Abstract
Background
The antitoxin EndoAI is a TA system component that directly inhibits EndoA activity in vitro. The targeted activation of a TA system represents a potentially novel antimicrobial or antiviral strategy. However, whether the antitoxin functions alone and can induce plant disease resistance remain unknown.
Results
An endoAI was previously identified in the genome of Paenibacillus terrae NK3-4. It underwent a bioinformatics analysis, cloned and expressed in Escherichia coli. Then the functions of EndoAI inducing plant resistance to diseases as an elicitor were evaluated. The results showed that, EndoAI is a stable, alkaline, and hydrophilic protein, with a J-shaped three-dimensional structure in the absence of a ligand. It was clustered on the same branch with an antitoxin from Paenibacillus polymyxa SC2. Ectopically expressed EndoAI triggered a reactive oxygen species burst and a positive hypersensitive response (HR) in tobacco leaves. Moreover, 2 μmol EndoAI induced HR activity in tomato leaf, and it remained active after a 15-min exposure at 4–50 °C, and pH 6–8. Additionally, EndoAI induced plant systemic resistance against Alternaria alternata and tobacco mosaic virus, and the up-regulated transcription of PR genes, including PR1a, PR1b, PR5, PDF1.2, COL1, NPR1, and PAL.
Conclusions
These results imply that EndoAI may enhance the disease resistance of tobacco by promoting a series of early defense responses and up-regulating PR gene expression. These findings are relevant for future investigations on the mechanism underlying the EndoAI–plant interaction that leads to enhanced disease resistance. Furthermore, the endoAI may be useful for developing effective biocontrol agents to protect plants from diseases.
Graphical Abstract

Similar content being viewed by others
Introduction
Toxin–antitoxin (TA) systems are encoded in the plasmids and chromosomes of bacteria and archaea. Toxins and antitoxins are co-expressed to form a stable complex. Chromosomally encoded TA systems help regulate cell growth, thereby allowing cells to adapt to various environmental stresses [1, 2]. Some antitoxins bind to homologous toxin molecules to protect cellular targets, but also contain a DNA-binding domain, which enables them to bind to promoters near TA operons, typically as TA complexes [3, 4]. The TA systems have been classified into five groups (types I–V) [5], of which the type II TA system, in which antitoxins inhibit toxin functions via protein–protein interactions, has been the most extensively studied. The MazE–MazF complex is one of the most thoroughly characterized type II TA systems [6, 7]. Four MazE–MazF complexes in Mycobacterium tuberculosis arrest cell growth when expressed in Escherichia coli [8], whereas another four reportedly cleave mRNAs, some of which encode proteins associated with M. tuberculosis immunity and pathogenesis [9]. In addition to pathogens, MazE–MazF complexes have been identified in non-pathogenic microorganisms, including Bacillus subtilis [10].
The antitoxin EndoAI (toxin MazF complexed with the cognate antitoxin MazE in B. subtilis), which is a TA system component, directly inhibits EndoA activity in vitro [11, 12]. The targeted activation of a TA system, resulting in the release of free toxin proteins in response to pathogens, including viruses, represents a potentially novel antimicrobial or antiviral strategy [13]. Whether antitoxin functions alone can induce plant disease resistance remains unknown.
A gene encoding EndoAI was identified in the genome of Paenibacillus terrae NK3-4. NK3-4 was recently studied as a potential biocontrol bacterium useful for protecting plants from diseases [14]. However, there are no reports describing the functions of EndoAI in Paenibacillus strains, and it is unclear whether EndoAI contributes to the biocontrol effects of Paenibacillus species. The objectives of this study were to clone the endoAI in the P. terrae NK3-4 genome for the subsequent expression in Escherichia coli cells and to determine whether EndoAI functions as an elicitor to induce plant systemic resistance. The results presented herein may be useful for clarifying the mechanism by which Paenibacillus strains, including NK3-4, induce plant systemic resistance to diseases as well as the potential utility of EndoAI for controlling plant diseases.
Materials and methods
Materials
Nicotiana tabacum L. cv. Samsun NN and Nicotiana benthamiana LAB (susceptible tobacco phenotype to TMV) plants were cultivated in a phytotron with a 12-h light (25 °C)/12-h dark (18 °C) cycle for 6 weeks. Additionally, N. tabacum cells were cultured in Murashige and Skoog medium (4.2 g L−1) supplemented with 0.2% KH2PO4, 100 mg mL−1 inositol, 3% sucrose, 0.2 mg mL−1 2,4-dichlorophenoxyacetic acid, and 1 mg L−1 VB1 (pH 5.2) at 25 °C (150 rpm) in darkness for 7 days.
Paenibacillus terrae NK3-4 harboring the EndoAI gene was maintained in our laboratory. It was cultured in potato dextrose broth at 28 °C (180 rpm) in darkness for 24 h, after which genomic DNA was extracted from the bacterial cells. Tobacco mosaic virus (TMV-GFP) and Alternaria alternata, which is a fungal pathogen of tobacco, were maintained in our laboratory. The pET28a plasmid used for protein expression was kept in our laboratory, whereas competent E. coli Trans1-T1 and BL21 cells were purchased from TransGen Biotech Co., Ltd. (Beijing, China).
Bioinformatics analysis of endoAI
The endoAI sequence was obtained by sequencing the P. terrae NK3-4 genome. The physicochemical properties of the encoded EndoAI, including the molecular weight and isoelectric point (pI), were determined using the ExPASy Proteomics Server (https://web.expasy.org/protparam/). The conserved domain was identified by analyzing the amino acid sequence using the NCBI BLASTp tools. The protein secondary structure was analyzed with the SOPMA online software (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.htmL), whereas the three-dimensional (3D) structure was predicted using the SWISS-MODEL online tool (https://www.swissmodel.expasy.org/). The amino acid sequence was aligned to sequences in the UniProt database (https://www.uniprot.org/) using the BLAST tool. Additionally, a phylogenetic tree was constructed based on protein sequences highly homologous to EndoAI using the neighbor-joining method of the MEGA X program.
Cloning and expression of endoAI
Genomic DNA was isolated from P. terrae NK3-4 using a Bacterial Genomic DNA Extraction kit (Beijing Solarbio Technology Co., Ltd., Beijing, China), and the DNA quality was examined using a ultra-micro ultraviolet spectrophotometer (NanoDrop one, Thermo Fisher Scientific, Waltham, Massachusetts, USA), according to the method described in our previously published article [15]. The full-length EndoAI coding gene sequence was amplified by PCR using the following gene-specific primers (Fig. 1), which included BamHI (GGATCC) and XhoI (CTCGAG) restriction sites and protective bases. The PCR amplification was completed in a 50-μL solution comprising 25 μL 2 × HiFiMix I, 20 μL ddH2O, 2 μL DNA template, and 1 μL each primer (10 μM). The PCR program was as follows: 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 60.4 °C for 40 s, and 72 °C for 1.5 min; 72 °C for 10 min. The PCR product was maintained at 4 °C until it was analyzed by agarose gel electrophoresis.
Purification of the PCR product
The PCR product was purified from the agarose gel using the QIAquick Gel Extraction Kit (Qiagen, Frankfurt, Germany). The quality and concentration of the recovered DNA were determined, after which the DNA was stored at − 20 °C.
Insertion of endoAI into the pET28a vector
The EndoAI gene and the pET28a vector were separately digested with BamHI and XhoI in 50-μL solutions consisting of 43 μL EndoAI gene (approximately 50 ng) or pET28a vector (approximately 300 ng), 2 μL BamHI and XhoI (1 U each), and 5 μL 10 × CutSmart buffer. The samples were digested at 37 °C for 2 h, after which the digested products were analyzed by 1% agarose gel electrophoresis and purified using the QIAquick Gel Extraction Kit (Qiagen). The digested pET28a vector and EndoAI gene were ligated in a 10-μL solution containing 3.5 μL pET28a (100 ng), 3.5 μL EndoAI gene (17 ng), 2 μL 5 × ligase buffer, and 1 μL T4 ligase (1 U). The ligation was completed during a 10-min incubation at 25 °C.
Transformation of competent E. coli Trans1-T1 cells with the pET28a-endoAI recombinant plasmid
A 10-μL aliquot of the ligation product (pET28a-endoAI) was added to competent E. coli Trans1-T1 cells (100 μL) on ice. The cells were incubated on ice for 30 min, after which they were heated at 42 °C for 45 s and then maintained in an ice bath for 2 min. Next, 200 μL LB medium was added to the cells, which were incubated at 37 °C (200 rpm) for 1 h. The cell suspension was spread onto LB agar medium containing kanamycin (1 mg L−1) in Petri plates, which were incubated overnight at 37 °C. Colonies were randomly selected for colony PCR to identify positive clones. The PCR product was sequenced to confirm the cells were transformed with pET28a-endoAI and to verify that the EndoAI sequence was accurate. The cells were used to inoculate LB medium containing kanamycin (1 mg L−1). The pET28a-endoAI recombinant plasmid was extracted from the bacterial culture using the EasyPure® Plasmid MiniPrep Kit (Beijing Quansi Gold Biotechnology Co. Ltd., Beijing, China) for the subsequent transformation of competent E. coli BL21 cells.
Transformation of E. coli BL21 cells and heterologous expression of endoAI
Competent E. coli BL21 cells were transformed and positive clones were selected and analyzed as described for the E. coli Trans1-T1 cells. The correctly transformed E. coli BL21 cells were used to inoculate LB medium, which was then incubated at 37 °C (200 rpm) for 4 h or until the optical density at 595 nm (OD595) of the bacterial culture reached 0.6–0.8. To induce endoAI expression, 0.1 mM (final concentration) isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the culture, which was then incubated at 25 °C for 18 h. Cultures comprising E. coli BL21 cells transformed with pET28a alone and induced with IPTG and E. coli BL21 cells transformed with pET28a-EndoAI but not induced with IPTG were used as controls.
Purification of the expressed product
To purify the recombinant EndoAI, the transformed E. coli BL21 cells were harvested by centrifugation at 12,000g for 5 min at 4 °C, after which the cells were re-suspended in buffer (50 mM Tris–HCl and 200 mM NaCl, pH 7.5) and then disrupted ultrasonically. This step was repeated twice. The lysed cells were centrifuged at 12,000g for 30 min at 4 °C and the protein-containing supernatant was collected and analyzed by SDS-PAGE to detect EndoAI. The protein samples for the cells transformed with pET28a alone and induced with IPTG and the cells transformed with pET28a-EndoAI but not induced with IPTG were analyzed as controls. The expressed EndoAI was purified using Ni–NTA (Sangon Biotech Co., Ltd., Shanghai, China). After desalting via dialysis (2000 D) in 50 mM PBS (pH 7.5), the purified EndoAI protein was analyzed by SDS-PAGE, with the supernatant (i.e., non-purified proteins) as the control. Protein concentrations were determined using the Pierce® BCA Protein Assay Kit (Thermo, MA, USA). The purified protein samples were stored at − 80 °C.
Induction of the plant reactive oxygen species (ROS) accumulation and hypersensitive response (HR) by EndoAI
Induction of the oxidative burst and H 2 O 2 accumulation
Reactive oxygen species were qualitatively detected by 3,3′-diaminobenzidine (DAB) staining method [16]. Leaves of 6-week-old tobacco plants were infiltrated with EndoAI or bovine serum albumin (BSA; control) through the stomata. After 8 h, the leaves above the EndoAI-infiltrated and BSA-infiltrated leaves were collected and rinsed with ddH2O, dried, and immersed in 1 mg mL−1 DAB solution (pH 3.8). After an overnight incubation in darkness, the DAB solution was discarded and the leaves were decolorized in a solution comprising ethanol and glycerin [9:1 (v/v)] in a boiling water bath. After the chlorophyll was completely decolorized, the leaves were placed on separate glass slides and 75% glycerin was added dropwise to flatten the leaves, which were then examined with a microscope (1000× magnification). This analysis was repeated three times.
The accumulation of H2O2 in tobacco cells was quantitatively analyzed based on chemiluminescence using luminol and the GloMax-96 luminometer (Promega, Madison, WI, USA) [17, 18]. Briefly, tobacco suspension cells cultured for 7 days were harvested and re-suspended in hydroxyethyl piperazine ethylsulfonic acid buffer (10 mmol L−1 hydroxyethyl piperazine ethylsulfonic acid, 175 mmol·L−1 mannitol, 0.5 mmol·L−1 CaCl2, and 0.5 mmol·L−1 K2SO4, pH 5.75) and then incubated at 24 °C (150 rpm) for 1 h. After adding 50 μL 0.3 mmol L−1 luminol to the tobacco suspension cells, EndoAI or BSA (control) was added for a final concentration of 1 μmol L−1. The chemiluminescence intensity was measured using an enzyme labeling instrument Bio-Tek synergy 4 (Bio-Tek Instruments Co., LTD, Burlington, Vermont, USA). This analysis was repeated three times.
Induction of the HR in tobacco leaves
Six-week-old tobacco plants were used as the test plants. Specifically, HR test was conducted according a method described in a previously published article [14]. Briefly, 20 μL EndoAI solution (30 μmol mL−1) was infiltrated into one side of each leaf, whereas bovine serum albumin (BSA) was injected into the other side as a control treatment. Additionally, another tobacco leaf injected with BSA served as a control for the ROS burst test. Analyses were repeated three times. 48 h after infiltration, the HR response in leaves was observed.
To determine the EndoAI concentration required to induce the HR, 20 μL EndoAI at various concentrations (1, 2, 5, 10, 15, 20, and 30 μmol mL−1) was injected into tobacco leaves. Additionally, EndoAI solutions (30 μmol mL−1) were incubated at different temperatures (4, 30, 40, 50, 60, 80, or 100 °C) for 15 min to evaluate the effect of temperature on the ability of EndoAI to induce the HR. Furthermore, the pH of the protein solution was adjusted (4, 5, 6, 7, 8, 9, or 10). The HR was analyzed 48 h after the infiltrations. The experiments were conducted three times, with each as one replicate. The HR lesion area in leaf was measured using AutoCAD 2019.
Induction of tobacco resistance to Alternaria alternata and TMV by EndoAI
Alternaria alternata, which causes tobacco brown spot disease, was cultured on PDA medium in Petri plates at 28 °C for 7 days to allow the mycelia to cover the medium surface. Next, PDA plugs (6 mm diameter) containing mycelia were used to inoculate tobacco (N. tabacum) systemic leaves, which were collected from 6-week-old plants in which the fourth reciprocal leaf was infiltrated with 20 μL EndoAI solution (30 μmol mL−1) or BSA (control) 24 h before the inoculation with A. alternata. Three A. alternata-inoculated leaves were incubated at 28 °C for 5 days under humid conditions. The PDA plugs were removed and the incubation continued for an additional 3 days. Disease severity was assessed by measuring the lesion area. The fungal inhibition rate was calculated using the following formula:
where Ac and AT represent the lesion areas on the control and EndoAI-treated leaves, respectively.
The fourth reciprocal leaf of 6-week-old tobacco (N. benthamiana) plants was infiltrated with 20 μL EndoAI solution (30 μmol mL−1) or BSA (control). After 24 h, systemic leaves were inoculated with TMV-GFP. Briefly, leaves that were previously infected with TMV-GFP were ground to a homogenate in PBS (150 mmol L−1, pH 7.4; 15 mL g−1 leaves) and then centrifuged at 3600g at 4 °C for 5 min. The supernatant was collected and spread on systemic leaves to inoculate tobacco plants. The inoculated plants were incubated at 25 °C with a 12-h light/12-h dark cycle. The TMV-GFP lesions were counted under a UV lamp at 4 days after the TMV-GFP inoculation. Three leaves from three plants were examined. The viral inhibition rate was calculated using the following formula:
where Nc and NT represent the number of lesions on the control and EndoAI-treated leaves, respectively.
Analysis of defense gene transcription in EndoAI-treated tobacco leaves
The fourth reciprocal leaf of nine 6-week-old tobacco (N. tabacum) plants was infiltrated with 20 μL EndoAI solution (30 μmol mL−1) or BSA (control). After 0, 1, 2, 3, 4, and 5 days, total RNA was extracted from the systemic leaves of three EndoAI-treated and BSA-treated plants using the RNAprep Pure Plant Kit (TIANGEN Biotech, Beijing, China). First-strand cDNA was synthesized from 1.0 μg total RNA using the ReverTra Ace qPCR RT Master Mix with gDNA Remover [TOYOBO Biotech (Shanghai) Co., Ltd., Shanghai, China]. A quantitative real-time PCR (RT-qPCR) assay was performed using the IQ-5 real-time system (Bio-Rad Laboratories, Hercules, California, USA) and the ReverTra Ace qPCR RT Kit [TOYOBO Biotech (Shanghai) Co., Ltd.]. An N. tabacum actin gene (Accession: X63603.1, GI: 22608) was used as an internal reference. The relative gene expression levels were calculated using the 2−ΔΔCT method [19]. The analysis was repeated three times. Details regarding the primers are listed in Table 1.
Statistical analysis
The SPSS 13.0 software (Chicago, USA) was used to evaluate the significance of the difference between two variables (one-way ANOVA).
Results
Properties, structures, and phylogenetic relationships of EndoAI
The endoAI coding sequence was submitted to the GenBank database (Accession: MW603604). The EndoAI gene open reading frame was revealed to comprise 279 bp, encoding a protein consisting of 93 amino acids with a chemical formula of C456H750N136O141S7. The predicted molecular weight was 10,618.22 Da and the theoretical pI was 8.28. The extinction coefficient was 4470 M−1 cm−1. The grand average of hydropathicity index was − 0.435, the instability index was 39.79, and the aliphatic index was 83.98. These results indicate EndoAI is a stable, alkaline, and hydrophilic protein. The EndoAI sequence includes the HicB domain, similar to members of the antitoxin and NikR superfamilies. Modeling predictions revealed that EndoAI consists of two α-helices connected by a random coil, with an extended strand–random coil–beta turn–random coil–extended strand–random coil at the N-terminal of the short α-helix (Fig. 2A). The predicted 3D structure indicated the EndoAI amino acid sequence most closely matches a module protein named antitoxin EndoAI (crystal structure similar to that of the cognate antitoxin MazE in B. subtilis) in the SWISS-MODEL template library (version 2021-01-20; PDB release 2021-01-15). The sequence identity was 46.74%, and the similarity between the submitted protein sequence and the module was 0.42. On the basis of the predicted 3D structure, EndoAI is a J-shaped protein in the absence of a ligand. Moreover, both the N-terminal (positively charged) and C-terminal (negatively charged) are exposed (Fig. 2B).
Structural analysis of antitoxin EndoAI of in Paenibacillus terrae NK3-4. A Predicted EndoAI secondary structure. h: alpha helix; e: extended strand; c: random coil; t: beta turn. B Predicted EndoAI 3D structure. a and c present EndoAI; b and d present the calculated surface potential maps (blue, positive charge; orange, negative charge; green, neutral); c and d present the images of a and b rotated 180°, respectively
The constructed phylogenetic tree was separated into two sub-trees, with a big branch and a small branch. The P. terrae NK3-4 EndoAI (marked with red dots) was positioned at the end of the small branch, and was highly similar to an antitoxin from Paenibacillus polymyxa SC2 (sequence identity of 98.9%) (Fig. 3). The phylogenetic tree indicated the antitoxins are relatively conserved at the Paenibacillus genus level, with the exception of a CopG family transcriptional regulator/antitoxin EndoAI (A0A369BFZ7) in Fontibacillus phaseoli and a putative CopG family transcriptional regulator (D3EFV2) in Geobacillus sp. Y412MC10.
EndoAI gene was cloned and expressed in prokaryotic cells
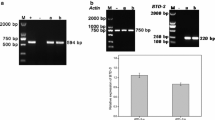
The full-length endoAI amplified from genomic DNA by PCR (Fig. 4A, B) was purified by extracting the band with the expected size from the agarose gel (Fig. 4C). The purified sequence was inserted into the pET28a vector (Fig. 4D). The resulting recombinant plasmid was used to transform E. coli BL21 cells (Fig. 4E). After inducing the production of EndoAI, the recombinant protein was extracted and purified using Ni–NTA and then desalted. The purified His-tagged protein was detected as a single band in an SDS-PAGE gel, with an apparent molecular mass of approximately 13 kDa (Fig. 5). This was consistent with the predicted size of EndoAI.
Electrophoretic analysis of the endoAI of Paenibacillus terrae NK3-4. A 1–2: Paenibacillus terrae NK3-4 genomic DNA. B 3–4: PCR amplification of the EndoAI gene. C 5: recovered EndoAI gene from the agarose gel. D 6–8: vector preparation. E 9–10: PCR amplification of the EndoAI gene in transformed E. coli BL21 cells; M: marker (bp)
SDS-PAGE analysis of the expressed and purified EndoAI. M: protein molecular weight marker; 1, 2, and 5: crude extracts of cells harboring pET28a-endoAI with IPTG induction; 3: crude extracts of cells harboring pET28a-endoAI without IPTG induction; 4: crude extracts of cells harboring pET28a with IPTG induction; 6: protein purified from the crude extracts presented in 1 and 2 using Ni–NTA Sepharose
EndoAI-induced HR and ROS accumulation in tobacco leaves
At 24 h after tobacco leaves were infiltrated with EndoAI, clearly defined HR necrotic areas were detected in the infiltrated area, whereas chlorotic lesions were observed outside of the infiltrated area (Fig. 6A). At 5 days after the infiltration, the chlorosis had extended to the leaf edge (Fig. 6B). The BSA injected into the other side of the leaf did not induce any detectable HR (Fig. 6A, B).
Hypersensitive response (HR) and reactive oxygen species (ROS) accumulation in tobacco leaves induced by EndoAI. A, B HR in tobacco leaves at 24 h and 5 days post-infiltration with EndoAI, respectively. C, D Bleached systemic leaves from EndoAI-treated and control plants, respectively. E, F Microscopy images of EndoAI-treated and control systemic leaves, respectively. G The chemiluminescence intensity after EndoAI treatment which reflected the ROS content, values represent mean ± standard errors of three independent assays
The DAB tissue staining analysis revealed significant brown DAB-stained precipitates in the EndoAI-treated tobacco leaves at 5 h post-infiltration (Fig. 6C, E), whereas there were no obvious stained precipitates in the BSA-treated (control) leaves (Fig. 6D, F). The EndoAI-induced H2O2 production in tobacco cells was quantitatively analyzed based on chemiluminescence. Following the treatment of tobacco suspension cells with EndoAI, the chemiluminescence intensity, which reflected the ROS content, increased rapidly after 20 min and peaked at 30 min, after which it decreased, reaching baseline levels at 55 min. In contrast, the ROS level in the control cells was relatively unchanged at all time-points (Fig. 6G). These results suggested that EndoAI induced the ROS burst in tobacco cells.
Leaves were also infiltrated with serially diluted recombinant EndoAI solutions to analyze the HR. The results indicated that the minimum concentration of EndoAI inducing HR was 1 μmol mL−1 and 2 μmol mL−1, and 5 μmol mL−1 induced clear HR, while 30 μmol mL−1 had the largest HR lesion area in leaf (Fig. 7A, D). EndoAI retained its ability to induce clear HR in leaves after a 15-min exposure at 4, 30, 40, and 50 °C (Fig. 7B, E). The EndoAI was able to induce a distinct HR at pH 6–8 (Fig. 7C, F).
EndoAI-induced plant systemic resistance to Alternaria alternata and tobacco mosaic virus (TMV)
At seven days after plants were infected with A. alternata, the leaf lesion area was 2.0 cm2 for the EndoAI-treated tobacco (N. tabacum) plants. In contrast, the control plants infected with A. alternata had a leaf lesion area of 4.0 cm2, and the leaf tissue surrounding the lesions was yellow. Thus, the fungal inhibition rate for EndoAI was 50.1% (Fig. 8A–C).
Induction of tobacco resistance to Alternaria alternata (A–C) and tobacco mosaic virus (TMV) by EndoAI (D–F). A–C Nicotiana tabacum. D–F Nicotiana benthamiana. B, E BSA-treated leaves (control). C, F EndoAI-treated leaves. a and b in B: 0 h and 7 days after the Alternaria alternata inoculation, respectively. a and b in C: 0 h and 3 days after the TMV-GFP inoculation, respectively. Values represent mean ± standard errors of three independent assays, ** represents difference between columns was significant at P = 0.01 level (A)
At three days after the infection with TMV, the EndoAI-treated tobacco (N. benthamiana) plants had 21.0 lesions per leaf, which was fewer than the 45.3 lesions per leaf of the control plants. The viral inhibition rate for EndoAI was 53.3% (Fig. 8D–F).
EndoAI-induced up-regulated expression of pathogenesis-related (PR) genes
The expression levels of PR genes PR1a, PR1b, PR5, and PAL, all of which are marker genes for the salicylic acid (SA)-dependent defense pathway, were significantly up-regulated in response to EndoAI. However, the expression levels decreased after day 3 or 4 (post-treatment) (Fig. 9). More specifically, the PR1a and PR1b transcription levels were, respectively, 18.5-times and 15.7-times higher in the EndoAI-treated plants than in the control plants on day 3. The expression of PR5 peaked on day 4, with a transcript level that was 9.6-times higher than that of the control plants. The PAL transcription level increased significantly after the EndoAI treatment and remained high for the duration of the test period.
Transcription levels of pathogenesis-related genes after the EndoAI treatment. At each time-point, the transcription levels of normally growing plants were set as the baseline. 0 d, 1 d, 2 d, 3 d, 4 d, and 5 d represent the days after the treatment of EndoAI. Values represent mean ± standard errors of three independent assays. * and ** represent differences were significant at P = 0.05 level and P = 0.01 level, respectively
The PDF1.2 transcription level was 12.7-times higher in the EndoAI-treated plants than in the control plants on day 3. The expression of NPR1 increased rapidly from day 1 to day 4, which is required for the activation of PR gene expression. Furthermore, EndoAI triggered the up-regulated expression of the coronatine insensitive 1 gene (COI1), with transcript levels that were 19.8-times higher in the EndoAI-treated plants than in the control plants on day 4.
Discussion
Characterizing and developing new elicitors is a promising strategy for generating viable alternatives to conventional biocides used for plant disease management. More and more protein elicitors that enhance plant disease resistance have been identified, and the underlying mechanisms have been investigated in depth. Systemic acquired resistance induced by elicitors protects plants from invading pathogens is one of the most recognized mechanisms [20,21,22,23]. Previous studies have explored protein elicitors from plant growth-promoting rhizobacteria, such as Bacillus amyloliquefaciens [18], P. polymyxa [24], and B. subtilis [25], but there are no reports describing research focused on protein elicitors from P. terrae. In this study, the elicitor activity of the antitoxin EndoAI from P. terrae NK3-4 was demonstrated. It induced the HR, ROS burst, and PR gene expression in the treated tobacco plants, which are features of induced systemic resistance.
The HR contributes to plant innate immunity [26]. The effects of the HR on stress resistance-related gene expression as well as the association between the HR and death-related signals have been elucidated, such as calcium, ROS, NO, SA, and sphingolipids [27]. One of the early events during the HR is the generation of ROS that activate downstream cellular processes [28]. The ROS burst is the main source of ROS in plants in response to pathogens or pathogen-derived elicitors, which are also known as PAMPs [29]. The ROS regulate multiple cellular activities in plants, including redox reactions, that directly affect specific transcription factors and antimicrobial properties [30, 31]. For example, ROS metabolism may provide plant cells with the redox conditions required for regulating the function of NPR1. The expression of NPR1 was up-regulated by EndoAI in this study (Fig. 9). The importance of NPR1 as a mediator of systemic plant responses to disease was confirmed in an earlier study [32]. Reactive oxygen species can also activate defense gene expression, leading to the formation of defense-related papillae and the production of antimicrobial secondary metabolites, which protect plants from pathogens [26, 33, 34]. Accordingly, ROS, which accumulated in EndoAI-treated tobacco plants (Fig. 6), induce plant resistance through diverse pathways. For example, ROS strengthen host cell walls via cross-linking glycoproteins, while also functioning as intercellular or intracellular signaling molecules that interact with various molecules, including lipids, Ca2+, NO, mitogen-activated protein kinases, and wounding-induced protein kinases. These signaling molecules are involved in defense signaling pathways [35, 36].
There is evidence that crosstalk among the SA, jasmonic acid (JA), and ethylene (ET) pathways modulates plant defense responses to various pathogens [37]. In this study, we proved that EndoAI enhances the transcription of several SA-responsive genes, including PR1a, PR1b, PR5, and PAL, and JA/ET-responsive genes, including PDF1.2 and COI1, as well as NPR1 (Fig. 9). Of these genes, COI1 encodes a protein that is essential for SA-mediated systemic acquired resistance and an important contributor to rhizobacterium-triggered induced systemic resistance [38]. Several studies confirmed NPR1 is required for the induction of PR gene expression because of its interactions with TGA transcription factors [39,40,41]. Moreover, it differentially regulates defense responses, depending on the signals that are elicited during the induction of disease resistance [42]. The PDF1.2 gene encodes a phytoalexin that mediates plant resistance to fungi, viruses, and environmental stresses [43]. Additionally, accumulating secondary metabolites can enhance plant defenses against pathogens by establishing mechanical barriers. The PAL enzyme is a key component of the phenylpropanoid metabolic pathway, wherein it synthesizes various phenolic compounds related to plant disease resistance [44]. Thaumatin-like proteins, such as PR5, are associated with plant responses to many biotic and abiotic stresses. They can enhance plant resistance to fungal pathogens [45] including A. alternata [46]. A recent study confirmed the interaction between PR5 and PR1 influences wheat defense responses to the leaf rust fungus [47]. Another study demonstrated that PR1 (subtilisin-like protease isoform) can mediate defense activities by releasing C-terminal peptides, which are targeted by fungal pathogen effectors [48]. The expression of signaling-related genes (e.g., NPR1 and PR1) is reportedly up-regulated in Brassica napus plants infected with Xanthomonas campestris pv. Campestris [49]. These PR genes play important roles in SA-dependent and JA/ET-dependent pathways, which may be involved in the EndoAI-induced systemic defense responses in tobacco.
Not all PR genes up-regulated expression in tolerant plants suffering biological and abiotic stresses, and different PR genes are differently expressed in various plants in response to same or different stress, although many PR genes are essential for plant defense. One example showed that one PR gene LsHPL1 was not up-regulated expressed in tolerant Jahrom lettuce accessions compared with susceptible Mazandaran line 1 by the infection of Sclerotinia sclerotiorum in Lactuca sativa–Sclerotinia sclerotiorum interaction [50]; another example showed that PR1 was not up-regulated expressed in one tolerant wheat (Triticum aestivum) phenotype and all of the susceptible wheat phenotype which involved in defense against Puccinia striiformis f [51]. It can be seen that the mechanisms of plant confronting with stress are diverse, and the mechanisms of elicitors activating plant defense are also different because they trigger different signal pathway among which different PR genes are involved [31, 35, 52, 53]. In this study, we confirmed the function of EndoAI in inducing tobacco disease resistance, although we examined only a few PR genes.
Conclusions
In this study, we analyzed the antitoxin EndoAI, which is encoded by genes in Paenibacillus species, including P. terrae NK3-4. The functional analysis of P. terrae NK3-4 EndoAI revealed it is an elicitor that activates early defense-related events in plants, including the HR and ROS burst. Furthermore, EndoAI treatments up-regulated the expression of PR genes and enhanced the resistance of tobacco plants to TMV and A. alternata. These findings may be useful for elucidating the mechanisms underlying the inductive effects of EndoAI on plant disease resistance. The data presented herein are relevant for the development of new biocontrol agents to protect agriculturally important plants from pathogens.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- A. alternate :
-
Alternaria alternata
- BSA:
-
Bovine serum albumin
- 3D structure:
-
Three-dimensional structure
- DAB:
-
3,3′-Diaminobenzidine
- ET:
-
Ethylene
- GFP:
-
Green fluorescent protein
- H2O2 :
-
Hydrogen peroxide
- HR:
-
Hypersensitive response
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- JA:
-
Jasmonic acid
- rpm:
-
Revolutions per minute
- PDB:
-
Program database
- P. terrae :
-
Paenibacillus terrae
- PAL:
-
Phenylalanine ammonia lyase
- PR gene:
-
Pathogenesis-related gene
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- SA:
-
Salicylic acid
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- ROS:
-
Reactive oxygen species
- TA system:
-
Toxin–antitoxin system
- TMV:
-
Tobacco mosaic virus
- PAMPs:
-
Pathogen-associated molecular pattern molecules
References
Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79.
Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol. 2011;9:779–90.
Brown BL, Wood TK, Peti W, Page R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J Biol Chem. 2011;286:2285–96.
Maté MJ, Vincentelli R, Foos N, Raoult D, Cambillau C, Ortiz-Lombardía M. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Res. 2012;40:3245–58.
Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett. 2013;340:73–85.
Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–23.
Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–53.
Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, et al. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem. 2006;281:18638–43.
Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol. 2008;69:559–69.
Park JH, Yamaguchi Y, Inouye M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. Febs Lett. 2011;585:2526–32.
Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol. 2005;56:1139–48.
Simanshu DK, Yamaguchi Y, Park J, Inouye M, Patel DJ. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol Cell. 2013;52:447–58.
Harms A, Brodersen DE, Mitarai N, Gerdes K. Toxins, Targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell. 2018;70:768–84.
Yu WQ, Zheng GP, Qiu DW, Yan FC, Liu WZ, Liu WX. Biocontrol of rice blast by Paenibacillus terrae NK3-4, a β-1, 3-glucanase producing strain. Biol Control. 2019;129:92–101.
Yu WQ, Wang X, Tang YC, Yan FC, Liu WZ, Zheng GP, et al. Cloning and expression of the EsxA gene and the growth-promoting effects of the encoded protein on rice seedlings. AMB Expr. 2021;11:72. https://doi.org/10.21203/rs.3.rs-97808/v1.
Frías M, Gonzalez C, Brito N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011;192:483–95.
Chen M, Zeng H, Qiu D, Guo L, Yang X, Shi H, et al. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE. 2012;7:e37654.
Wang N, Liu M, Guo L, Yang XY, Qiu D. A novel protein elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 induces systemic resistance in tobacco. Int J Biol Sci. 2016;12:757–67.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Wang Y, Xu Y, Sun Y, Wang H, Qi J, Wan B, et al. Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nature Commun. 2018;9:594.
Li L, Wang S, Yang X, Francis F, Qiu D. Protein elicitor PeaT1 enhanced resistance against aphid (Sitobion avenae) in wheat. Pest Manag Sci. 2020;76:236–43.
Cheplick S, Sarkar D, Bhowmik PC, Shetty K. Improved resilience and metabolic response of transplanted blackberry plugs using chitosan oligosaccharide elicitor treatment. Can J Plant Sci. 2018;98:717–31.
Jisha S, Gouri PR, Anith KN, Sabu KK. Piriformospora indica cell wall extract as the best elicitor for asiaticoside production in Centella asiatica (L.) Urban, evidenced by morphological, physiological and molecular analyses. Plant Physiol Bioch. 2018;125:106–15.
Lee SH, Cho YE, Park SH, Balaraju K, Park JW, Lee SW, et al. Fusaricidin-a cyclic depsipeptide from Paenibacillus polymyxa E681 as an elicitor of induced systemic resistance against phytophthora blight of red-pepper. Phytoparasitica. 2013;41:49–58.
Shen Y, Li J, Xiang J, Wang J, Liu Q. Isolation and identification of a novel protein elicitor from a Bacillus subtilis strain bu412. AMB Expr. 2019;9:117. https://doi.org/10.1186/s13568-019-0822-5.
Atkinson MM, Keppler LD, Orlandi EW, Mischke BCF. Involvement of plasma membrane calcium influx in bacterial induction of the K+/H+ and hypersensitive responses in tobacco. Plant Physiol. 1990;92:215–21.
Mur L, Kenton P, Lloyd A, Ougham H, Prats E. The hypersensitive response, The centenary is upon us but how much do we know? J Exp Bot. 2008;59:501–20.
Torres MA, Jones JD, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141:373–8.
Torres MA. ROS in biotic interactions. Physiol Plantarum. 2009;138:414–29.
Loon L, Bakker P, Heijdt W, Wendehenne D, Alain P. Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. MPMI. 2009;21:1609–21.
Zhao J, Davis L, Verpoorte R. Elicitor signal transduction leading to the production of plant secondary metabolites. Biotechnol Adv. 2005;23:283–333.
Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–44.
Bradley DJ, Kjellbom P, Lamb CJ. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30.
Hammond-Kosack K, Parker J. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotech. 2003;14:177–93.
Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Pugin A. Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe In. 2006;19:711–24.
Srivastava N, Gonugunta VK, Puli MR, Raghavendra AS. Nitric oxide production occurs downstream of reactive oxygen species in guard cells during stomatal closure induced by chitosan in abaxial epidermis of Pisum sativu. Planta. 2009;229:757–65.
Moon J, Park JM. Cross-talk in viral defense signaling in plants. Front Microbiol. 2016;7:2068.
Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–52.
Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–50.
Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. MPMI. 2000;13:191–202.
Kim HS, Delaney TP. Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J. 2002;32:151–63.
Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–80.
Liu ZX, Zhou Z, Cai CW, Cheng J, Ren CM. Construction and transformation of recombinant plasmid of plant defensins gene PDF1.2 promoter and GUS gene in Arabidopsis thaliana. Crop Res. 2018;32:131–4 (Chinese article with an English abstract).
Hano C, Addi M, Fliniaux O, Bensaddek L, Duverger E, Mesnard F, et al. Molecular characterization of cell death induced by a compatible interaction between Fusarium oxysporum f. sp linii and flax (Linum usitatissimum) cells. Plant Physiol Bioch. 2008;46:590–600.
Jesús-Pires CD, Ferreira-Neto JRC, Bezerra-Neto JP, Kido EA, Silva RLDO, Pandolfi V, et al. Plant thaumatin-like proteins: function, evolution and biotechnological applications. Curr Protein Pept Sc. 2020;21:36–51.
Velazhahan R, Muthukrishnan S. Transgenic tobacco plants constitutively overexpressing a rice thaumatin-like protein (PR-5) show enhanced resistance to Alternaria alternata. Biol Plantarum. 2003;47:347–54.
Wang F, Yuan S, Wu W, Yang Y, Liu D. TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PLoS Genet. 2020;16:e1008713.
Sung YC, Outram M, Breen S, Wang C, Dagvadorj B, Kobe B, et al. PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 2021. https://doi.org/10.1111/nph.17128.
Islam MT, Mamun MA, Lee BR, La HV, Jung WJ, Bae DW, et al. Role of salicylic acid signaling in the biotrophy-necrotrophy transition of Xanthomonas campestris pv. campestris infection in Brassica napus. Physiol Mol Plant Pathol. 2021;113:101578. https://doi.org/10.1016/j.pmpp.2020.101578.
Almatwari A, Hassandokht M, Soltani F, Gohari AM, Javan-Nikkhah M. Temporal expression profiles of defense-related genes involved in Lactuca sativa-Sclerotinia sclerotiorum interactions. J Plant Pathol. 2021;103:61–9.
Esmail SM, Aboulila AA, El-Moneim DA. Variation in several pathogenesis-related (PR) protein genes in wheat (Triticum aestivum) involved in defense against Puccinia striiformis f. sp. tritici. Physiol Mol Plant Pathol. 2020;112:101545. https://doi.org/10.1016/j.pmpp.2020.101545.
Liu Q, Shen Y, Yin K. The antimicrobial activity of protein elicitor AMEP412 against Streptomyces scabiei. J Microbiol Biotechn. 2020;36:18. https://doi.org/10.1007/s11274-019-2794-7.
Li S, Nie H, Qiu D, Shi M, Yuan Q. A novel protein elicitor PeFOC1 from Fusarium oxysporum triggers defense response and systemic resistance in tobacco. Biochem Bioph Res Co. 2019. https://doi.org/10.1016/j.bbrc.2019.05.018.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (32060633), the Natural Science Foundation of Heilongjiang Province, China (LH2021C086), the China Postdoctoral Foundation (2021T140187), and the Innovation and Entrepreneurship Training Program for college Students in Jiangxi Province, China (S202110416007).
Author information
Authors and Affiliations
Contributions
BWY: performed experiments, writing–original draft preparation, data analysis, methodology; WZL: contributed reagents and materials, data acquisition, article writing and later modification; WQY: performed experiments, writing–original draft preparation, data analysis, funding acquisition, resources, writing–review, editing, and revised the manuscript; PL: supervision, validation; GJW, contributed analysis tools, LGZ: provided suggestions on the experimental design; FCY: project administration, supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yan, B.W., Liu, W.Z., Yu, W.Q. et al. Antitoxin EndoAI can induce disease resistance in tobacco as a protein elicitor. Chem. Biol. Technol. Agric. 8, 66 (2021). https://doi.org/10.1186/s40538-021-00271-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-021-00271-2