Abstract
Background
Leishmania infantum is the causative agent of human visceral leishmaniasis (VL) and sporadic human cutaneous leishmaniasis (CL) in the Mediterranean region. The genetic variation of the Leishmania parasites may result in different phenotypes that can be associated with the geographical distribution and diversity of the clinical manifestations. The main objective of this study was to explore the genetic polymorphism in L. infantum isolates from human and animal hosts in different regions of Morocco.
Methods
The intraspecific genetic variability of 40 Moroccan L. infantum MON-1 strains isolated from patients with VL (n = 31) and CL (n = 2) and from dogs (n = 7) was evaluated by PCR-RFLP of nagt, a single-copy gene encoding N-acetylglucosamine-1-phosphate transferase. For a more complete analysis of L. infantum polymorphism, we included the restriction patterns of nagt from 17 strains available in the literature and patterns determined by in-silico digestion of three sequences from the GenBank database.
Results
Moroccan L. infantum strains presented a certain level of genetic diversity and six distinct nagt-RFLP genotypes were identified. Three of the six genotypes were exclusively identified in the Moroccan population of L. infantum: variant M1 (15%), variant M2 (7.5%), and variant M3 (2.5%). The most common genotype (65%), variant 2 (2.5%), and variant 4 (7.5%), were previously described in several countries with endemic leishmaniasis. Phylogenetic analysis segregated our L. infantum population into two distinct clusters, whereas variant M2 was clearly distinguished from both cluster I and cluster II. This distribution highlights the degree of genetic variability among the Moroccan L. infantum population.
Conclusion
The nagt PCR-RFLP method presented here showed an important genetic heterogeneity among Moroccan L. infantum strains isolated from human and canine reservoirs with 6 genotypes identified. Three of the six Moroccan nagt genotypes, have not been previously described and support the particular genetic diversity of the Moroccan L. infantum population reported in other studies.
Similar content being viewed by others
Multilingual abstracts
Please see Additional file 1 for translation of the abstract into the five official working languages of the United Nations.
Background
Leishmania infantum, a flagellated protozoan in the Trypanosomatidae family, is the causative agent of human visceral leishmaniasis (VL) and sporadic human cutaneous leishmaniasis (CL) in the Mediterranean region [1,2,3]. In Morocco, and all around the Mediterranean basin, L. infantum MON-1 is the predominant causative agent of VL, with the domestic dog as the main reservoir and Phlebotomus perniciosus, Phlebotomus ariasi, and Phlebotomus longicuspis as the vectors [4,5,6,7]. The zymodeme MON-24, mainly considered to cause CL, has been detected in both humans and dogs in Morocco [8]. VL is endemic in the northern part of the country, but sporadic cases have been reported in the south [9]. An average of 128 VL cases were reported every year from 2008 to 2013 with a predominance of cases in children aged under 10 years [10]. The annual incidence rate of VL is estimated to be 0.4 cases per 100 000 people [11]. In recent years, changes in the epidemiology of L. infantum have been reported in Morocco, including a southward spread of the parasite, a new focus of CL, and the appearance of treatment-resistant L. infantum [12, 13]. Several studies investigating the potential contribution of the parasite to the clinical pleomorphism of leishmaniasis have shown a correlation between specific Leishmania genotypes and clinical forms, and demonstrate that parasite genotyping plays an essential role in both clinical and epidemiological risk assessment [14]. A sound and consensual taxonomical background based on the knowledge of the phylogenetic diversity of Leishmania is needed for a better understanding of epidemiological changes [15]. The broad epidemiological and clinical diversity of L. infantum raises the need of analysis of genetic variability. To date, several methods using powerful molecular tools have been employed for typing Leishmania strains [16, 17]. Multilocus enzyme electrophoresis (MLEE) is the most commonly used technique for Leishmania typing. However, this technique requires cultivation of the parasite and cannot always discriminate between closely related strains. Different PCR-based methods have been employed to ascertain the intraspecific genetic variability of Leishmania and have contributed to the development of epidemiological studies [16, 17]. PCR amplification of kinetoplast DNA (kDNA) minicircles followed by analysis of restriction fragment length polymorphism (RFLP) has shown the genetic diversity between closely related strains of L. infantum MON-1 [18,19,20]. Further genetic diversity within Leishmania strains has been elucidated by random amplification of polymorphic DNA (RAPD) [21, 22], PCR-RFLP of the antigen-encoding genes gp63 and cpb [23, 24], sequence analysis of intergenic spacer regions (ITS) [25, 26], multilocus microsatellite typing (MLMT) [18, 27,28,29], and multilocus sequence typing (MLST) [30,31,32]. PCR-RFLP of the single-copy gene, nagt, which encodes N-acetylglucosamine-1-phosphate transferase (NAGT), has been used for intraspecies divergence analyses of Leishmania spp. [33, 34]. Analysis of the nagt sequence revealed the existence of five different genotypes within a population of 86 L. infantum isolates from distinct regions [34]. The nagt gene is highly conserved and functionally indispensable. Therefore, we have used PCR-RFLP of nagt gene as a molecular method to analyze genetic variability within a population of Moroccan L. infantum isolated from humans and canine reservoirs.
Methods
Leishmania infantum strains
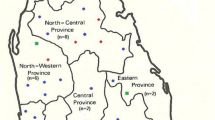
A total of 40 clinical isolates of L. infantum were used. The strains were isolated in Novy-MacNeal-Nicolle culture medium. Thirty-three were isolated from immunocompetent patients with VL (n = 31) and CL (n = 2); seven strains were isolated from canine reservoirs. The samples were collected from the northern VL endemic foci of Morocco in the provinces of Fes (n = 27), Taounate (n = 10) and Al Hoceima (n = 1), as well as from some VL sporadic areas in the southern provinces of Ouarzazate (n = 1) and Taroudant (n = 1). The clinical form, geographical distribution and the number of the studied isolates are presented in Fig. 1. All VL and canine leishmaniasis (CanL) strains were previously characterized by MLEE as belonging to zymodeme MON-1, except one CL strain collected from Taounate, which belongs to zymodeme MON-24. The CL strain from Ourzazate province was not characterized.
DNA extraction
DNA was extracted from promastigote pellets using the PureLink® Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. A final volume of 100 μl of DNA was obtained and stored at − 20 °C until use.
PCR amplification and RFLP analysis
The ~ 1.4-kb nagt gene amplifications were performed with the primer pair L1 (5’-TCATGACTCTTGGCCTGGTAG-3′) and L4 (5’-CTCTAGCGCACTTCATCGTAG-3′), as previously described by Akman et al. [33], with some modifications. PCR mixtures consisted of between 50 and 100 ng of DNA, 1.25 U of GoTaq® DNA polymerase (Promega, Madison, WI, USA), 1X PCR buffer, 0.4 μmol/L of each primer, 1.5 mmol/L MgCl2 and 0.2 mmol/L of each dNTP. The thermocycler settings were an initial denaturation at 95 °C for 5 min, 30 cycles at 94 °C for 60 s, 58 °C for 60 s, and 72 °C for 90 s, and a final extension step at 72 °C for 5 min. Further RFLP analysis of the PCR-amplified nagt gene was performed separately using three restriction enzymes: NaeI, AlwI, and NciI (New England Biolabs, Ipswich, MA, USA). After conditions optimization, digestion reactions were carried out in a final volume of 10 μl including 5 μl of PCR product, 10 U of restriction enzyme, and 1× recommended buffer for each enzyme. All restriction reactions were incubated overnight at 37 °C. The restriction fragments were resolved by electrophoresis for 2–3 h on a 3% agarose gel containing ethidium bromide (0.5 μg/ml) and visualized under UV illumination.
Selection of nagt sequences from GenBank database and in-silico RFLP analysis
The Primer-BLAST tool from NCBI was used to search nagt sequences corresponding to the L. infantum species. Briefly, the sequences of the primer pair L1/L4 were blasted, and only nagt sequences with nearly 100% homology with the primers and belonging to L. infantum species were chosen. Three nagt sequences, approximately 1405 bp, were selected and analyzed by in-silico digestion with the same restriction enzymes using the option “Find restriction sites” in Unipro UGENE 1.25 [35]. Characteristics of the three selected sequences are shown in Table 1.
Phylogenetic analysis
The restriction patterns resulting from the digests of the nagt PCR products by the 3 tested endonucleases and those obtained through in-silico digestion were used to identify genotypes of the Moroccan L. infantum isolates and the strains selected from GenBank databse, respectively. The genotypes identified in this work were named according to Waki et al. [34]. To analyze the phylogenetic relationships, the 40 Moroccan isolates of this study (Fig. 1) and the 3 samples selected from Genbank database (Table 1) as well as 17 strains previously described by Waki et al. [34] (Additional file 2: Table S1), forming a database of 60 samples. Subsequently, the nagt restriction patterns from all samples were inserted into a binary matrix, with the restriction sites coded as present (1) or absent (0). Phylogenetic analysis was performed using the package PHYLIP 3.69 [36]. The binary matrix was converted into a distance matrix with the Restdist tool and the resulting distance matrix was used to construct a rooted tree based on the Unweighted Pair Group Method with Arithmetic mean (UPGMA) algorithm with the Neighbor tool. The robustness of the phylogenetic analysis and significance of the branch order were determined by bootstrap analysis carried out on 100 replicates using SEQBOOT program, provided with the PHYLIP package.
Results
PCR-RFLP analysis of nagt
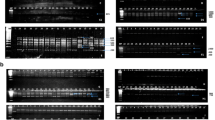
PCR amplification of nagt from 40 Moroccan L. infantum strains produced a DNA fragment with the expected size of about 1.4 kb (Fig. 2a). nagt-RFLP analysis of PCR products with three endonucleases revealed genetic heterogeneity among the Moroccan L. infantum population. Digestion with AlwI and NaeI produced two and four different patterns, respectively. The endonuclease NciI resulted in a monomorphic pattern (Table 2). To discriminate genotypes, the RFLP patterns of each strain were compared with L. infantum nagt restriction maps [34]. The L. infantum strain JPCM5 (Accession number: FR796468.1) was used as a reference sequence to identify the nagt restriction sites of the three tested enzymes and to confirm the size of restriction fragments. The cut positions on the reference sequence corresponding to each enzyme are shown in additional Additional file 3: Table S2. Representative RFLP patterns for AlwI, NciI, and NaeI are shown in Fig. 2b, c, and d.
a PCR amplification of nagt gene (1.4 kb) from Moroccan L. infantum strains. b, c, d PCR-amplified nagt digested with NciI, NaeI, and AlwI, respectively. The numbers above and below the digested fragments indicate their sizes. Lanes 1–5: DNA samples of Moroccan L. infantum isolates 1 to 5; NC: Negative control; M: Molecular marker HyperLader™ 100 bp plus (Bioline, Taunton, MA, USA). e RFLP maps illustrating the nagt genotype identified in this work and those identified by Waki et al. [34]*. The vertical numbers indicate the cut position of each enzyme
Identification of genotypes
Analysis of nagt-RFLP restriction patterns allowed grouping of the 40 Moroccan isolates of L. infantum into six distinct genotypes (Table 3) when compared to nagt genotypes previously established for the L. donovani and L. infantum species [34]. Of the six genotypes identified, common L. infantum and L. infantum variants 2 and 4 had been previously described [34]. The other three genotypes were exclusively identified in this study of Moroccan L. infantum and named M1, M2, and M3. The predominant L. infantum in this study was the common genotype with a proportion of 65% (26/40), followed by L. infantum variant M1 at 15% (6/40), variant 4 and variant M2 at 7.5% each (3/40 each), and one each of variant 2 and variant M3 (2.5% each). The 32 L. infantum isolates causing VL were grouped into five genotypes, whereas strains causing CanL (n = 7) belonged to the common genotype. The dermotropic strain isolated from the south of Morocco had a specific nagt genotype and was named variant M3. However, the dermotropic strain originating from the north of Morocco had the common nagt genotype. An additional nagt genotype was identified among three nagt sequences retrieved from the GenBank database by in-silico RFLP analysis (Table 2). Of the three sequences, the variant causing VL is from Iran and was designated as the “Iranian variant” (Table 3). The other two L. infantum sequences correspond to the predominant nagt genotype. Restriction maps were constructed to clearly illustrate the differences between the nagt genotypes identified here and in the 2007 study by Waki et al. [34] (Fig. 2e).
Phylogenetic analysis
The UPGMA dendrogram, inferred from the genetic distances calculated from the nagt PCR-RFLP data, allowed an overall and clear visualization of the relationships between the nine nagt genotypes identified (Fig. 3). The UPGMA tree topology was supported by highly significant bootstrap values (> 90%). Phylogenetic analysis of the 60 samples showed that most of the isolates (37/60) correspond to the common L. infantum genotype, regardless of geographical origin, host, or clinical forms. Except for variant M2, the L. infantum variants were grouped into three clusters (Fig. 3). The largest, cluster I, included strains from Mediterranean countries (Morocco, Spain, France, Tunisia, Greece, and Turkey), East African countries (Sudan and Kenya), Asian countries (Iran and China), and Brazil. Cluster II was composed of strains isolated from Morocco, Sudan, and China. However, cluster III consisted of strains solely originating from Asia: two L. donovani strains each isolated from India and Sri Lanka and three Chinese L. infantum variant 7 strains, exclusively identified in Chinese isolates. There was no structuring of the strains according to their hosts (human or dog) or clinical forms (CL or VL).
UPGMA tree constructed from nagt PCR-RFLP data of L. infantum (n = 58) and L. donovani (n = 2). The strains numbered from 1 to 40 correspond to Moroccan L. infantum isolates; aIsolates retrieved from GenBank database; bIsolates described by Waki et al. [34]. Bootstrap values (out of 100 replicates) are mentioned below the nodes
Discussion
nagt is a highly conserved, single-copy gene, which encodes the endoplasmic reticulum trans-membrane protein, N-acetylglucosamine-l-phosphate transferase. The stability of the Leishmania virulence factor, zinc-metalloprotease GP63, is associated with N-glycosylation, which is dependent on N-acetylglucosamine-1-phosphate transferase catalyzing the first step [37].
In this study, we investigated the genetic variability of 40 Moroccan L. infantum strains isolated from canine reservoirs and immunocompetent patients with VL and CL. PCR-RFLP analysis of nagt showed important genetic heterogeneity among Moroccan L. infantum strains. Six different nagt genotypes were identified and the common nagt L. infantum genotype was predominant in our samples (65%, 26/40). The common genotype was also reported as the most frequent genotype (73%, 63/86) in a worldwide population of L. infantum collected from Brazil, southern European countries, Tunisia, Iran, and China [34]. Taken together, these data indicate that the L. infantum common nagt variant is the most widespread genotype in different foci of VL worldwide. Moreover, this predominant genotype included samples isolated from human CL and VL patients and canine reservoirs, illustrating the zoonotic cycle of transmission of L. infantum [38, 39]. One of the isolated strains was characterized as nagt variant 2 and three strains were characterized as variant 4. Those genotypes were previously reported in small numbers in Sudan and China for variant 2 and in Kenya for variant 4 [34]. These findings suggest that variant 4 may be an African genotype as opposed to L. infantum variant 7, which seems to be exclusive to Chinese isolates [34].
The Moroccan L. infantum genotypes, variants M1, M2, and M3, have not been previously described. Variants M1 and M2 were isolated from VL patients and variant M3 was from a CL patient. Phylogenetic analysis segregated variants M1 and M3 into two distinct clusters (I and II, respectively). However, variant M2 was clearly distinguished from the three clusters obtained. This distribution highlights the important degree of genetic variability among the Moroccan L. infantum population. Among the three clusters, there was no association between geographical origin of isolates or the disease forms (VL/CL). The lack of association between Leishmania nagt gene-based grouping and VL/CL disease phenotype of isolates had been previously reported [34]. We also identified a new nagt genotype, the “Iranian variant.” This unique variant, belonging to cluster I, is the most homologous to the predominant genotype. Additionally, L. donovani species presented a unique nagt restriction profile, which corroborates that L. donovani and L. infantum are genetically different [40]. Chinese L. infantum variant 7 isolates are grouped in cluster III with L. donovani from India and Sri Lanka, two Asian neighboring countries. These results align with a previous report that a group of Chinese L. infantum strains were closely related to L. donovani strains from India [41]. Other studies have also reported that some L. infantum strains are more closely related to L. donovani than other L. infantum strains [42].
Conclusions
This work showed important intraspecific genetic variability among Moroccan L. infantum strains. Three of the six nagt variants are exclusive to Moroccan isolates. The nagt PCR-RFLP method used here seems to have a good resolving power and supports the substantial genetic diversity of the Moroccan L. infantum population reported in other studies [20, 27], thus confirming its usefulness. However, further studies should be carried out by extending the nagt PCR-RFLP method to a larger number of strains representing different hosts (human, dogs and phlebotomine sand fly), geographical areas and zymodemes types, to better understand the molecular epidemiology of L. infantum in Morocco and the other endemic countries.
Abbreviations
- CanL:
-
Canine leishmaniasis
- CL:
-
Cutaneous leishmaniasis
- nagt :
-
N-acetylglucosamine-1-phosphate transferase
- VL:
-
Visceral leishmaniasis
- WHO:
-
World Health Organization
References
Özbel Y, Turgay N, Özensoy S, Özbilgin A, Alkan M, Özcel M, et al. Epidemiology, diagnosis and control of leishmaniasis in the Mediterranean region. Ann Trop Med Parasitol. 1995;89(sup1):89–93.
Rioux J-A, Mahjoub J, Gallego M, Dereure J, Perieres J, Lahmrani A, et al. Leishmaniose cutanée humaine à Leishmania infantum MON-24 au Maroc. Bulletin de la Société française de parasitologie. 1996;14(2):179–83.
WHO: Report of a meeting of the WHO Expert Committee on the Control of Leishmaniases, Geneva 22–26 March 2010. 2010.
Benabdennbi I, Pesson B, Cadi-Soussi M, Morillas Marquez F. Morphological and isoenzymatic differentiation of sympatric populations of Phlebotomus perniciosus and Phlebotomus longicuspis (Diptera: Psychodidae) in northern Morocco. J Med Entomol. 1999;36(1):116–20.
Es-Sette N, Ajaoud M, Laamrani-Idrissi A, Mellouki F, Lemrani M. Molecular detection and identification of Leishmania infection in naturally infected sand flies in a focus of cutaneous leishmaniasis in northern Morocco. Parasit Vectors. 2014;7:305.
Guessous-Idrissi N, Riyad M, Chiheb S. Les leishmanioses au Maroc: Actualités épidémiologiques et diagnostiques. Bulletin SMSM. 1996:31–5.
Pesson B, Ready JS, Benabdennbi I, Martin-Sanchez J, Esseghir S, Cadi-Soussi M, et al. Sandflies of the Phlebotomus perniciosus complex: mitochondrial introgression and a new sibling species of P. Longicuspis in the Moroccan Rif. Med Vet Entomol. 2004;18(1):25–37.
Haralambous C, Dakkak A, Pratlong F, Dedet J-P, Soteriadou K. First detection and genetic typing of Leishmania infantum MON-24 in a dog from the Moroccan Mediterranean coast: genetic diversity of MON-24. Acta Trop. 2007;103(1):69–79.
Rhajaoui M. Human leishmaniases in Morocco: a nosogeographical diversity. Pathol Biol (Paris). 2011;59(4):226–9.
MMH. Santé en chiffres 2013. Direction de la Planification et des Ressources Financière, Division de la Planification et des Etudes, Service des Etudes et de l’Information Sanitaire. 2014.
Sadeq M. Spatial patterns and secular trends in human leishmaniasis incidence in Morocco between 2003 and 2013. Infect Dis Poverty. 2016;5(1):48.
Mniouil M, Fellah H, Amarir F, Et-touys A, Bekhti K, Adlaoui EB, et al. Epidemiological characteristics of visceral leishmaniasis in Morocco (1990–2014): an update. Acta Trop. 2017;170:169–77.
Rhajaoui M, Nasereddin A, Fellah H, Azmi K, Amarir F, Al-Jawabreh A, et al. New clinico-epidemiologic profile of cutaneous leishmaniasis, Morocco. Emerg Infect Dis. 2007;13(9):1358–60.
Schonian G, Kuhls K, Mauricio IL. Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania. Parasitology. 2011;138(4):405–25.
Banuls AL, Hide M, Tibayrenc M. Molecular epidemiology and evolutionary genetics of Leischmania parasites. Int J Parasitol. 1999;29(8):1137–47.
Schönian G, Mauricio I, Cupolillo E. Is it time to revise the nomenclature of Leishmania? Trends Parasitol. 2010;26(10):466–9.
Van der Auwera G, Dujardin JC. Species typing in dermal leishmaniasis. Clin Microbiol Rev. 2015;28(2):265–94.
Botilde Y, Laurent T, Quispe Tintaya W, Chicharro C, Canavate C, Cruz I, et al. Comparison of molecular markers for strain typing of Leishmania infantum. Infect Genet Evol. 2006;6(6):440–6.
Cortes S, Mauricio I, Almeida A, Cristovao JM, Pratlong F, Dedet JP, et al. Application of kDNA as a molecular marker to analyse Leishmania infantum diversity in Portugal. Parasitol Int. 2006;55(4):277–83.
El Hamouchi A, Ejghal R, Hida M, Lemrani M. Intraspecific genetic variability in a population of Moroccan Leishmania infantum revealed by PCR-RFLP of kDNA minicircles. Acta Trop. 2017;169:142–9.
Hide M, Banuls A, Tibayrenc M. Genetic heterogeneity and phylogenetic status of Leishmania (Leishmania) infantum zymodeme MON-1: epidemiological implications. Parasitology. 2001;123(05):425–32.
Zemanova E, Jirku M, Mauricio IL, Miles MA, Lukes J. Genetic polymorphism within the leishmania donovani complex: correlation with geographic origin. Am J Trop Med Hyg. 2004;70(6):613–7.
Mauricio I, Gaunt M, Stothard J, Miles M. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology. 2001;122(04):393–403.
Tintaya KWQ, Ying X, Dedet J-P, Rijal S, De Bolle X, Dujardin J-C. Antigen genes for molecular epidemiology of leishmaniasis: polymorphism of cysteine proteinase B and surface metalloprotease glycoprotein 63 in the Leishmania donovani complex. J Infect Dis. 2004;189(6):1035–43.
Ajaoud M, Es-sette N, Hamdi S, El-Idrissi AL, Riyad M, Lemrani M. Detection and molecular typing of Leishmania tropica from Phlebotomus sergenti and lesions of cutaneous leishmaniasis in an emerging focus of Morocco. Parasit Vectors. 2013;6:217.
Parvizi P, Ready PD. Nested PCRs and sequencing of nuclear ITS-rDNA fragments detect three Leishmania species of gerbils in sandflies from Iranian foci of zoonotic cutaneous leishmaniasis. Tropical Med Int Health. 2008;13(9):1159–71.
Amro A, Hamdi S, Lemrani M, Mouna I, Mohammed H, Mostafa S, et al. Moroccan Leishmania infantum: genetic diversity and population structure as revealed by multi-locus microsatellite typing. PLoS One. 2013;8(10):e77778.
Kuhls K, Chicharro C, Cañavate C, Cortes S, Campino L, Haralambous C, et al. Differentiation and gene flow among European populations of Leishmania infantum MON-1. PLoS Negl Trop Dis. 2008;2(7):e261.
Ochsenreither S, Kuhls K, Schaar M, Presber W, Schonian G. Multilocus microsatellite typing as a new tool for discrimination of Leishmania infantum MON-1 strains. J Clin Microbiol. 2006;44(2):495–503.
El Baidouri F, Diancourt L, Berry V, Chevenet F, Pratlong F, Marty P, et al. Genetic structure and evolution of the Leishmania genus in Africa and Eurasia: what does MLSA tell us. PLoS Negl Trop Dis. 2013;7(6):e2255.
Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F, Zemanova E, et al. Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int J Parasitol. 2006;36(7):757–69.
Zemanova E, Jirku M, Mauricio IL, Horak A, Miles MA, Lukes J. The Leishmania donovani complex: genotypes of five metabolic enzymes (ICD, ME, MPI, G6PDH, and FH), new targets for multilocus sequence typing. Int J Parasitol. 2007;37(2):149–60.
Akman L, Aksu H, Wang RQ, Ozensoy S, Ozbel Y, Alkan Z, et al. Multi-site DNA polymorphism analyses of Leishmania isolates define their genotypes predicting clinical epidemiology of leishmaniasis in a specific region. J Eukaryot Microbiol. 2000;47(6):545–54.
Waki K, Dutta S, Ray D, Kolli BK, Akman L, Kawazu S, et al. Transmembrane molecules for phylogenetic analyses of pathogenic protists: Leishmania-specific informative sites in hydrophilic loops of trans- endoplasmic reticulum N-acetylglucosamine-1-phosphate transferase. Eukaryot Cell. 2007;6(2):198–210.
Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–7.
Felsenstein J. PHYLIP Version 3.695. University of Washington: Department of Genome Sciences Seattle, USA; 2013.
McGwire BS, Chang KP. Posttranslational regulation of a Leishmania HEXXH metalloprotease (gp63). The effects of site-specific mutagenesis of catalytic, zinc binding, N-glycosylation, and glycosyl phosphatidylinositol addition sites on N-terminal end cleavage, intracellular stability, and extracellular exit. J Biol Chem. 1996;271(14):7903–9.
Guessous-Idrissi N, Hamdani A, Rhalem A, Riyad M, Sahibi H, Dehbi F, et al. Epidemiology of human visceral leishmaniasis in Taounate, a northern province of Morocco. Parasite. 1997;4(2):181–5.
Nejjar R, Lemrani M, Malki A, Ibrahimy S, Amarouch H, Benslimane A. Canine leishmaniasis due to Leishmania infantum MON-1 in northern Morocco. Parasite. 1998;5(4):325–30.
Moreno GG, Lanotte G, Riou JA, Maazoun R. Leishmania infantum Nicolle, 1908, complexe systematique? Taxonomie et Phylogènese des Leishmania. Montpellier, 1984: Colloque International du CNRS; 1986. p. 1984.
Zhang C-Y, Lu X-J, Du X-Q, Jian J, Shu L, Ma Y. Phylogenetic and evolutionary analysis of Chinese Leishmania isolates based on multilocus sequence typing. PLoS One. 2013;8(4):e63124.
Bañuls AL, Hide M, Tibayrenc M. Molecular epidemiology and evolutionary genetics of Leishmania parasites. Int J Parasitol. 1999;29(8):1137–47.
Acknowledgements
We would like to gratefully acknowledge all the people who actively participated in this research work. The authors would like to thank also Enago (www.enago.com) for the English language review.
Funding
This publication is based on work supported by a grant (GTRX-14-60403-0) from the U.S. Civilian Research & Development Foundation (CRDF Global) with funding from the United States Department of State. The opinions, findings, and conclusions stated herein are those of the author(s) and do not necessarily reflect those of CRDF Global or the United States Department of State.
Availability of data and materials
All relevant data can be found within this paper.
Author information
Authors and Affiliations
Contributions
ML conceived the study. RE collected samples. AEH, RE and SEK carried out the laboratory experiments. AEH analyzed the data and wrote the manuscript. ML reviewed the draft paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from all adults in the study. Consent for inclusion of young children was obtained from parents or guardians. The study and all protocols were approved by the Ethics Committee for Biomedical Research at the Faculty of Medicine and Pharmacy Rabat, Morocco.
Competing interests
The authors declare that they have no competing interests.
Additional files
Additional files 1:
Multilingual abstracts in the five official working languages of the United Nations. (PDF 1009 kb)
Additional files 2:
Table S1. List of strains of L. infantum and L. donovani previously genotyped as nagt variant by Waki et al. [34] and used in this work. (DOCX 13 kb)
Additional files 3:
Table S2. Cut positions of restriction enzymes tested by in-silico digestion of nagt (1405 bp) of L. infantum JPCM5. (DOCX 12 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
El Hamouchi, A., El Kacem, S., Ejghal, R. et al. Genetic polymorphism in Leishmania infantum isolates from human and animals determined by nagt PCR-RFLP. Infect Dis Poverty 7, 54 (2018). https://doi.org/10.1186/s40249-018-0439-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40249-018-0439-y