Abstract
Background
Early diagnosis of esophageal squamous cell carcinoma (ESCC) remains a challenge due to the lack of specific blood biomarkers. We aimed to develop a serum multi-protein signature for the early detection of ESCC.
Methods
We selected 70 healthy controls, 30 precancerous patients, 60 stage I patients, 70 stage II patients and 70 stage III/IV ESCC patients from a completed ESCC case-control study in a high-risk area of China. Olink Multiplex Oncology II targeted proteomics panel was used to simultaneously detect the levels of 92 cancer-related proteins in serum using proximity extension assay.
Results
We found that 10 upregulated and 13 downregulated protein biomarkers in serum could distinguish the early-stage ESCC from healthy controls, which were validated by the significant dose-response relationships with ESCC pathological progression. Applying least absolute shrinkage and selection operator (LASSO) regression and backward elimination algorithm, ANXA1 (annexin A1), hK8 (kallikrein-8), hK14 (kallikrein-14), VIM (vimentin), and RSPO3 (R-spondin-3) were kept in the final model to discriminate early ESCC cases from healthy controls with an area under curve (AUC) of 0.936 (95% confidence interval: 0.899 ~ 0.973). The average accuracy rates of the five-protein classifier were 0.861 and 0.825 in training and test data by five-fold cross-validation.
Conclusions
Our study suggested that a combination of ANXA1, hK8, hK14, VIM and RSPO3 serum proteins could be considered as a potential tool for screening and early diagnosis of ESCC, especially with the establishment of a three-level hierarchical screening strategy for ESCC control.
Similar content being viewed by others
Background
The International Agency for Research on Cancer estimated that there were 572,034 new esophageal cancer cases and 508,585 deaths from esophageal cancer worldwide in 2018, and the mortality-to-incidence ratios in most countries were more than 0.8 [1]. The 5-year overall survival rate of esophageal cancer ranges from 15 to 25% because most patients are diagnosed at an advanced stage with a dismal prognosis [2, 3]. Evidence demonstrates that the population-based screening programs for upper gastrointestinal cancers in East Asia could efficiently identify precancerous lesions and early cancers which leads to improved prognosis due to timely treatment [4, 5]. The government-sponsored endoscopic screening program is also conducted for asymptomatic adults in high incidence area for esophageal cancer in China, but the ongoing program introduced a large burden for public health: only a small part of residents could participate in the gastroscopy screening program and even among this small proportion of population long-term follow-up for high-risk subjects is becoming increasingly burdensome as regards endoscopic management [6]. Thus, a cost-effective and fast blood-based screening test (liquid biopsy) is an ideal solution for risk stratification in order to identify a truly high-risk population for endoscopy [7].
At present, various blood biomarkers including mutations and methylation status in cell-free DNA, cell-free RNA, noncoding RNAs, proteins, and so on, have been explored to fulfill the purpose of early detection of multiple cancer types via different detection platforms [8,9,10,11]. Esophageal squamous cell carcinoma (ESCC) is the dominant histological subtype of esophageal cancer (> 80% proportion), especially in Asia and Eastern African, [12] showing contrasting risk factors and molecular features with esophageal adenocarcinoma. To date, few effective biomarkers for screening early ESCC are established in clinical applications, because the area under curve (AUC) of most potential biomarkers is usually lower than 80% [8, 13].
In order to identify novel protein biomarkers, the development of proximity extension assays (PEA) has enabled simultaneous quantification of multiple targeted protein biomarkers for a bunch of samples in every experiment, thereby enabling quick screening of possible biomarkers. PEA innovatively combines the specificity of antibody-linked detection methods with the sensitivity of the polymerase chain reaction (PCR), permitting multiplex biomarker detection and quantification with reliable assay precision using only microliter quantities of sera [14]. Based on a prospectively designed population-based case-control study of upper gastrointestinal cancer in Taixing (with a population of about 1.13 million), a high-incidence area in China, this study applied the PEA technology to identify candidate serum protein biomarkers for early-stage ESCC.
Methods
Study design and participants
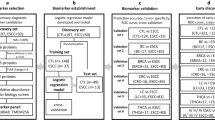
The research design of this large population-based case-control study has been delineated in our previous studies [15,16,17,18]. In brief, we attempted to recruit all newly diagnosed esophageal cancer cases from October 2010 to September 2013 in Taixing, and the inclusion criteria were limited to 40–85 year-old participants who had lived in Taixing at least 5 years. In the endoscopic units of the local four largest hospitals (covering almost 90% of local clinical diagnoses), the participants were invited to complete a questionnaire by trained interviewers and provided biological samples, if they were suspected of having an upper gastrointestinal tumor. Moreover, we further enrolled missed esophageal cancer patients in the endoscopy units by matching with the local Cancer Registry system. We finally recruited 1401 suspected esophageal cases from the hospitals’ endoscopy units and 280 reported esophageal cases via the local Cancer Registry system during 3 y. After reviewing the pathological sections and surgical pathology reports for those without pathological sections, 33 patients with precancerous lesions (high grade intraepithelial neoplasia, in-situ carcinoma, and high grade dysplasia) and 1418 ESCC patients were included in this study. Through evaluating the tumor stage of the 1418 ESCC patients via inpatient medical records based on the American Joint Commission on Cancer Staging Manual, 8th edition [19], we found additional 4 precancerous patients (reclassified from ESCC), 84 patients with stage I, 333 patients with stage II, 158 patients with stage III, 145 patients with stage IV and remaining 694 patients with unknown tumor stage. During the same period, we applied a frequency-matched method by sex and 5-year age groups to select control participants for the cases of upper gastrointestinal cancers. Finally, 1992 eligible controls participated in our study (participation rate: 70.4%).
The significant level of the hypothesis test was set as 0.001 for 92 proteins and the statistical power was set as 90%. As this study was dedicated to identifying high-efficiency serum protein biomarkers, it was estimated that the difference for significant biomarkers between patient group and control group should be at least 0.8 times of the standard deviation. The sample size of each group was calculated as 69, and we planned to select 70 subjects for each group.
For this study, we further limited suitable blood samples as those collected before clinical treatment and without moderate hemolysis. After excluding hemolytic samples and samples after treatment, the remaining 30 precancerous patients and 60 stage I patients were included. We then first randomly selected 70 patients from stage II ESCC patients, and randomly selected 70 advanced patients (stage III or stage IV) and 70 healthy controls by matching sex and 5-year age groups with stage II ESCC patients. If the sample size of patients in an age group was insufficient, it was supplemented from an adjacent age group.
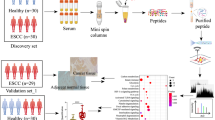
We finally enrolled 30 precancerous patients, 60 stage I patients, 70 stage II patients, 70 stage III/IV patients and 70 healthy controls in the biomarker study (Fig. 1). The early-stage ESCC was defined as precancerous lesions and stage I cancer in our study because of mini-invasive treatment, better prognosis and small sample size, and early screening requirement in community-based practice [20]. Without external validation, we performed a dose-response relationship between serum protein levels and ESCC pathological progression to further illustrate the reliability of the identified biomarkers.
Olink multiplex oncology II targeted proteomics panel
Serum proteins were analyzed using Olink Oncology II 96-well in which 92 oligonucleotide-labeled antibody probe pairs bind to their specific targeted proteins based on PEA technology [14, 21]. The precision, reproducibility and scalability of the PEA assay have been documented by the manufacturer (http://www.olink.com) and relevant articles [14, 21]. The protein names, gene names, and abbreviations for the 92 proteins of the Olink Oncology II panel are delineated in Table S1.
Sample processing and detection
Blood samples have been stored in the − 80 °C refrigerator before shipment. The serum samples were shipped to Olink Proteomics AB (Uppsala, Sweden) using cold chains and the samples were randomly placed in four 96-well plates. On each plate, we included three “Inter-plate controls” for data normalization between plates and three “Negative controls” to establish background levels. Data generated from the plates were analyzed, including normalization and linearization, per manufacturer’s protocol. The protein levels were expressed as Normalized Protein eXpression (NPX) values, a relative quantification on a log scale, which are cycle threshold values normalized by the subtraction of values for the extension control. All assay characteristics including detection limits and measurements of assay performance and validation are available from the manufacturer’s website (http://www.olink.com/products/).
Statistical methods
Chi-squared test or one-way ANOVA test were performed for testing the difference of the distributions of categorical or continuous variables in subgroups. An exploratory multivariate analysis (principal component analysis, PCA) was applied to test for potential clustering of study groups. The association between each protein NPX value and early ESCC was investigated using unconditional logistic regression, and the P value was adjusted by the Benjamini-Hochberg method for controlling the false discovery rate (FDR < 0.01). For the potential protein biomarkers, we further applied Spearman correlation to assess the dose-response relationship between protein levels and stages of ESCC, and the P value was also adjusted by the Benjamini-Hochberg method. For all preliminarily verified proteins, unsupervised clustering methods were applied to the data to identify clusters of proteins and visually evaluate their association with disease status. The Protein-Protein Interactions Network analysis of identified proteins was performed using the STRING database (https://string-db.org/). The online ConsensusPathDB-human interaction network database (http://cpdb.molgen.mpg.de/) was used for gene ontology (GO) enrichment analysis and pathway enrichment analysis of identified protein biomarkers. Three GO enrichment categories were checked, i.e. biological process, cellular component and molecular function.
For developing a multi-biomarker classifier to discriminate early ESCC cases from healthy controls, we used the least absolute shrinkage and selection operator (LASSO) regression to select optimal proteins. Moreover, we further used the backward elimination logistic regression model to build a more concise and efficient classification model. The specificity and sensitivity of the classifier were evaluated using the receiver operating characteristic (ROC) curve and the optimal cutoff points were selected using Youden’s index, which maximizes the sum of sensitivity and specificity. The AUC was applied to summarize the classification accuracy of diagnostic models and 95% confidence intervals (CI) were estimated by the non-parametric bootstrap. Five-fold cross-validation was used to estimate the validity of our multiple-protein model on the same data that was used to build the classifier. All statistical analyses and figure drawing were conducted using R (version 3.6.2).
Results
Patient overview and assay performance characteristics
The age and gender distribution were homogeneous among healthy controls and four groups of ESCC cases with different cancer stages in Table S2. The average intra-assay and inter-assay coefficient of variation (CV) based on quality control samples were 5 and 23% across the four plates, respectively.
Prinicipal component analysis
The results from principal component analysis of 70 healthy controls, 30 precancerous patients, 60 stage I patients, 70 stage II patients and 70 stage III/IV patients are shown in Fig. 2a. The numbers on the axes represent the variation captured by each principal component. The levels of 92 serum proteins were explained 40.2% by the first two principal components (PC1 27.9%, PC2 12.3%, respectively), and healthy controls were separated from ESCC patients with PC2. Thus, the PC2 distribution across various groups is illustrated in Fig. 2b, and ANOVA analysis found a significant difference among five groups (P = 2.5e-10) and pairwise comparison showed the healthy controls were significantly distinct from each ESCC group.
Evaluation of diagnostic efficiency of each protein
To identify potential protein biomarkers of early ESCC, the P value adjusted by FDR (Q value) and AUC of each protein for distinguishing healthy controls from early ESCC cases are shown in Fig. 3. According to the criterion of Q value less than 0.01, 26 potential proteins were discerned preliminarily, namely (sorting by Q value), ANXA1, hK8, CDKN1A, ABL1, SCAMP3, EGF, LYN, MetAP2, PVRL4, KLK13, ADAMTS15, hK14, VIM, TXLNA, GPC1, RSPO3, hK11, TRAIL, 5NT, CPE, FADD, TGFR2, SEZ6L, CD160, FCRLB, and ESM 1. The largest and smallest AUC of the 26 proteins were 0.770 for ANXA1 and 0.652 for ESM 1, respectively.
For assessing the dose-response relationship between the levels of protein and the progression from healthy controls to advanced ESCC, the violin plots of different groups and P value of Spearman correlation adjusted by FDR (Q value) for these 26 proteins are displayed in Fig. 4. According to the criterion of Q values less than 0.01, remaining 10 upregulated (ANXA1, CDKN1A, ABL1, SCAMP3, EGF, LYN, MetAP2, VIM, TXLNA and FADD) and 13 downregulated (hK8, KLK13, ADAMTS15, hK14, GPC1, RSPO3, hK11, TRAIL, 5NT, CPE, TGFR2, SEZ6L and CD160) protein biomarkers in serum were authenticated as potential ESCC biomarkers.
Protein interaction network and GO enrichment analyses
The protein interaction network analysis (Fig. S1) of 23 preliminarily authenticated proteins showed that 5NT, ABL1, ANXA1, CDKN1A, EGF, GPC1, hK11, hK14, hK8, KLK13, LYN, TGFR2, and VIM shared potential interactions. GO enrichment analysis further revealed that signaling receptor binding and catalytic activity, were the top ontologies for the ‘molecular function’ category, while extracellular space and extracellular organelle were the top ontologies for the ‘cellular component’ category, and negative regulation of response to stimulus and regulation of response to stimulus were the top enriched ontologies for the ‘biological process’ category (Table S3). Pathway enrichment analysis revealed that TP53 Network and Glypican 1 network were the top two enriched pathways (Table S4). An unsupervised hierarchical clustering analysis of 23 preliminarily authenticated proteins showed a significant distinction for early ESCC cases from healthy controls (Fig. S2).
Creation of multi-protein diagnostic model
Considering the complex relationship of these 23 proteins and clinical feasibility for using these biomarkers, LASSO regression was performed to select optimal proteins based on dimensionality reduction in order to develop a compact multi-protein classifier (Fig. S3). Remaining 11 proteins, namely, ANXA1, hK8, CDKN1A, MetAP2, hK14, VIM, GPC1, RSPO3, TRAIL, 5NT, and SEZ6L were used to construct a multiple logistic regression model with an AUC of 0.950 (95%CI:0.918 ~ 0.982, Fig. 5 red line). Because the model still had the problem of multicollinearity and redundant protein biomarkers, a backward elimination algorithm was further applied to construct a brief and efficient multi-protein model. Finally, ANXA1, hK8, hK14, VIM and RSPO3 were kept to discriminate early ESCC cases from healthy controls with an AUC of 0.936 (95%CI:0.899 ~ 0.973, Fig. 5 black line). The specificity and sensitivity of the classifier were 78.6 and 96.7% at optimal Youden’s index, and the classified accuracy was 0.888. Besides, the average accuracy rates of the five- protein model were 0.861 and 0.825 in training and test data by 5-fold cross-validation. For intuitive understanding, the results from logistic regression analysis for protein levels (quartiles) are shown in Table 1, and an easy-to-use predictive nomogram tool was created to evaluate the individual ESCC risk based on the five-protein panel (Fig. S4).
Receiver operating characteristic (ROC) curve for selected multiple protein classifier. The red line and area under the curve (AUC) value was fitted by 11 proteins model, namely, ANXA1, hK8, CDKN1A, MetAP2, hK14, VIM, GPC1, RSPO3, TRAIL, 5NT, SEZ6L. The black line with 95% confidence interval (CI) and AUC value was fitted by 5 compact proteins model, namely ANXA1, hK8, hK14, VIM and RSPO3. Diagnosis models were built by using logistic regression
The overall results did not change substantially, after conducting a sensitivity analysis by adjusting for age and sex in logistic regression models.
Discussion
Protein signatures comparing with existent studies
Proteomic studies have been conducted to explore potential biomarkers for ESCC diagnosis by using different biological samples, such as body fluids (plasma, serum, etc.), tumor tissues (fresh frozen tissues or formalin-fixed-paraffin-embedded tissues) and cells in vitro. In 2016, Harada et al. summarized 18 non-targeted proteomic studies with limited sample sizes for ESCC diagnosis based on mass spectrometry technology using serum, tissue and cell line samples, and identified several novel ESCC diagnostic markers, such as Apolipoprotein A-I, Tubulin beta chain filamin A alpha, HSP70, and so on [22]. Blood-based diagnostic studies have been extensively used as a cost-effective and fast screening tool for understanding diseases and medication treatment efficiency over the years, and organ-specific proteins in plasma could mirror organ dysfunction [23]. Development of a liquid biopsy method for early ESCC detection would significantly improve the efficiency of subsequent gastroscopy examination, especially for asymptomatic high-risk population.
Recently, a study identified 13 protein biomarkers in serum using the protein chip AAH-BLG-507 from RayBiotech for discriminating 10 early ESCC patients from 10 healthy controls in China [24]. Liao et al. reported that a combination of plasma FAPα plus traditional biomarker (CEA, CYFRA211, SCCA) using ELISA could significantly discriminate (AUC = 0.745) ESCC (n = 151, stage I: 29 + stage II: 59 + stage III/IV: 63) from non-malignancy controls (n = 230, healthy: 194 + benign:36) [25]. Huang et al. reported an AUC of 0.725 for serum IGFBP7 based on a study including 107 controls and 37 early ESCC patients [26]. Xu et al. reported the serum autoantibody panel (p53, MMP-7, HSP70, Prx VI and Bmi-1) could distinguish early-stage ESCC patients (n = 76) from normal controls (n = 134) with sensitivity of 45% and specificity of 96% in a validation cohort [13]. In our study, 23 proteins, namely, ANXA1, hK8, CDKN1A, ABL1, SCAMP3, EGF, LYN, MetAP2, KLK13, ADAMTS15, hK14, VIM, TXLNA, GPC1, RSPO3, hK11, TRAIL, X5NT, CPE, FADD, TGFR2, SEZ6L and CD160, showed potential diagnostic utility for distinguishing early ESCC from controls and their serum levels showed a significant dose-response relationship with ESCC stages. However, few overlapped proteins were found in the above-mentioned studies, which may be due to differences of candidate protein signatures, sample sizes, ESCC stages, biological nature of samples (plasma vs. serum) and detection methods (PEA vs. protein chip vs. ELISA) used in various studies.
This is the first study to estimate the efficiency of Olink Oncology II panel for the early diagnosis of ESCC. Although this panel was not designed specifically for identifying ESCC patients, the majority of the proteins on the Oncology II panel are secreted proteins that show abnormal expression in the tissues or sera of multiple types of cancer [21, 27, 28]. Especially, several proteins, such as, ANXA1, CEACAM5 (aka CEA), VIM, ALB1 and IL6, have been reported to be potential biomarkers in the diagnosis of ESCC, [24, 25, 29,30,31,32] however, most proteins on the Oncology II panel have not yet been examined for their expression in ESCC blood samples.
Model performance
In order to avoid overfitting and consider the clinical feasibility for early diagnosis of ESCC, a concise multi-protein classifier containing ANXA1, hK8, hK14, VIM and RSPO3 was created. The AUC of the five-protein classifier for differentiating early ESCC from controls was 0.936 (95%CI:0.899 ~ 0.973). The specificity and sensitivity were 78.6 and 96.7% at optimal Youden’s index, and the classification accuracy was 0.888. We used five-fold cross-validation to estimate the average accuracy rate of the five-protein classifier, and the corresponding figure was 0.861 and 0.825 in the discovery set and validation set, respectively. Overall, the differentiation efficiency of our multi-protein classifier was relatively superior to other studies [13, 25, 26, 33].
Biological functions
In our study, 92 tumor-related candidate proteins were detected in serum from various stage ESCC patients and healthy controls to predict cancer status, and 23 proteins were preliminarily identified as potential diagnostic protein biomarkers for ESCC. Functional enriched pathway analyses of these 23 proteins showed that they were involved in signaling receptor binding, extracellular space, regulation of response to stimulus and TP53 network implicated in development of ESCC. Thus, their compositions in serum could mirror the pro-tumorigenic ESCC microenvironment and can be used to monitor the progression of ESCC.
In our final diagnostic classifier for early stage ESCC, ANXA1, hK8, hK14, VIM and RSPO3 were selected. The serum levels of ANXA1 and VIM were over-expressed in ESCC patients, on the contrary, the serum levels of hK8, hK14 and RSPO3 were decreased.
ANXA1 (annexin A1), known as an endogenous anti-inflammatory protein, has now been recognized to be closely related to tumor cell proliferation, invasion, differentiation, apoptosis, metastasis and chemotherapy sensitivity via modulation of various cancer-associated pathways [34, 35]. Moreover, ANXA1 shows contrasting expression profiles in various cancer types: over-expressed in lung cancer, colorectal cancer, and pancreatic cancer, and so on, by the contrary, lack of expression in cervical cancer, prostate cancer, nasopharyngeal carcinoma, etc. [34, 36] We found a high level of ANXA1 in serum of ESCC patients, which is consistent with the finding of a previous study showing upregulated levels of ANXA1 in ESCC tissues versus matching normal tissues [30]. However, most previous studies reported that ANXA1 expression was significantly downregulated in cell lines and tissues from ESCC patients compared with adjacent normal tissues [29, 32, 37,38,39]. Further studies are needed to examine the correlation of tumor ANXA1 expression with serum level.
VIM (vimentin), one of class-III intermediated filament proteins, is involved with cytoskeletal integrity, cell adhesion and cell migration via epithelial-mesenchymal transition, [40, 41] and upregulated VIM levels in tissues have been reported as a potential diagnostic and prognostic marker of multiple types of cancers, such as prostate cancer, breast cancer, malignant melanoma and lung cancer [42]. The over-expressed VIM was reported in ESCC tissues compared with adjacent normal tissues, [30] which was somewhat consistent with the results of our study. The biological expression of vimentin is regulated by the transcription factors Twist, Zeb1, Snail, and Slug, which are induced by TGF-β signal transduction [43].
Dysregulation of kallikrein-related peptidases (KLKs) is related to differential expression signatures in various types of cancers, [44, 45] but little is known about its role in ESCC development. Four proteins from kallikrein-related peptidase family, namely, hK8(kallikrein-8), hK11(kallikrein-11), KLK13(kallikrein-13) and hK14(kallikrein-14), were detected by Olink Oncology II panel, and we found all of them had low levels in serum in ESCC patients regardless tumor stage, compared with healthy controls. KLKs, the largest secreted serine protease family, are involved in cancer cell growth, migration, invasion, and chemo-resistance by activation of PARs, the release of active growth factors, modulation of the proteolytic network, and activation of androgen receptor signaling [45, 46].
RSPO3 (R-spondin-3), an activator of the canonical Wnt signaling pathway and PI3K/AKT pathway as a key regulator of angiogenesis and epithelial-mesenchymal transition, has shown low expression in colorectal cancer, squamous cell carcinoma of the lung and prostate cancer, but upregulated expression in bladder cancer, ovarian cancer and lung adenocarcinoma [47,48,49,50]. Our study showed that RSPO3 level in serum was inversely associated with ESCC progression.
Limitations and future perspectives
The results of our models should be interpreted with caution. First, the study was conducted in an ESCC high-risk area of China, which might weaken the generalization of our five-protein prediction classifier to other relatively normal-risk areas. Second, although we found the overall good dose-response relationship between the serum levels of identified biomarkers and ESCC stages, the trends of certain proteins were not perfect, which recommends that external, independent studies are needed to validate and generalize our findings. Moreover, the identified protein biomarkers for ESCC were generally universal biomarkers for multiple types of tumors. Further work is needed to determine the specificity of our five-protein classifier for ESCC diagnosis versus other cancer types. Considering a three-level hierarchical screening strategy, i.e. “environment exposure + blood biopsy + esophagogastroduodenoscopy”, to be established in ESCC high-incidence area, our serum multi-protein classifier with high sensitivity and specificity would have a promising application value in high-risk population. The identified ESCC biomarkers are also involved in ESCC progression, which highlights their possible application also as prognostic biomarkers.
In summary, we identified and established a multi-protein classifier for discriminating early ESCC patients from healthy controls, which might contribute to improving the three-level hierarchical screening strategy for decreasing the ESCC burden in high-incidence areas. However, the results need to be further validated in prospective cohort studies.
Availability of data and materials
All data that support the findings of this study are available from the corresponding authors upon a reasonable request.
Abbreviations
- ESCC:
-
Esophageal squamous cell carcinoma
- LASSO:
-
Least absolute shrinkage and selection operator
- ANXA1:
-
Annexin A1
- hK8:
-
Kallikrein-8
- hK14:
-
Kallikrein-14
- VIM:
-
Vimentin
- RSPO3:
-
R-spondin-3
- AUC:
-
Area under curve
- PEA:
-
Proximity extension assays
- PCR:
-
Polymerase chain reaction
- NPX:
-
Normalized Protein eXpression
- PCA:
-
Principal component analysis
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- ROC:
-
Receiver operating characteristic
- CI:
-
Confidence intervals
- CV:
-
Coefficient of variation
- PC:
-
Principal components
- hK11:
-
Kallikrein-11
- KLK13:
-
Kallikrein-13
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Njei B, McCarty TR, Birk JW. Trends in esophageal cancer survival in United States adults from 1973 to 2009: a SEER database analysis. J Gastroenterol Hepatol. 2016;31(6):1141–6.
Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer. 2015;136(8):1921–30.
Shen L, Shan YS, Hu HM, Price TJ, Sirohi B, Yeh KH, et al. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol. 2013;14(12):e535–47.
Chiu Y, Uedo N, Singh R, Gotoda T, Ng W, Yao K, et al. An Asian consensus on standards of diagnostic upper endoscopy for neoplasia. Gut. 2019;68(2):186–97.
Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9(5):278–87.
di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the esophagus: screening, diagnosis, and therapy. Gastroenterology. 2018;154(2):421–36.
Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018.
Mader S, Pantel K. Liquid biopsy: current status and future perspectives. Oncol Res Treat. 2017;40(7–8):404–8.
Brucher BL, Li Y, Schnabel P, Daumer M, Wallace TJ, Kube R, et al. Genomics, microRNA, epigenetics, and proteomics for future diagnosis, treatment and monitoring response in upper GI cancers. Clin Transl Med. 2016;5(1):13.
Chen X, Gole J, Gore A, He Q, Lu M, Min J, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11(1):3475.
Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–7.
Xu YW, Peng YH, Chen B, Wu ZY, Wu JY, Shen JH, et al. Autoantibodies as potential biomarkers for the early detection of esophageal squamous cell carcinoma. Am J Gastroenterol. 2014;109(1):36–45.
Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9(4):e95192.
Yang X, Chen X, Zhuang M, Yuan Z, Nie S, Lu M, et al. Smoking and alcohol drinking in relation to the risk of esophageal squamous cell carcinoma: a population-based case-control study in China. Sci Rep. 2017;7(1):17249.
Yang X, Ni Y, Yuan Z, Chen H, Plymoth A, Jin L, et al. Very hot tea drinking increases esophageal squamous cell carcinoma risk in a high-risk area of China: a population-based case-control study. Clin Epidemiol. 2018;10:1307–20.
Yang X, Zhang T, Yin X, Yuan Z, Chen H, Plymoth A, et al. Adult height, body mass index change, and body shape change in relation to esophageal squamous cell carcinoma risk: a population-based case-control study in China. Cancer Med. 2019;8(12):5769–78.
Ekheden I, Yang X, Chen H, Chen X, Yuan Z, Jin L, et al. Associations between gastric atrophy and its interaction with poor Oral health and the risk for esophageal squamous cell carcinoma in a high-risk region of China: a population-based case-control study. Am J Epidemiol. 2020;189(9):931–41.
Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the esophagus and Esophagogastric junction: an eighth edition staging primer. J Thorac Oncol. 2017;12(1):36–42.
Rice TW, Gress DM, Patil DT, Hofstetter WL, Kelsen DP, Blackstone EH. Cancer of the esophagus and esophagogastric junction-major changes in the American joint committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(4):304–17.
Skubitz APN, Boylan KLM, Geschwind K, Cao Q, Starr TK, Geller MA, et al. Simultaneous measurement of 92 serum protein biomarkers for the development of a multiprotein classifier for ovarian Cancer detection. Cancer Prev Res (Phila). 2019;12(3):171–84.
Harada K, Mizrak Kaya D, Shimodaira Y, Song S, Baba H, Ajani JA. Proteomics approach to identify biomarkers for upper gastrointestinal cancer. Expert Rev Proteomics. 2016;13(11):1041-53.
Malmström E, Kilsgård O, Hauri S, Smeds E, Herwald H, Malmström L, et al. Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nat Commun. 2016;7:10261.
Tong Q, Wang XL, Li SB, Yang GL, Jin S, Gao ZY, et al. Combined detection of IL-6 and IL-8 is beneficial to the diagnosis of early stage esophageal squamous cell cancer: a preliminary study based on the screening of serum markers using protein chips. Onco Targets Ther. 2018;11:5777–87.
Liao Y, Xing S, Xu B, Liu W, Zhang G. Evaluation of the circulating level of fibroblast activation protein alpha for diagnosis of esophageal squamous cell carcinoma. Oncotarget. 2017;8(18):30050–62.
Huang X, Hong C, Peng Y, Yang S, Huang L, Liu C, et al. The diagnostic value of serum IGFBP7 in patients with esophageal squamous cell carcinoma. J Cancer. 2019;10(12):2687–93.
Bhardwaj M, Weigl K, Tikk K, Benner A, Schrotz-King P, Brenner H. Multiplex screening of 275 plasma protein biomarkers to identify a signature for early detection of colorectal cancer. Mol Oncol. 2020;14(1):8–21.
Berggrund M, Enroth S, Lundberg M, Assarsson E, Stalberg K, Lindquist D, et al. Identification of candidate plasma protein biomarkers for cervical Cancer using the multiplex proximity extension assay. Mol Cell Proteomics. 2019;18(4):735–43.
Zhang J, Wang K, Zhang J, Liu SS, Dai L, Zhang JY. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res. 2011;10(6):2863–72.
Chen JY, Xu L, Fang WM, Han JY, Wang K, Zhu KS. Identification of PA28β as a potential novel biomarker in human esophageal squamous cell carcinoma. Tumour Biol. 2017;39(10):1010428317719780.
Mao YS, Zhang DC, Zhao XH, Wang LJ, Qi J, Li XX. Significance of CEA, SCC and Cyfra21-1 serum test in esophageal cancer. Chin J Oncol. 2003;25(5):457–60.
Moghanibashi M, Jazii FR, Soheili ZS, Zare M, Karkhane A, Parivar K, et al. Proteomics of a new esophageal cancer cell line established from Persian patient. Gene. 2012;500(1):124–33.
Xu YW, Liu CT, Huang XY, Huang LS, Luo YH, Hong CQ, et al. Serum autoantibodies against STIP1 as a potential biomarker in the diagnosis of esophageal squamous cell carcinoma. Dis Markers. 2017:5384091.
Foo SL, Yap G, Cui J, Lim LHK. Annexin-A1 - a blessing or a curse in Cancer? Trends Mol Med. 2019;25(4):315–27.
Ganesan T, Sinniah A, Ibrahim ZA, Chik Z, Alshawsh MA. Annexin A1: A Bane or a Boon in Cancer? A Systematic Review. Molecules. 2020;25:16.
Fu Z, Zhang S, Wang B, Huang W, Zheng L, Cheng A. Annexin A1: a double-edged sword as novel cancer biomarker. Clin Chim Acta. 2020;504:36–42.
Xia S-H, Hu L-P, Hu H, Ying W-T, Xu X, Cai Y, et al. Three isoforms of annexin I are preferentially expressed in normal esophageal epithelia but down-regulated in esophageal squamous cell carcinomas. Oncogene. 2002;21(43):6641–8.
Chung JY, Braunschweig T, Hu N, Roth M, Traicoff JL, Wang QH, et al. A multiplex tissue immunoblotting assay for proteomic profiling: a pilot study of the normal to tumor transition of esophageal squamous cell carcinoma. Cancer Epidemiol Biomark Prev. 2006;15(7):1403–8.
Hu N, Flaig MJ, Su H, Shou J-Z, Roth MJ, Li W-J, et al. Comprehensive characterization of annexin I alterations in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10(18 Pt 1):6013–22.
Sharma P, Alsharif S, Fallatah A, Chung BM. Intermediate filaments as effectors of Cancer development and metastasis: a focus on keratins, Vimentin, and nestin. Cells. 2019;8(5):497.
Chen Z, Fang Z, Ma J. Regulatory mechanisms and clinical significance of vimentin in breast cancer. Biomed Pharmacother. 2021;133:111068.
Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–46.
Strouhalova K, Přechová M, Gandalovičová A, Brábek J, Gregor M, Rosel D. Vimentin Intermediate Filaments as Potential Target for Cancer Treatment. Cancers (Basel). 2020;12:1.
Tailor PD, Kodeboyina SK, Bai S, Patel N, Sharma S, Ratnani A, et al. Diagnostic and prognostic biomarker potential of kallikrein family genes in different cancer types. Oncotarget. 2018;9(25):17876–88.
Kryza T, Silva ML, Loessner D, Heuzé-Vourc'h N, Clements JA. The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie. 2016;122:283–99.
Filippou PS, Karagiannis GS, Musrap N, Diamandis EP. Kallikrein-related peptidases (KLKs) and the hallmarks of cancer. Crit Rev Clin Lab Sci. 2016;53(4):277–91.
Wu L, Zhang W, Qian J, Wu J, Jiang L, Ling C. R-spondin family members as novel biomarkers and prognostic factors in lung cancer. Oncol Lett. 2019;18(4):4008–15.
Mesci A, Lucien F, Huang X, Wang EH, Shin D, Meringer M, et al. RSPO3 is a prognostic biomarker and mediator of invasiveness in prostate cancer. J Transl Med. 2019;17(1):125.
Chen Z, Zhou L, Chen L, Xiong M, Kazobinka G, Pang Z, et al. RSPO3 promotes the aggressiveness of bladder cancer via Wnt/β-catenin and hedgehog signaling pathways. Carcinogenesis. 2019;40(2):360–9.
Gu H, Tu H, Liu L, Liu T, Liu Z, Zhang W, et al. RSPO3 is a marker candidate for predicting tumor aggressiveness in ovarian cancer. Ann Transl Med. 2020;8(21):1351.
Acknowledgements
The authors would like to acknowledge the interviewers and technicians at Fudan University Taizhou Institute of Health Sciences, for their invaluable contribution to data collection and sample preparation, the staff at the Taixing Center for Disease Control and Prevention for their help in organization of field work, and the staff at Taixing People’s Hospital for their assistance with sample collection.
Funding
This work was supported by the National Natural Science Foundation of China (81973116, 82073637, 91846302 and 81573229), National Key Research and Development program of China (2017YFC0907002 and 2017YFC0907003), International S&T Cooperation Program of China (2015DFE32790), European Research Council (682663), and Shandong Provincial Natural Science Foundation (ZR2020QH302).
Author information
Authors and Affiliations
Contributions
XRY and CS: collected research datasets, analyzed data and drafted the manuscript; TCZ, XLY, JYM and ZYY: participated in samples collection and medical records assessment; JRY and WMY organized the protein detection in Sweden; ML, XDC, WMY and LJ initiated, organized and supervised the study; ML, XDC and WMY: critically revised the manuscript; ML and XDC: provided technical support. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Institutional Review Boards of the School of Life Sciences, Fudan University (date: February 19, 2009), Qilu Hospital, Shandong University (date: March 8, 2010), and Stockholm Ethical Vetting Board (2018/357–31). The study was carried out in accordance with the approved protocol, and all participants provided written informed consent.
Consent for publication
We have received consents from all participants involving in this study. The consent forms will be provided upon request.
Competing interests
The authors declare no potential conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
92 proteins from the Olink multiplex Oncology II panel. Table S2. The general information of selected participants, controls and cases based on different cancer stages. Figure S1. The protein interaction of 23 preliminarily authenticated proteins. Each node represents a protein, and the gene name is marked at the top right of the node. Table S3. Gene ontology enrichment analysis of the identified 23 proteins that were differentially expressed between early ESCC and controls, covering three categories, i.e. molecular function, cellular component, and biological process. Top 5 gene ontologies in each enrichment category were selected. Data were obtained from the online ConsensusPathDB- human interaction network database http://cpdb.molgen.mpg.de/. Table S4. Pathway enrichment analysis of the identified 23 proteins that were differentially expressed between early ESCC and controls. Top 7 enriched pathway were selected. Data were obtained from the online ConsensusPathDB- human interaction network database http://cpdb.molgen.mpg.de/. Figure S2. An unsupervised hierarchical clustering analysis of 23 preliminarily authenticated proteins for discriminating early esophageal squamous cell carcinoma (ESCC) from healthy controls. Figure S3. The selection feature of least absolute shrinkage and selection operator (LASSO) via tenfold cross-validation based on area under the ROC curve (AUC). Selection of the tuning parameter (λ) in the LASSO model was via tenfold cross-validation based on minimum standard error. The y-axis indicates AUC. The lower x-axis indicates the log(λ). Numbers along the upper x-axis represent the average number of predictors. Red dots indicate average AUC values for each model with a given λ, and vertical bars through the red dots show the upper and lower values of AUC. The vertical black lines define the optimal values of λ, where the model provides its best fit to the data. Figure S4. A nomogram to predict individual ESCC risk based on the identified five-protein panel.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, X., Suo, C., Zhang, T. et al. Targeted proteomics-derived biomarker profile develops a multi-protein classifier in liquid biopsies for early detection of esophageal squamous cell carcinoma from a population-based case-control study. Biomark Res 9, 12 (2021). https://doi.org/10.1186/s40364-021-00266-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40364-021-00266-z