 0.001) more copious in OA sites.
Relative abundance of five sugar beet growth-promoting rhizobacteria (PGPR) detected by qPCR. The plot shows the comparison between the Threshold Cycle (Ct) values for all the tested genera. Ct values are inversely related to the target abundance, meaning that a lower Ct value corresponds to a higher abundance of the investigated amplicon. ***Significance level p < 0.001
Discussion
In this study, despite observing variations in several chemical soil characteristics and recording soil nutrients concentrations that, in accordance with Seufert et al. [59], supported OA farms’ production rate, as well as an over twenty-year-long difference in the organic versus conventional management regimes, the six studied sites exhibited similar alpha-diversity profiles for the total bacterial communities, as far as the 16S metabarcoding approach is concerned. The data confirm those from prior analyses of ours in a survey that included ten horticultural farms [60], as well as with the findings of a study covering 25 years of conversion from conventional to organic cropping in the Netherlands, in which authors concluded that the shift to organic management did not increase bacterial community diversity, and for fungi the increase occurred only in some soils [61].
The absence of significant differences in richness, diversity and evenness indices suggests that the bacterial community structures, in terms of absolute diversity per se, are determined by a combination of soil structure and chemistry [62], and the crop’s rhizosphere [63, 64], or by a passive atmospheric discharge of cells [65] rather than by a deterministic selection brought about by each of the two farming systems. Despite the diverse soil management practices implemented over the past decades, the observed similarity in overall soil bacteria composition may be attributed to a long-lasting legacy effect of a much older common cropping regime before conversion to organic farming of part of the sites [66]. The beta-diversity analysis partially clustered the two farming systems separately. Although the PERMANOVA analysis, oscillating between significant and non-significant results, reports an uncertain level of differences in the overall community composition, PERMDISP2 analysis describes a significant difference in the dispersion of the samples, meaning that there is a somewhat detectable difference in the heterogeneity in the community composition [67, 68]. On the other hand, the ANOSIM analysis reports exiguous but still significant differences in the microbial communities' composition. The core microbiome evaluation reported an enhanced number of detected taxa within the CA soils compared to the OA soils.
The higher number of detected taxa can depend on the combination of environmental processes, for instance, the airborne immigration of foreign DNA from different sites as described by Rosselli et al. [65], and the mechanical incorporation of that migrated DNA due to the frequent tillage the CA sites are subjected to. On the other hand, the qPCR results instead spotlight a remarkably different situation. Within a two-fold increase of the whole bacterial community shown by the 16S primers in OA soils, the single functional genes targets reported in Table 4 unraveled major changes. All these changes are pointed towards the same direction, as results from the ten-fold increase of the cellulase and nitrite reductase genes in OA soils, from the more than 100-fold increase of both nitrification genes, for the synthesis of the non-ribosomal peptide II and polyketide II, and for the terminal gene of the denitrification pathway nosZ, which eliminates nitrous oxide, which is one of the worst gases involved in global warming. Even more pronounced in OA soils as well, are the increases in the number of copies of other determinants as the nearly thousand-fold rise of the nitrogen fixation gene nifH, and of the polyketide synthesis I, and the almost 10,000-fold over-representation of the non-ribosomal peptide I. At the same time, the five genera Asticcacaulis, Mesorhizobium, Nocardioides, Sphingobium and Sphingomonas, which were all known as promoters of the sugar beet crop, that was in place at the sampling time, were found enriched in the OA fields. All the above-mentioned tested genes, all of which were significantly more abundant in the organic farms in comparison to the conventional ones, not only qualify as proxies to tell apart the effects of the two farming systems but are also positive indicators of actively ongoing cycles for C and N and of the abundance of proficient plant-growth promoting guilds. As a general consideration, the 16S is a multicopy gene implying that some species may possess more copies of the gene than others. However, as always in these types of studies involving metabarcoding, it is important to note that the number of microbial gene copies does not accurately reflect the actual number of genomes in the system. This is due to the variable gene dosage, which introduces a certain level of overestimation [69,70,71]. Nonetheless, this situation is averagely occurring across various habitats, and there are no indications or reasons to suggest that either of our compared soil types (organic vs. conventional management) would unequivocally exhibit an overabundance of bacteria with higher numbers of 16S gene copies. Therefore, we hypothesize that any differences should balance each other out.
Conclusions
The novel findings presented herein are consistent with earlier studies that compared soil properties between conventional and organic cropping systems using various approaches, such as microbial biomass estimation and plate counts [72], soil respiration and enzymatic assays [73], and phospholipid fatty acid (PLFA) profiles [74]. In the current report, the combination of the 23-fold more abundant total soil DNA, the 2.6 times more plentiful 16S gene, the significantly more copious amount of the considered functional genes in OA sites, and the consistently higher presence of all five PGPR taxa investigated, leads to the consideration that the OA management appears to have a positive effect on the functioning of the agroecosystem which is tightly connected to soil fertility.
As concerns the approaches employed, the metagenomics-based barcoding of the 16S via sequencing was extremely valuable in providing a detailed taxonomical view of the community structure and allowed to achieve the very important finding that conventional soils feature a bacterial alpha diversity which does not record signs of decline when compared with the one occurring in organic soils. However, the approach was not providing clues to identify the impacts of each management as it yielded non-significant differences in the bacterial biodiversity stemming from each of the indices. It appears that a high taxonomic richness, equally characterizing CA, and OA sites does not reveal elements of functional biodiversity that instead occur in the soil and can be unraveled by qPCR. The lack of concordance between metabarcoding and qPCR results strongly suggests that the analysis of the absolute microbial biodiversity cannot adequately differentiate agricultural ecosystems as a function of their different managing systems. However, while the CA and OA soils show a similar level of diversity with the diversity indices, deeper investigation, as shown by Venn diagrams in Fig. 5, shows a set of taxa present uniquely in each soil type, which can perhaps lead to relevant functional attributes of the two soil types. Consequently, bacterial community sequencing, although extremely informative, is not a suitable stand-alone proxy to achieve the functional differentiation of the two farming systems. In this respect, it is necessary to state that this is not due to any limitation inherent to the technique, which remains one of the deepest tools for soil biota analysis, but rather on two distinct facts. The efficiency of taxa detection from environmental DNA strongly depends on the number of replicates, sequencing depth and quality, PCR conditions, and the characteristics of the environment itself. A lower number of biological replicates, as the one characterizing this project, can be sufficient for the assessment of the presence/absence of the main taxa but, at the same time, can lead to a biased analysis of rare taxa due to DNA degradation and sequencing errors. Moreover, soil as a whole is a poor reporter source of its ongoing true activity due to the way soils accumulate bacteria and preserve their DNA, i.e., to the fact that a vast majority of soil bacterial DNA is a relic or belongs to passive inmates, with no physiologically functional roles in the system. This limit, however, may be more specific to the prokaryotic component and likely, fungal ITS-based metabarcoding could be more suited to put in evidence function-related differences [75] as future studies on the same soil could reveal. In any event, the present data show that the microbial profiling of soils, constituting a fundamental step in the evaluation of their quality, health and sustainability, should consider pairing DNA sequencing, also with other highly informative analyses, such as qPCR targeting meaningful bio-indicator genes as the ones hereby covered along with functional biodiversity prediction from taxonomy data, and metatranscriptomics, to untangle the underlying mechanisms regulating the soil ecosystem functioning for its long-term stability and resilience.
Availability of data and materials
The dataset generated for this study can be found in the EMBL Nucleotide Sequence Database (ENA), ID PRJEB60430, https://www.ebi.ac.uk/ena/browser/view/PRJEB60430.
Abbreviations
- AOA:
-
Ammonia oxidising archaea
- AOB:
-
Ammonia oxidising bacteria
- ARPAV:
-
Regional Agency for Prevention and Environmental Protection of Veneto
- ASV:
-
Amplicon sequence variant
- CA:
-
Conventional agriculture
- Ct:
-
Cycle threshold
- NRPs:
-
Non-ribosomal peptide synthetase
- OA:
-
Organic agriculture
- OTU:
-
Operational Taxonomic Unit
- PCA:
-
Principal Component Analysis
- PCoA:
-
Principal Coordinates Analysis
- PERMANOVA:
-
Permutational Multivariate Analysis of Variance
- PERMDISP2:
-
Permutational Multivariate Analysis of Dispersion
- PGPR:
-
Plant Growth-Promoting Rhizobacteria
- PKs:
-
Polyketide synthase
- QIIME2:
-
Quantitative Insight Into Microbial Ecology 2
- qPCR:
-
Quantitative Real-Time Polymerase Chain Reaction
- RER:
-
Emilia-Romagna Region
References
United Nations Department of Economic and Social Affairs, Population Division. World Population Prospects 2022: Summary of Results. UN DESA/POP/2022/TR/NO. 3.
Pelletier N, Tyedmers P. Forecasting potential global environmental costs of livestock production 2000–2050. Proc Natl Acad Sci. 2010;107:18371–4. https://doi.org/10.1073/pnas.1004659107.
Foley JA, Ramankutty N, Brauman KA, Cassidy ES, Gerber JS, Johnston M, Mueller ND, O’Connell C, Ray DK, West PC, Balzer C, Bennett EM, Carpenter SR, Hill J, Monfreda C, Polasky S, Rocksström J, Sheehan J, Siebert S, Tilman D, Zaks DPM. Solutions for a cultivated planet. Nature. 2011;478:337–42. https://doi.org/10.1038/nature10452.
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir J, Pretty J, Robinson S, Thomas SM, Toulmin C. Food security: the challenge of feeding 9 billion people. Science. 2010;327(5967):812–8. https://doi.org/10.1126/science.1185383.
Dunwell JM. Global population growth, food security and food and farming for the future. In: Bennett DJ, Jennings RC, editors. Successful agricultural innovation in emerging economies. Cambridge: Cambridge University Press; 2013. p. 23–38. https://doi.org/10.1017/CBO9781139208475.003.
Reganold JP, Wachter JM. Organic agriculture in the twenty-first century. Nat Plants. 2016;2:15221. https://doi.org/10.1038/nplants.2015.221.
Gasteyer SP. Agricultural transitions in the context of growing environmental pressure over water. Agric Hum Values. 2008;25:469–86. https://doi.org/10.1007/s10460-008-9137-x.
Gomiero T, Paoletti MG, Pimentel D. Energy and environmental issues in organic and conventional agriculture. Crit Rev Plant Sci. 2008;27:239–54. https://doi.org/10.1080/07352680802225456.
Bommarco R, Kleijn D, Potts SG. Ecological intensification: harnessing ecosystem services for food security. Trends Ecol Evol. 2013;28:230–8. https://doi.org/10.1016/j.tree.2012.10.012.
Duru M, Therond O, Martin G, Martin-Clouaire R, Magne MA, Justes E, Journet ET, Aubertot JN, Savary S, Bergez JE, Sarthou JP. How to implement biodiversity-based agriculture to enhance ecosystem services: a review. Agron Sustain Dev. 2015;35:1259–81. https://doi.org/10.1007/s13593-015-0306-1.
Heckman JR, Weil R, Magdoff F. Practical steps to soil fertility for organic agriculture. In: Francis C, editor. Agronomy monographs. Madison: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 2015. p. 137–72. https://doi.org/10.2134/agronmonogr54.c7.
Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schultz B, Stadler PF, Schmidt T, Gabaldón T, Lehrach H, Weisshaar B, Himmelbauer H. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature. 2014;505:546–9. https://doi.org/10.1038/nature12817.
Nieberl P, Ehrl C, Pommerrenig B, Graus D, Marten I, Jung B, Ludewig F, Koch W, Harms K, Flügge UI, Neuhaus HE, Hedrich R, Sauer N. Functional characterisation and cell specificity of BvSUT1, the transporter that loads sucrose into the phloem of sugar beet (Beta vulgaris L.) source leaves. Plant Biol. 2017;19:315–26. https://doi.org/10.1111/plb.12546.
Stevanato P, Chiodi C, Broccanello C, Concheri G, Biancardi E, Pavli O, Skaracis G. Sustainability of the sugar beet crop. Sugar Tech. 2019;21:703–16. https://doi.org/10.1007/s12355-019-00734-9.
Eurostat database. 2020. https://ec.europa.eu/eurostat/data/database
Jordan N, Boody G, Broussard W, Glover JD, Keeney D, McCown BH, McIsaac G, Muller M, Murray H, Neal J, Pansing C, Turner RE, Warner K, Wyse D. Sustainable development of the agricultural bio-economy. Science. 2007;316:1570–1. https://doi.org/10.1126/science.1141700.
Chabert A, Sarthou JP. Conservation agriculture as a promising trade-off between conventional and organic agriculture in bundling ecosystem services. Agric Ecosyst Environ. 2020;292:106815. https://doi.org/10.1016/j.agee.2019.106815.
Blum WEH. Functions of soil for society and the environment. Rev Environ Sci Biotechnol. 2005;4:75–9. https://doi.org/10.1007/s11157-005-2236-x.
Larkin RP. Soil health paradigms and implications for disease management. Annu Rev Phytopathol. 2015;53:199–221. https://doi.org/10.1146/annurev-phyto-080614-120357.
Vester JK, Glaring MA, Stougaard P. Improved cultivation and metagenomics as new tools for bioprospecting in cold environments. Extremophiles. 2015;19:17–29. https://doi.org/10.1007/s00792-014-0704-3.
Dreier M, Meola M, Berthoud H, Shani N, Wechsler D, Junier P. High-throughput qPCR and 16S rRNA gene amplicon sequencing as complementary methods for the investigation of the cheese microbiota. BMC Microbiol. 2022;22:48. https://doi.org/10.1186/s12866-022-02451-y.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. https://doi.org/10.1038/nmeth.3869.
Joos L, Beirinckx S, Haegeman A, Debode J, Vandecasteele B, Baeyen S, Goormachtig S, Clement L, De Tender C. Daring to be differential: metabarcoding analysis of soil and plant-related microbial communities using amplicon sequence variants and operational taxonomical units. BMC Genomics. 2020;21:733. https://doi.org/10.1186/s12864-020-07126-4.
Regional Agency for Prevention and Environmental Protection of Veneto (ARPAV). Carta dei suoli 2020, https://gaia.arpa.veneto.it/maps/778
Regione Emilia-Romagna, Area Geologia, Suoli e Sismica, Settore Difesa del Territorio (RER). Carta dei suoli della Regione Emilia-Romagna 2021, https://mappe.regione.emilia-romagna.it/
U.S. Department of Agriculture. Keys to Soil Taxonomy. Soil Survey Division Staff; Natural Resources Conservation Service, 2010, 11th Edition.
IUSS Working Group. World Reference Base (WRB) for Soil Resources. World Soil Resources Reports 2006, 103, FAO.
Meier U, Bachmann L, Buhtz E, Hack H, Klose R, Märländer B. Phänologische Entwicklungsstadien der Beta-Rüben (Beta vulgaris L. ssp.). Nachrichtenblatt des Deutschen Pflanzenschutzdienstes (Braunschweig). 1993;45(2):37–41.
Brown A. A review of soil sampling for chemical analysis. Aust J Exp Agric. 1993;33:983. https://doi.org/10.1071/EA9930983.
Wang F, Che R, Deng Y, Wu Y, Tang L, Xu Z, Wang W, Liu H, Cui X. Air-drying and long time preservation of soil do not significantly impact microbial community composition and structure. Soil Biol. 2021;157:108238. https://doi.org/10.1016/j.soilbio.2021.108238.
Bittelli M, Pellegrini S, Olmi R, Andrenelli MC, Simonetti G, Borrelli E, Morari F. Experimental evidence of laser diffraction accuracy for particle size analysis. Geoderma. 2022;409:115627. https://doi.org/10.1016/j.geoderma.2021.115627.
Olsen SR, Cole CV, Watanabe FS, Dean LA. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA circular 939, 1954, U.S. Gov. Print. Office, Washington D.C.
Maretto L, Deb S, Ravi S, Chiodi C, Manfredi P, Squartini A, Concheri G, Renella G, Stevamato P. Microbial diversity of reconstituted, degraded, and agricultural soils assessed by 16S rDNA multi-amplicon sequencing. Front Environ Sci. 2022;9:807889. https://doi.org/10.3389/fenvs.2021.807889.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. https://doi.org/10.1093/bioinformatics/btp352.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–2. https://doi.org/10.14806/ej.17.1.200.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, Van Der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, Von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. https://doi.org/10.1038/s41587-019-0209-9.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. https://doi.org/10.1093/nar/gks1219.
Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016. https://ggplot2.tidyverse.org
Posit team. RStudio: Integrated Development Environment for R. Posit Software, PBC, Boston, MA, 2022. https://www.posit.co/
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2022. https://www.R-project.org/
Müller K, Wickham H. tibble: Simple Data Frames, 2023. https://tibble.tidyverse.org/, https://github.com/tidyverse/tibble
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. https://doi.org/10.1371/journal.pone.0061217.
Xu S, Zhan L, Tang W, Wang Q, Dai Z, Zhou L, Feng T, Chen M, Wu T, Hu E, Yu G. MicrobotaProcess: a comprehensive R package for deep mining microbiome. Innovation. 2023;4:100388. https://doi.org/10.1016/j.xinn.2023.100388.
Dror B, Wang Z, Brady SF, Jurkevitch E, Cytryn E. Elucidating the diversity and potential function of nonribosomal peptide and polyketide biosynthetic gene clusters in the root microbiome. mSystems. 2020;5(6):e00866-e920. https://doi.org/10.1128/mSystems.00866-20.
Okazaki K, Tsurumaru H, Hashimoto M, Takahashi H, Okubo T, Ohwada T, Minamisawa K, Ikeda S. Community analysis-based screening of plant growth-promoting bacteria for sugar beet. Microbes Environ. 2021. https://doi.org/10.1264/jsme2.ME20137.
Zanardo M, Rosselli R, Meneghesso A, Sablok G, Stevanato P, Altissimo A, Perserico L, Dezuani V, Concheri G, Schloter M, Squartini A. Dynamics of soil prokaryotes catalyzing nitrification and denitrification in response to different fertilizers in a greenhouse experiment with Cynodon dactylon. Eur J Soil Biol. 2016;76:83–91. https://doi.org/10.1016/j.ejsobi.2016.08.002.
Wickham H, François R, Henry L, Müller K, Vaughan D. dplyr: A Grammar of Data Manipulation, 2023. R package version 1.1.0. https://CRAN.R-project.org/package=dplyr
Wilcoxon F. Individual comparisons by ranking methods. Biometr Bull. 1945;1(6):80–3. https://doi.org/10.2307/3001968.
Johnson TA, Stedtfeld RD, Wang Q, Cole JR, Hasham SA, Looft T, Zhu YG, Tiedje JM. Clusters of antibiotic resistance genes enriched together stay together in swine agriculture. MBio. 2016. https://doi.org/10.1128/mBio.02214-15.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102(41):14683–8. https://doi.org/10.1073/pnas.0506625102.
Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63(12):4704–12. https://doi.org/10.1128/aem.63.12.4704-4712.1997.
Rösch C, Mergel A, Bothe H. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol. 2002;68(8):3818–29. https://doi.org/10.1128/AEM.68.8.3818-3829.2002.
Henry S, Baudoin E, López-Gutiérrez JC, Martin-Laurent F, Brauman A, Philippot L. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Methods. 2004;59(3):327–35. https://doi.org/10.1016/j.mimet.2004.07.002.
De Menezes AB, Prendergast-Miller MT, Poonpatana P, Farrell M, Bissett A, Macdonald LM, Toscas P, Richardson AE, Thrall PH. C/N ratio drives soil actinobacterial cellobiohydrolase gene diversity. Appl Environ Microbiol. 2015;81(9):3016–28. https://doi.org/10.1128/AEM.00067-15.
Amos GC, Borsetto C, Laskaris P, Krsek M, Berry AE, Newsham KK, Calvo-Bado L, Pearce DA, Vallin C, Wellington EM. Designing and implementing an assay for the detection of rare and divergent NRPS and PKS clones in European, Antarctic and Cuban Soils. PLoS ONE. 2015;10(9):e0138327. https://doi.org/10.1371/journal.pone.0138327.
Wawrik B, Kerkhof L, Zylstra GJ, Kukor JJ. Identification of unique type II polyketide synthase genes in soil. Appl Environ Microbiol. 2005;71:2232–8. https://doi.org/10.1128/AEM.71.5.2232-2238.2005.
Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49:10–24. https://doi.org/10.1007/s00248-004-0249-6.
U.S. Department of Agriculture. Soil Survey Manual. Soil Survey Division Staff; Soil Conservation Service 1951, Volume Handbook 18.
Seufert V, Ramankutty N, Foley JA. Comparing the yields of organic and conventional agriculture. Nature. 2012;485:229–32. https://doi.org/10.1038/nature11069.
Fusaro S, Squartini A, Paoletti MG. Functional biodiversity, environmental sustainability and crop nutritional properties: a case study of horticultural crops in north-eastern Italy. Appl Soil Ecol. 2018;123:699–708. https://doi.org/10.1016/j.apsoil.2017.06.023.
Van Rijssel SQ, Veen GFC, Koorneef GJ, Bakx-Schotman JMTT, Hooven FCT, Geisen S, Van Der Putten WH. Soil microbial diversity and community composition during conversion from conventional to organic agriculture. Mol Ecol. 2022;31:4017–30. https://doi.org/10.1111/mec.16571.
Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol. 2003;69:1800–9. https://doi.org/10.1128/AEM.69.3.1800-1809.2003.
Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, Osbourn A, Grant A, Poole PS. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013;7:2248–58. https://doi.org/10.1038/ismej.2013.119.
Botnen S, Vik U, Carlsen T, Eidesen PB, Davey ML, Kauserud H. Low host specificity of root-associated fungi at an Arctic site. Mol Ecol. 2014;23:975–85. https://doi.org/10.1111/mec.12646.
Rosselli R, Fiamma M, Deligios M, Pintus G, Pellizzaro G, Canu A, Duce P, Squartini A, Muresu R, Cappuccinelli P. Microbial immigration across the Mediterranean via airborne dust. Sci Rep. 2015;5:16306. https://doi.org/10.1038/srep16306.
Buckley DH, Schmidt TM. The structure of microbial communities in soil and the lasting impact of cultivation. Microb Ecol. 2001;42:11–21. https://doi.org/10.1007/s002480000108.
Heino J, Grönroos M, Ilmonen J, Karhu T, Niva M, Paasivirta L. Environmental heterogeneity and β diversity of stream macroinvertebrate communities at intermediate spatial scales. Freshw Sci. 2013;32:142–54. https://doi.org/10.1899/12-083.1.
White JC, Hill MJ, Bickerton MA, Wood PJ. Macroinvertebrate taxonomic and functional trait compositions within lotic habitats affected by river restoration practices. Environ Manage. 2017;60:513–25. https://doi.org/10.1007/s00267-017-0889-1.
Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE. 2013;8:e57923. https://doi.org/10.1371/journal.pone.0057923.
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12(1):118–23. https://doi.org/10.1111/j.1462-2920.2009.02051.x.
Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61(7):2798–801. https://doi.org/10.1128/aem.61.7.2798-2801.1995.
Fraser DG, Doran JW, Sahs WW, Lesoing GW. Soil microbial populations and activities under conventional and organic management. J Environ Qual. 1988;17:585–90. https://doi.org/10.2134/jeq1988.00472425001700040011x.
Gunapala N, Scow KM. Dynamics of soil microbial biomass and activity in conventional and organic farming systems. Soil Biol Biochem. 1998;30:805–16. https://doi.org/10.1016/S0038-0717(97)00162-4.
Bossio DA, Scow KM, Gunapala N, Graham KJ. Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb Ecol. 1998;36:1–12. https://doi.org/10.1007/s002489900087.
Wang H, Chen D, Wu C, Gu L, Sun Z, Zhang S. Forest thinning alleviates the negative effects of precipitation reduction on soil microbial diversity and multifunctionality. Biol Fertil Soils. 2023. https://doi.org/10.1007/s00374-023-01716-6.
Acknowledgements
Not applicable.
Funding
The study was carried out within the Agritech National Research Center and received funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)—MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 17/06/2022, CN00000022). Our study represents a position paper related to (1) Spoke 1 “Plant and animal genetic resources and adaptation to climate changes” and a baseline for the fulfilment of the milestones within task 1.2.4 titled “Profiling plant-microbial associations and modulating these interactions by biostimulant treatments to enhance the ability of plants to cope with environmental stressors”. (2) Spoke 7 “Integrated models for the development of marginal areas to promote multifunctional production systems enhancing agroecological and socio-economic sustainability” and a baseline for the fulfilment of the milestones within Task 7.1.2 titled “Strategies for development of the agricultural and forestry systems, plant and animal biodiversity enhancement also at landscape level in marginal areas”. This manuscript reflects only the authors’ views and opinions, neither the European Union nor the European Commission can be considered responsible for them. This study was also funded by Veneto Region in the framework of the PSR 2014–2020 (Project: “Implementation and validation of innovative plant protection methods to increase the environmental sustainability of organic and sugar beet production”). Authors SR and MB were supported by Cariparo Foundation and PON Research &. Competitiveness MIUR-CUP C93H20000320007, respectively. This manuscript reflects only the authors’ views and opinions, neither the European Union nor the European Commission can be considered responsible for them.
Author information
Authors and Affiliations
Contributions
PS, GCa, and LM designed the study; LM, SD, and MCDL collected the samples; LM performed the experiment and collected the data; GCo classified the soil samples; LM, SD, SR, and MB analyzed the data; LM drafted the manuscript; all authors critically revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Author Giovanni Campagna is employed by COPROB.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Sugar beet productivity traits. Significant differences among the mean values in the vertical comparison were evaluated with the Student t-test followed by post hoc analysis (Duncan’s test). Significance was estimated at the p < 0.05 level.
Additional file 2: Figure S1.
Alpha-diversity rarefaction plot. The curve is based on the number of observed amplicon sequence variants (ASVs) as a function of the sequencing depth.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Maretto, L., Deb, S., Ravi, S. et al. 16S metabarcoding, total soil DNA content, and functional bacterial genes quantification to characterize soils under long-term organic and conventional farming systems. Chem. Biol. Technol. Agric. 10, 78 (2023). https://doi.org/10.1186/s40538-023-00450-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-023-00450-3