Abstract
The definition of the number and nature of the signal transduction pathways involved in the pathogenesis and the identification of the molecules promoting metastasis spread might improve the knowledge of the natural history of osteosarcoma, also allowing refine the prognosis and opening the way to novel therapeutic strategies. Phosphatydil inositol (4,5) bisphosphate (PIP2), belonging to the Phosphoinositide (PI) signal transduction pathway, was related to the regulation of ezrin, an ezrin–radixin–moesin protein involved in metastatic osteosarcoma spread. The levels of PIP2 are regulated by means of the PI-specific Phospholipase C (PLC) enzymes. Recent literature data suggested that in osteosarcoma the panel of expression of PLC isoforms varies in a complex and unclear manner and is related to ezrin, probably networking with Ras GTPases, such as RhoA and Rac1. We analyzed the expression and the subcellular localization of PLC enzymes in cultured human osteosarcoma MG-63 cells, commonly used as an experimental model for human osteoblasts, using U-73122 PLC inhibitor, U-73343 inactive analogue, and by silencing ezrin. The treatment with U-73122 significantly reduces the number of MG-63 viable cells and contemporarily modifies the expression and the subcellular localization of selected PLC isoforms. U-73122 reduces the cell growth in cultured MG-63 ostesarcoma cell line involving PI-specific Phospholipases C.
Similar content being viewed by others
Background
Osteosarcoma comprises less than 1 % of cancers in the United States, occurring in less than 1000 patients per year. It is the most common primary bone tumour in childhood and adolescence (Mirabello et al. 2009a, b). The presence of metastasis confers worse prognosis for osteosarcoma affected patients (Meyers et al. 2005). Metastasizing tumours partially respond to current therapies and represent the primary cause of cancer related mortality (Meyers et al. 2005).
Osteosarcoma includes several pathological entities, comprising of different clinical, radiological, and histological features (Mirabello et al. 2009a, b; Gatta et al. 2005). Osteosarcoma arises from a mesenchymal cell that owns or can acquire the ability to produce osteoid (Gorlick et al. 2003; Marina et al. 2010). The complex pathogenesis of osteosarcoma includes numerous factors (Gorlick et al. 2003), both genetic abnormalities and environmental exposures, in experimental models as well as in humans (Gorlick 2009). Many efforts to identify a unifying recurrent event were made, bringing multiple genetic risk factors together with epidemiologic and association data. However, despite the considerable improvement in the knowledge, the pathogenesis of osteosarcoma is largely unknown.
The definition of the number and nature of the signal transduction pathways involved in the pathogenesis and the identification of the molecules promoting metastasis spread might improve the knowledge of the natural history of osteosarcoma, allowing refine the prognosis and opening the way to novel therapeutic strategies. Recently, great interest arose about ezrin a protein belonging to the ezrin–radixin–moesin (ERM) family, and related signal transduction pathways (Khanna et al. 2004; Ferrari et al. 2008; Hunter 2004, Dard et al. 2004; Zhu et al. 2007; Yang et al. 2012; Zhao et al. 2011; Tan and Yang 2010). Ezrin protein is codified by Vil2 gene (OMIM *123900). The Protein 4.1, ezrin, radixin, moesin (FERM) domain (Chishti et al. 1998) of ezrin is involved in the recognition of Phosphatydil inositol (4,5) bisphosphate (PIP2), a crucial molecule belonging to the Phosphoinositide (PI) signal transduction pathway (Gautreau et al1999; Martin 2003; Pujuguet et al. 2003; Zhao et al. 2004; Hao et al. 1997; Fievet et al. 2004, 2007). The actin binding activity of Ezrin (Defacque et al. 2000, 2002) largely depends on the membrane PIP2 levels (Hao et al. 2009). ERM proteins simultaneously bind actin and, by means of their N-terminal domains, PIP2 located at the membrane (Niggli and Rossy 2008; Gilmore and Burridge 1996; Isenberg and Niggli 1998; Nakamura et al. 1999; Eberle et al. 1990; Dobos et al. 1992; Apgar 1995; Hartwig et al. 1995; Gachet et al. 1997; Gratacap et al. 1998). Beside phosphorylation, activation of ERM proteins, was suggested to occur after interaction with PIP2, which induces the conformation to open (Gilmore and Burridge 1996). Both PIP2 binding and phosphorylation are thought to allow the stabilization of ERM proteins or a more efficient binding to their own receptors (Hirao et al. 1996; Heiska et al. 1998; Legg and Isacke 1998; Nakamura et al. 1999). Increasing evidences indicated that ezrin is involved in osteosarcoma progression and metastasis and that the levels of PIP2 play a critical role for its activation.
PIP2, a phosphorylated derivative of phosphatydil inositol mainly located in the inner half of the plasma membrane lipid bilayer, is critical for many cellular activities, such as endo- and exocytosis, ion channel activity and cell motility. The levels of PIP2 are regulated by means of PI-specific Phospholipase C (PLC) family of enzymes (Berridge and Dupont 1994; Divecha and Irvine 1995; Hisatsune et al. 2005; Rhee 2001; Bunney and Katan 2011; Fukami et al. 2010).
Activated PLC cleaves PIP2 into inositol trisphosphate (IP3) and diacylglycerol (DAG), both crucial molecules in signal transduction (Rhee et al. 1991). IP3 induces calcium release. DAG can be further cleaved to release arachidonic acid (Tang et al. 2005) or can activate serine/threonine calcium-dependent protein kinase C enzymes (PKC), also influenced by the IP3-induced calcium increase.
The mammalian PLC family comprises a related group of complex, modular, multi-domain enzymes which cover a broad spectrum of regulatory interactions, including direct binding to G protein subunits, small GTPases from Rho and Ras families, receptor and non-receptor tyrosine kinases and lipid components of cellular membranes (Rhee et al. 1991). PLC enzymes are thirteen isoforms classified on the basis of amino acid sequence, domain structure and mechanism of recruitment into six subfamilies: β(1–4), γ(1–2), δ(1, 3, 4), ε(1), ζ(1), and η(1–2) (Suh et al. 2008).
The activity of PLC is required for chemokine mediated dissociation of ERM proteins from the membrane (Brown et al. 2011). Previous studies had placed selected PLC enzymes at the convergence point for the broad range of signalling pathways that promote Rho and Ras GTPase mediated signalling (Hao et al. 2009; Lo Vasco et al. 2015), which also contributes to the regulation of ezrin metabolism. In our previous reports we suggested that the RasGTPases network ezrin involving the PLC enzymes (Lo Vasco et al. 2015).
In our previous reports, we identified the panel of expression of PLC enzymes (Lo Vasco et al. 2013) and analyzed the effect of ezrin silencing or PLCE isoform silencing upon selected osteosarcoma cell lines (Lo Vasco et al. 2014a, b).
In the present experiments, we analyzed the PLC signal transduction system in cultured human osteosarcoma MG-63 cells. MG-63 cell line is commonly used as an experimental model for human osteoblasts, presenting with low levels of alkaline phosphatase activity, and PTH unresponsive adenylate cyclase (Fukayama and Tashjian Jr. 1990).
We treated MG-63 cells with U-73122 (1-[6-[[17b-3-methoxyestra-1,3,5(10)-trien-17-yl]amino]exyl]-1H-pyrrole-2,5-dione), a widely used PLC enzyme inhibitor that probably also acts upon the gene regulation (Lo Vasco et al. 2010a, 2013 os1). U-73343 [1-(6-((17-beta-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-2,5-pyrrolidinedione], a structural analogue of U73122, with negligible activity as a PLC inhibitor, was used as a control compound (Heemskerk et al. 1997). We silenced Vil2 using siRNA methodology (Lo Vasco et al. 2014a). We evaluated all those treatments upon the MG-63 viability, in order to investigate the role of PLC inhibition upon cell growth and survival. As U-73122 is not water soluble and it used to be dissolved in dimethyl sulfoxide (DMSO), we evaluated the effects of DMSO alone upon cultures. We also evaluated the morphological changes occurring in MG-63 after different treatments and investigated the localization and sub-cellular distribution of PLC isoforms within untreated and U-73122 treated MG-63 cell.
Methods
Cell culture
MG-63 human osteosarcoma cell line was obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were counted using a Neubauer haemocytometer and a phase contrast microscope. Cells were grown at 37 °C with 5 % of CO2 in Dulbecco’s minimum essential medium (Sigma) supplemented with 10 % fetal bovine serum (GIBCO), penicillin (100 μg/ml), streptomycin (100 U/ml) and sodium pyruvate. Cells were grown for 24 h, reaching a confluence of around 40–60 %, and until confluence (reached from 72 to 96 h). U-73122 (Sigma-Aldrich) was addicted solved in DMSO (Takenouchi et al. 2005; Lo Vasco et al. 2011).
The initial number of cells (time 0) was 250.000 for each experiment of growth curve assessment. The number of cells for molecular biology experiments was 1 × 106 cells/each experiment. Each experiment was repeated at least three times.
Cells were grown under different conditions. Cells were grown respectively 24 h: (1st) without treatment, with addiction to the culture medium of respectively (2nd) DMSO (3th) 10 μΜ of U-73122, (4th) 10 μM U-73343 (Sigma-Aldrich), (5th) 1 μM U-73122. Untreated cells were also grown (6th) until the confluence was reached (range 72–96 h). Analyses of cultures was performed, both in treated and untreated cells, in the beginning of experiment (time 0), after 1, 3, 6, 24 h and at confluence for PCR experiments; after 18 and 24 h for morphology experiments. Experiments were independently repeated at least 3 times for each isoform.
Cell survival Trypan blue test
Cells suspension was diluted 1:1 in 0.4 % Trypan blue staining (Sigma Aldrich, Dorset, UK) for survival quantification. Viable cells were counted using a Neubauer haemocytometer and a phase contrast microscope. The following equation was used to calculate the total number of viable cells in 1 ml suspension: number of total viable cells in 1 ml (TC) = \(\bar{x}*2*10^{4}\) (\(\bar{x}\) = average of the cell counts from the squares of the haemocytometer grid, 2 = dilution factor 1:1). The number of live cells was used to determine the growth rate and experiments were repeated three times.
Cells transfection for ezrin silencing
MG63 cells were transiently transfected with ezrin silencing RNA using METAFECTENE SI+ (Biontex Laboratories GmbH, Munich, Germany). siRNA sequences targeting Ezrin and negative control siRNA, were designed and synthesized by Invitrogen (Life Technologies, Foster City, CA, USA). The siRNA was designed according to Ezrin complementary DNA (cDNA) sequence (EZR Gene ID: 7430). Briefly, 2.2 ml cell suspension were prepared in complete cell culture medium with a concentration of 1.5 × 105 cells/ml. Cells were seeded, in 6-well plates, shortly before the addition of the lipoplex, according to the manufacturer’s instructions. Then cells were incubated under normal culture conditions (37 °C in CO2–containing atmosphere) until the lipoplex addition. Before transfection, 150 µl of 1× SI+ buffer were mixed with 72 µl of METAFECTENE® SI+ and 540 pMol of RNA stock solution. The mixture was incubated for 15 min at room temperature and then added to the cells in 1 h from seeding. Cells were incubated 72 h. Functional siRNA was measured by reverse transcription–polymerase chain reaction (RT-PCR) and western blot analysis 24, 48 and 72 h after tranfection.
RNA extraction
After all the above indicated treatment procedures and times, cells were detached and suspended using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA). Total RNA was extracted with a SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The concentration and purity of the obtained RNA was checked using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Inc. USA).
RT-PCR
RNA was reverse-transcribed into cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Foster City, CA, USA) following manufacturer’s indications. The RNA mix was then amplified for 10 min at 25 °C, 120 min at 37 °C and 5 min at 85 °C in a Gene Amp® PCR System 9700 (Applied Biosystems) thermocycler.
Glyceraldehyde 3 phosphate dehydrogenase (GAPDH) was used as positive control (Bio Basic Inc, Amherst, New York, USA). The primer pairs (Bio Basic Inc, Amherst, New York, USA) for each PLC isoform, GAPDH and Vil2 gene are listed in Table 1. The specificity of the primers was verified by searching in the NCBI database for possible homology to cDNAs of unrelated proteins. RNA samples were also amplified by PCR without RT to exclude possible contamination.
Standard analytical PCR reaction was performed with GoTaq Master Mix (Promega) following manufacturer’s instructions. Cycling conditions were performed with 95 °C initial denaturation step for 1 min was followed by 40 cycles consisting of 95 °C denaturation (30 s), annealing (30 s) at the appropriate temperature for each primer pair and 72 °C extension (1 min) in Gene Amp® PCR System 9700 (Applied Biosystems) thermocycler. Amplified PCR products were analysed by 1.5 % TAE ethidium bromide-stained agarose gel electrophoresis (Agarose Gel Unit, Bio-Rad Laboratories S.r.l., Segrate, IT). A PC-assisted CCD camera (GelDoc 2000 System/Quantity One Software; Bio-Rad) was used for gel documentation and quantification. Optical densities were normalized to the mRNA content of GAPDH.
RNA samples were also amplified by PCR without RT. No band was observed, excluding DNA contamination during the procedure (data not shown). Experiments were independently repeated at least 3 times for each isoform.
Real-time PCR
Gene expression of Ezrin, PLC ε, PLC γ2, PLC δ4 were determined by real-time PCR using the 7500 Real-Time PCR instrument from Applied Biosystems™. TaqMan® primers and probes for each gene, as well as the GAPDH reference gene, were obtained from Applied Biosystems™.
Transfected MG63 cells and normal controls were harvested 24, 48 and 72 h after transfection. The messenger RNA (mRNA) expression of Ezrin, PLC ε, PLC γ2, PLC δ4 was determined by Real-Time PCR. Total RNA was extracted with a SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. We confirmed purity and quantity of RNA by NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Inc. USA). The RNA was reverse transcribed into cDNA with High Capacity cDNA Reverse Transcription Kit (Life Technologies, Foster City, CA, USA).
PCR products were detected using gene-specific primers and probes labeled with reporter day FAM which yielded a predicted amplicons of 82, 84, 61, 78, 64, 93 and 62 base pairs respectively; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal standard, which yielded a predicted amplicon of 58 base pairs. Reaction mixtures for all gene expression assays contained: 5 μl TaqMan® mastermix (2×; Applied Biosystems™), 0.5 μl gene of interest primer/probe mix and 1 μl PCR grade water. To each reaction, 3.5 μl of the diluted cDNA (35 ng) were added. All samples were assayed in triplicate. PCR reaction was carried out in triplicate on 96-well plate with 10 μl per well using 1× TaqMan Master Mix. After an incubation for 2 min at 50 °C and 10 min at 95 °C, the reaction continue for 40 cycles at 95 °C for 15 s and 60 °C for 1 min. At the end of the reaction, the results were evaluated using the ABI PRISM 7500 software. For each sample, ΔCt value was calculated as Ct of the target gene minus Ct of the endogenous gene. Subsequently, for each sample, ΔΔCt value was calculated as ΔCt of the sample minus ΔCt of the control sample. Relative quantification was obtained as the mathematical function 2 − ΔΔCt. Based on these calculations, the control sample has a value of 1, taken as 100 %.
Western Blot
Whole-cell lysates (106 cells for each experiment) were prepared by lysing cells in RIPA buffer (50 mM Tris pH = 7.5, NP-40, 0.1 % SDS, 100 mM NaCl, 50 mM NaF, 1 mM EDTA) supplemented with a set of protease inhibitors: 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, 1 mM sodium benzamidine, and 1 mM phenylmethylsulfonyl fluoride. Proteins (50 μg) were separated on 12 % polyacrylamide, 0.1 % SDS gel. Then, incubation with a monoclonal antibody specific for each PLC isoform (Santa Cruz, CA) followed. Immunoreactive bands were visualized using the enhanced chemiluminescence method. Experiments were independently repeated at least 2 times for each isoform.
For PLC β2 and PLC η2, positive controls were used to test the efficacy of primers in RT-PCR analyses and of antibodies in Western Blot analyses. For PLC β2 human leucocytes were used as positive controls. For PLC η2, nervous tissue was used as positive control.
Immunofluorescence analysis of subcellular distribution of target molecules
Immunofluorescence localization of all PLC isoforms was performed on coverslips cultured cells with 18 and 24 h U73122 treated cells and untreated control cells. Cells were washed three times with PBS and fixed with 4 % paraformaldehyde (PFA) in phosphate buffer saline (PBS) for 10 min at 4 °C, followed by three washes with PBS. Cells were incubated with primary antibodies diluted in PBS for 1 h at room temperature. Cover-slips were then incubated with the specific secondary antibody Texas Red or fluorescein-conjugated for 1 h at room temperature. Cells were washed twice with 1X PBS 5 min, then counterstained with 4′,6-diamidino-2-phenylindole (DAPI) fluorescent staining. The slides were visualized images were visualized and captured with an Olympus IX50 inverted fluorescence microscope (Olympus, Tokyo, Japan) and processed using Adobe Photoshop 7.0 software.
Statistical analysis
For in vitro studies, differences were determined either with two-way repeated measures analysis of variance (ANOVA) (http://www.physics.csbsju.edu/stats/anova_NGROUP_NMAX_form.html) with Bonferroni’s multiple comparison test, or by student’s one tailed t test, using Prism 5.0a software (GraphPad Software, San Diego, CA, USA). A p value <0.05 was considered significant.
Results
Cells morphology and growth
Untreated cells reached the confluence after 72–96 h, acquiring the expected morphology: cells were oval to spindle-shaped, without branching cell processes. The number of Cells treated with 1 μΜ U-73122 was decreased and intercellular adhesion was subsequently reduced; moreover, a little percentage of round cells were countered (5 %) after 6 h from treatment. In cultures treated with 10 μΜ U-73122, marked morphology changes were observed in MG-63 cells, acquiring round shape.
Untreated cells linearly grew with doubling time about 15 h. After 24 h the number of viable cells increased 2.7 folds (Table 2; Fig. 1).
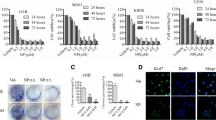
MG-63 cultures. a Phase contrast microscopy of MG-63 cells. Cell morphology cultured 24 h: (upper line) untreated control cells (CTRL) and cells treated with U-73122 1 μΜ (centre line) and 10 μΜ (lower line) after 1, 3, 6 h from seeding b, c MG-63 cells after 1, 3, 6 h from seeding: untreated (NT), DMSO, U-73122 1 μΜ, U-73122 10 μΜ and U-73343 10 μΜ treated cells. c Comparison histogram of cell growth with error bars
In cultures in which DMSO was added to the culture medium, cells showed a little growth slowing down after 1 h from administration of DMSO. However, the number of viable cells rapidly increased after 6 h. The number of viable cells increased 1.9 and 2.8 folds respectively after 1 μΜ and 10 μΜ DMSO. After 24 h from DMSO administration, the doubling time was not completely reached with 1 μΜ DMSO, while it was reached after 13.5 h (Table 2; Fig. 1).
In cultures in which different concentrations of U-73122 in DMSO was added to the culture medium, we observed a significant reduction of the number of viable cells, more evident with the highest U-73122 concentration. The number of viable cells after treatment with 1 μΜ U-73122 was reduced after from 1 to 6 h, and increased 1.7 folds after 24 h (Table 2; Fig. 1).
In cultures in which 10 μΜ U-73343 in DMSO was added to the culture medium, the growth linearly increased from 6 to 24 h. After 24 h, the number of live cells increased 2.5 folds; the doubling time was reached after 16 h (Table 2; Fig. 1).
Statistical analysis
Mean, standard deviation (SD) and standard error of the mean (SEM) for each cells group are indicated in Table 2.
The unpaired t-test was results are listed in Table 3. Comparing UT versus 10 μΜ U-73122 treated cells resulted statistically significant. Comparing UT versus 1 μΜ U-73122 treated cells resulted not quite statistically significant. Comparing UT versus 10 μΜ U-73343 treated cells, versus 10 μΜ DMSO treated cells and versus 1 μΜ DMSO treated cells resulted not statistically significant (Table 3).
RT-PCR
Confluence cultured cells: mRNA for PLCB1, PLCB3, PLCB4, PLCG1, PLCD1, PLCD3, PLCE, GAPDH and Vil2 was detected. For PLCG2 the presence of mRNA was variably detected in 50 % of experiments. No mRNA for PLCB2, PLCD4, PLCH1 and PLCH2 was detected (Table 4).
Untreated controls: mRNA for PLCB1 was present at time 0, absent at 1, 3, 6 h and detected after 24 h. mRNA of PLCB3, PLCB4, PLCG1, PLCG2, PLCD1, PLCD3, PLCE, GAPDH and Vil2 was detected in all the analyzed times (1.3, 6 and 24 h). mRNA of PLCB2, PLCD4, PLCH1 was never detected. mRNA of PLCH2 was not present at time 0 and after 1 h from cells seeding, while was detected after 3, 6 and 24 h (Table 4).
Control treatment with DMSO 10 μΜ: mRNA of PLCB1 was variably present after 1 h from treatment and detected after 3, 6 and 24 h. mRNA of PLCB3, PLCB4, PLCG1, PLCD1, PLCD3, GAPDH and Vil2 was detected after each considered interval. mRNA of PLCE was variably detected after 1 h from DMSO addiction in 25 % of experiments, while it was detected after 3, 6 and 24 h. mRNA of PLCH2 was detected after 1 h, variably present after 3 h in 25 % of experiments and present after 6 and 24 h from DMSO addiction. No mRNA of PLCB2, PLCD4 and PLCH1 was detected (Table 4).
Treatment with U-73122/DMSO 10 μΜ. PLCB1: after 1 h, mRNA was detected, absent after 3 h, present after 6 and 24 h. PLCB2, PLCD4, PLCH1: mRNA was never detected. PLCB3: mRNA was detected after 1 h in 50 % experiments, detected after 3 h, undetected after 6 h and detected after 24 h. PLCB4: mRNA was detected after 1 and 3 h in 50 % experiments, detected after 6 h and absent after 24 h. The mRNA of PLCG1, GAPDH and Vil2 transcription was detected at each interval. PLCG2: the mRNA was not detected excepting for detection in 50 % experiments after 6 h from U-73122 adding to the culture. PLCD1: mRNA was detected in 50 % experiments after 1 h, and was detected after 3 and 6 h; after 24 h was not detected. PLCD3: mRNA was not detected after 1 h, while was present in the remaining intervals. PLCE: mRNA was variably detected in 50 % experiments, was absent after 3 h, present after 6 and absent after 24 h. PLCH2: mRNA was not detected after 1 h, while was detected in the remaining intervals (Table 4).
Effectiveness of cells transfection and Real time PCR after ezrin silencing
Silencing of ezrin was validated by western blot, RT-PCR and gel electrophoresis of mRNA extracts and compared to non-targeting control siRNA. Ezrin transcription was compared in cells transfected with ezrin-silencing specific siRNA to controls, comprising untransfected cells and cells transfected with the carrier metefectamine.
Real Time PCR performed after ezrin silencing showed that Vil2 was not expressed, as expected. GAPDH was expressed, as expected. PLCG2 and PLCD4 were not expressed, while PLCE was quite almost reduced The results are listed in Table 5.
Immunofluorescence analysis of sub-cellular distribution of target molecules
PLB β1: present in the cytoplasm and weakly in the nucleus of untreated cells, in U-73122-treated cells the fluorescence signal was reduced and the protein was present in perinuclear area (Fig. 2).
Immunofluorescence images of MG-63 cells (controls and cells treated with 10 micrm U-73122). For each PLC isoform, on the left immunomarking with the corresponding antibody Red Texas (red) or FITC (green) conjugated; DAPI counterstain for nuclei in the centre; on the right merge (inverted fluorescence microscope, 40×)
PLC β2: a very weak signal was detected in untreated cells; the signal was almost undetectable in U-73122-treated cells (Fig. 2).
PLC β3: in untreated cells, the signal was weakly detected in the cytoplasm; in U-73122-treated cells the protein was distributed exclusively in the perinuclear area (Fig. 2).
PLC β4: in untreated cells, the protein was detected in the cytoplasm, where it was also in U-73122-treated cells, although the intensity seemed to be reduced with respect to controls (Fig. 2).
PLC δ1: in untreated cells, the enzyme was detected in the cytoplasm with evident submembrane reinforcement; in U-73122-treated cells, it was also detected in the cytoplasm, although the membrane reinforcement resulted less evident or absent (Fig. 2).
PLC δ3: in untreated cells, the enzyme was present and localized in the cytoplasm, while in U-73122-treated cells it was not detected (Fig. 2).
PLC δ4: absent both in untreated and U-73122 treated MG-63 cells (Fig. 2).
PLC γ1 and PLC γ2: in untreated cells it was detected in the cytoplasm, where it was detected also in U-73122-treated cells, although the intensity was reduced (Fig. 2).
PLC ε: in untreated cells the enzyme was detected in the cytoplasm, while it was not detected in U-73122-treated cells (Fig. 2).
PLC η1: in untreated cells, the enzyme was localized in the cytoplasm and probably accumulated in plasmatic vesicles; in U-73122-treated cells the intensity of the signal was reduced (Fig. 2).
PLC η2: in untreated cells, the enzyme was localized in the cytoplasm and in plasmatic vesicles; detected in U-73122-treated cells it was localized in the perinuclear area and the plasmatic vesicles resulted less numerous with respect to controls (Fig. 2).
Discussion
In previous reports, we described the panel of expression of PLC enzymes in MG-63 cells (Lo Vasco et al. 2013). The expressed PLC isoforms were PLC β1, β2, β3, β4, γ1, γ2, δ1, δ3 and ε. In the present experiments we analyzed the sub-cellular distribution of PLC enzymes and the expression panel of PLC isoforms in confluence cultures (72–96 h). In the present experiments, the PLC panel of expression of cells after reaching the confluence did not differ from the 24 h cultured control MG-63 cells. Interestingly, the expression of PLCG2 transcript was variably detected after confluence (50 % experiments).
In that previous work (Lo Vasco et al. 2013) we had used high concentrations of U-73122, a widely used PLC inhibitor (30 μΜ), observing lack of the PLCs’ transcription after 3 h. The transcripts for the expressed isoforms were observed after 24 h (Lo Vasco et al. 2013). In the present experiments, we confirmed previous observations using 30 μΜ U-73122 (Table 5). U-73122 is not water soluble and, as well as U-73343, needs to be dissolved in DMSO, an amphipathic molecule commonly used as a solvent and worldwide used as cryoprotectant. DMSO acts as a differentiating agent, playing multiple roles both on cellular functions (e.g., metabolism and enzymatic activity) and cell growth by affecting cell cycle and apoptosis (Santos et al. 2003). More specifically, DMSO was demonstrated to induce differentiation in stem and endothelial cells (Jasmin et al. 2010). We aimed to investigate whether DMSO might affect cell growth and/or PLC expression. Thus, we compared the effects of DMSO, U-73122 and U-73343, an inactive U-73122 analogue, upon cell growth, PLC expression and localization. MG-63 cell growth curve was significantly modified by U-73122 (Table 2; Fig. 1). After 24 h from seeding, the number of untreated cells increased 2.7 folds, with doubling time about 15 h. In cultures in which different concentrations of U-73122/DMSO were added to the culture medium, we observed reduction of the number of viable cells. The decrease of viable cells number was statistically significant using 10 μΜ U-73122 dosage. The differences induced in cell growth by the lowest 1 μΜ U-73122 dosage resulted quite not statistically significant. The differences in cell growth after use of DMSO and U-73343 did not result statistically significant compared to untreated cells (Tables 3, 4). Therefore, U-73122 acts upon the cell viability, significantly reducing the number of viable MG-63 cells within 24 h.
U-73122, amphiphilic alkylating aminosteroid homologue of the thiol reagent N-ethylmaleimide (Bleasdale et al. 1990), is the most known and archetypal inhibitor of PLC. U-73122 was described to inhibit the Ca2+ mobilization in a dose-dependent manner, consistent with a mechanism of action involving PLC inhibition. The inhibition of PLC after U-73122 treatment is supposed to be possibly due to an action at Gprotein coupling level (Smallridge et al. 1992). U-73122 was frequently used to define the role of PLC mediated elevation of intracellular calcium concentration, indirectly used as a tool to investigate the PLCs’ signal transduction. However, lack of selectivity of PLC inhibition by U-73122 was described, as it acts (Cenni and Picard 1999) upon a number of unrelated proteins (Feisst et al. 2005; Hughes et al. 2000; Walker et al. 1998; Berven and Barritt 1995; Pulcinelli et al. 1998). U-73122 PLC inhibiting activity is due to prevention of the turnover of PI, thus avoiding the formation of the second messengers IP3 and DAG (Vickers and Fisher 2004; Thomas et al. 2005). However, it is not fully known the mechanism leading to inhibition. Controversial reports described that U-73122 might be ineffective upon inhibition of PLC γ2 (Hellberg et al. 1996). The isomer of U-73122, namely U73343, is commonly used as a control. Although controversial reports might suggest a more complex relationship, U-73343 is considered inactive upon PLC enzymes (Heemskerk et al. 1997). The inhibitory effect of U-73122 appeared to be dependent on the presence of a pyrroledione group, as replacement of this with pyrrolidinedione (to form U-73343) abolished the inhibitory effect. (Hollywood et al. 2010). Both compounds play varying effects on cell signaling by activating nuclear estrogen receptors (Cenni and Picard 1999), acting as a protonophore in rabbit parietal cells, and activating ion channels (Mogami et al. 1997).
In the present experiments, MG-63 cell growth was reduced after U-73343 treatment, although results were not statistically significant (Tables 2, 3, 4; Fig. 1). The present results indicated that U-73122 reduced the cell growth of cultured MG-63 line, decreasing the number of cells from adding to 24 h. Comparisong to other treatments we performed, one might speculate that the cell growth is related to PLC enzymes and that viable cells number is reduced inhibiting PLC enzymes.
Beside the observed effects upon cell viability, previous evidences indicated that U-73122 might act upon the gene expression of PLC isoforms (Lo Vasco et al. 2010a, b, 2011, 2012, 2013). In the present experiments, molecular biology analyses indicated that a rearrangement of the panel of expression of PLC enzymes occurred after 1, 3, 6 and 24 h after U-73122 adding.
In cultured MG-63, PLCB1 transcript was absent after 1, 3 and 6 h and was detected after 24 h from seeding. In U-73122 treated cells, PLCB1 transcript was detected after 1 h, absent after 3 and detected after 6 and 24 h. In MG-63 cells grown adding DMSO to the culture medium, PLCB1 transcript was weakly detected after 1 h from seeding and detected in the remaining intervals. PLC β1 was described to be selectively increased during myoblast and adipocyte differentiation (Faenza et al. 2004; O’Carroll et al. 2009), and evidences suggested that deletion of PLCB1 favours cancer progression in the myeloid lineage (Lo Vasco et al. 2004; Kaminskas et al. 2005). Thus, increase in PLC β1 protein and PLCB1 transcript levels might be actually considered to unfavour cancer progression and/or favour differentiation. Our previous results in osteosarcoma cells corroborated the hypothesis that increase of PLCB1 expression might be opposite to cancer progression, confirming a differentiating role (Lo Vasco et al. 2013, 2014a, b, 2015). However, the increase in PLCB1 transcription we observed in the present experiments might be due to DMSO, and the role of U-73122 cannot be separated and understood. Moreover, while PLC β1 was localized in the nucleus and in the cytoplasm, after either treatment the protein was reduced and localized only in the peri-nuclear area. The differences in PLC β1 localization, due to DMSO, to U-73122 or to either compounds, might be related to differentiation.
In our present experiments, cultured MG-63 cells own PLCB3 transcripts and weak PLC β3 protein signal was localized in the cytoplasm, accordingly to our previous observations (Lo Vasco et al. 2014a, b). In U-73122 treated cells, no PLCB3 transcript was detected after 6 h. Interestingly, after U-73122 treatment, the PLC β3 protein was exclusively localized in the nucleus and peri-nuclear area. DMSO did not seem to affect the expression of PLCB3. PLC β isozymes are autoinhibited, and several proteins, including Gαq, Gβγ, and Rac1, directly engage distinct regions to release autoinhibition. High concentrations of Gαq or Gβ1γ2 selectively activate PLC β3 at membranes (Charpentier et al. 2014). The transcription factor Stat5, involved in various leukemias, is regulated by PLC β3-dependent manner (Xiao et al. 2010). Moreover, PLCB3 was claimed to be involved in a subset of endocrine tumours, where its expression was demonstrated to be decreased or absent (Stålberg et al. 2003).
Transcription of PLCB4 was detected after 1, 3 (in half experiments) and 6 h after U-73122 treatment, and was not detected after 24 h. PLC β4, just as PLC-β1, was specifically involved in the histamine-induced IP3 increases in HeLa cells (Ishida et al. 2014). Amplifications of PLCB4 were described in glioblastoma multiforme (Waugh 2016) and altered expression was reported in non-small cell lung cancer (Tan and Chen 2014) and endometrial cancer (Orchel et al. 2012). Therefore, PLC β4 might favour the progression of selected cancers. In the present experiments, in MG-63 untreated cells, the protein was detected in the cytoplasm, where it was also in U-73122-treated cells, although the intensity seemed to be reduced with respect to controls.
In our present experiments, PLCG2 was expressed in 24 h-cultured MG-63 cells, it was not expressed 1 and 3 h after adding DMSO to cultures, and detected in the remaining intervals. After adding U-73122 to MG-63 cultures, PLCG2 was very weakly detected exclusively after 6 h from treatment, while it was absent in the remaining intervals. The localization of PLC γ2 did not differ, being the protein detected in the cytoplasm, although the signal was dramatically reduced after U-73122 treatment. In our previous studies we had suggested a critical role for PLC γ2 in 143B cells (Lo Vasco et al. 2014a), probably related to their osteolytic features. Literature data indicated that the PLC γ subfamily enzymes used to be detected at higher level in tumour than normal tissues (Arteaga et al. 1991; Noh et al. 1995). Isoforms belonging to the PLC γ subfamily contain a unique region comprising two tandem SH2 domains and one SH3 domain adjacent to a split PH (Katan and Williams 1997), which allow the interaction with different molecules (Bunney and Katan 2011). PLC γ2 is required for early phase osteoclast differentiation (Kertész et al. 2012), and is involved both in actin cytoskeleton reorganization and Rac-activation in dendritic cells (Cremasco et al. 2010), as well as in the integrin-mediated processes of adhesion, migration and bone resorption in osteoclast (Epple et al. 2008). PLC γ2 was also suggested to represent a critical regulator of the cellular and molecular mechanisms occurring in bone and immune cells during autoimmune inflammation (Faccio and Cremasco 2010).
Although U-73122 is widely used as non isoform-specific PLC inhibitor (Lea et al. 2002), it was controversially considered to act upon PLC γ subfamily enzymes (Kim et al. 2012). In fact, isoforms belonging to the PLC γ subfamily are considered “udraggable” proteins (Lattanzio et al. 2013), with special regard to PLC γ1 isoform. However, U-73122 is used as a PLC γ inhibitor (Glassford et al. 2003). Our present results indicate that U-73122 affects the expression of PLCG genes.
The results of morphology experiments did not show accountable variations in the presence of the PLC γ2 protein within the U-73122 treated cells, excepting for slight reduction of fluorescence intensity. However, the transcription of PLC γ2 significantly differs after treatment with U-73122 compared with both untreated counterpart and DMSO adding to the cell cultures. Interestingly, ezrin silencing induced loss of PLCG2 expression according to previous findings in osteosarcoma which suggested a crucial role for this isoform (Lo Vasco et al. 2014a, b).
The transcription product of PLCD3 was not detected after 1 h from U-73122 treatment, while it was detected in the remaining intervals. PLC δ3, just like PLC δ1 and PLC β1, was localized to the cleavage furrow during cytokinesis. Activation of selected PLC isoforms at the cleavage furrow controls progression of cytokinesis through regulation of PIP2 levels (Naito et al. 2006). PLC δ3 is highly enriched in the cerebellum and cerebral cortex and was demonstrated to promote neurite extension negatively regulating RhoA expression (Kouchi et al. 2011). Moreover it was found altered in nasal polyps (Babeto et al. 2010). PLC δ isozymes, the most primitive and evolutionary conserved, are known to be the most sensitive to calcium (Suh et al. 2008). Our previous studies described the variation of PLC δ1 and PLC δ3 expression in LPS-induced inflammation and inflammatory diseases, such as endometriosis (Lo Vasco et al. 2010a, b, 2011, 2012). That suggested that PLC δ3 might be involved in the fine tuning and regulation of the inflammation cascade. Further studies are required in order to highlight the role of PLC δ3 isoform in the progression of the cell cycle, with special regard to the activity upon proliferation regulation.
In the present experiments, the PLCE mRNA was detected in 50 % experiments after 1 h from U-73122 treatment, was absent after 3 h, present after 6 and absent after 24 h, indicating a “fluctuating” behaviour. In untreated MG-63 cells, the enzyme was detected in the cytoplasm, while it was not detected in U-73122 treated cells. PLC ε, expressed in the outermost layer of the neural tube, was widely described in the central nervous system, probably related to neuron differentiation (Lo Vasco et al. 2012). Literature data indicated that PLC ε is important for heart development (Tadano et al. 2005; Wang et al. 2005), is involved in nephrotic syndrome (Hinkes et al. 2006), increases insulin secretion (Dzhura et al. 2011), and is highly expressed in the lung (Smrcka et al. 2012). PLC ε activates and is activated by small G protein Ras/mitogen-activated protein kinase (MAPK) signaling pathway (Lopez et al. 2001). The activity of PLC ε, regulated by association with Ras and Rap (Kelley et al. 2001; Schmidt et al. 2001; Song et al. 2001, 2002), might play a role in intracellular signalling from receptors for fibroblast growth factor (FGF) and various neurotrophic factors involved in the neural development (Vaccarino et al. 1999) and in PC12 pheochromocytoma cells (Qiu and Green 1991; Cowley et al. 1994; Zhu et al. 2002). PLC ε was frequently described to be involved in carcinogenesis. However, controversial observations were reported regarding its role. Evidences support the hypothesis that PLC ε might play a tumour suppressor role in Ras-triggered cancers (Martins et al. 2014). However, silencing of PLCɛ was suggested to induce apoptosis via modulation of bcl-2 and bax in bladder cancer (Zhang et al. 2013) and recent literature data suggest that PLC ε might bear cancer-suppression activity (Martins et al. 2014). Inhibition of PLC ε was suggested to prevent the inflammatory reactions associated with tumour development or inflammatory-related diseases (Wang et al. 2005). Plce knockout mice seemed to be resistant to intestinal tumour formation when crossed with Apc∓ mice (Li et al. 2009). Further studies associated selected PLCE polymorphisms with oesophageal squamous cell carcinoma (Abnet et al. 2012; Hao et al. 2013) and with gastric adenocarcinoma (Abnet et al. 2012). Knocking down of PLC ε both in vitro and in vivo was referred to inhibit the growth of bladder tumour cells (Cheng et al. 2011; Ou et al. 2010). Overexpression of PLCE gene was also reported in murine skin cancer (Bai et al. 2004) and Plce (−/−) mice exhibit marked resistance to tumour formation in two-stage skin chemical carcinogenesis (Oka et al. 2010). PLC ε was also claimed to promote progression in head and neck cancer (Bourguignon et al. 2006). Therefore, the mechanism of action and role of PLC ε in cancer promotion and/or progression is far to be highlighted. As several direct effectors of Ras, PLC ε owns RA domain that binds several Ras GTPases, including oncogenic Kras and Hras (Song et al. 2001). The binding involves different interaction surfaces and requires distinct specific recognition of Ras, Rap, or Rho GTPases by at least four different binding interfaces. Those differences have fundamental biological implications for Ras-effector signalling (Song et al. 2001, 2002; Fukami et al. 2010). Interestingly, suppression of ezrin protein expression by antisense transfection or stable expression of short hairpin RNA is associated with reduced Akt and MAPK activity (Khanna et al. 2001, 2004, Yu et al. 2004). In the present experiments, the transcription of PLCE gene was affected by ezrin silencing (Table 5), which almost reduced the corresponding transcript, according to previous findings (Lo Vasco et al. 2014a, b). That last observation might confirm the hypothesis that an extensive crosstalk among the PLC enzymes occurs in cells (Lo Vasco et al. 2012).
Conclusion
The present results confirm our previous observations in human osteosarcoma cell lines suggesting that each cell line owns a specific PLC panel of expression and that a complex organization of PLC enzymes occurs. Probably, PLC enzymes influence each other, networking in a complex manner, probably following a reciprocal hierarchy of control. However, further studies, addressed to identify the crosstalk and the ordered timing of cell line-specific PLC enzymes recruitment might help to highlight the specific role of each isoform, opening the way to novel insights in the progression of the disease, with special regard to metastatic spread.
Our present results indicate that adding of U-73122 to the MG-63 cultures can significantly reduce the number of viable cells, thus slowing the growth within 24 h intervals. Although the specific role played by U-73122 upon PLC enzyme inhibition is still controversial, our results suggest that the compound affects the expression of selected PLC genes, opening the way to novel adjuvant therapy perspectives.
Our present results indicate that the use of U-73122 in MG-63 cultured cells significantly reduces the growth rate contemporarily inducing different expression of PLC genes. That suggests that PLC expression might be related to the cell growth reduction. Interestingly, the expression of PLC genes which codify for PLC isoforms supposed to favour cancer progression was reduced, while the expression of PLCB1, which was thought to favour differentiation and/or apoptosis, was increased. Further studies are required in order to elucidate the specific role of PLC δ3 and PLC ε. Thus, our present results accord to previous literature data with respect to other cancer types. However, a number of PLC isoforms are differently expressed and lesser are differently localized within the MG-63 cells, so that no conclusive thesis about their specific role can be formulated. Further studies are required in order to highlight the role of PLCs upon cell viability and the extensive cross-talk among the isoforms that probably contributes to regulate and network the PLC enzymes.
References
Abnet CC, Wang Z, Song X et al (2012) Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Hum Mol Genet 21(9):2132–2141
Apgar JR (1995) Activation of protein kinase C in rat basophilic leukemia cells stimulates increased production of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: correlation with actin polymerization. Mol Biol Cell 6(1):97–108
Arteaga CL, Johnson MD, Todderud G, Coffey RJ, Carpenter G, Page DL (1991) Elevated content of the tyrosine kinase substrate phospholipase C-gamma 1 in primary human breast carcinomas. Proc Natl Acad Sci USA 88(23):10435–10439
Babeto E, De Fereitas Calmon M, Scarin Provazzi PJ, Colombo J, Cordeiro JA, Bonilha JL, Fernandes AM, Rahal P (2010) Gene expression analysis reveals novel altered genes in nasal polyps. Mol Med Rep 3(3):485–490
Bai Y, Edamatsu H, Maeda S, Saito H, Suzuki N et al (2004) Crucial role of phospholipase Ce in chemical carcinogen-induced skin tumor development. Cancer Res 64:8808–8810
Berridge MJ, Dupont G (1994) Spatial and temporal signalling by calcium. Curr Opin Cell Biol 6(2):267–274
Berven LA, Barritt GJ (1995) Evidence obtained using single hepatocytes for inhibition by the phospholipase-C inhibitor U73122 of store-operated Ca2+ inflow. Biochem Pharmacol 49:1373–1379
Bleasdale JE, Thakur NR, Gremban RS, Bundy GL, Fitzpatrick FA, Smith RJ, Bunting S (1990) Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. J Pharmacol Exp Ther 255:756–768
Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P (2006) Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem 281(20):14026–14040
Brown JB, Cheresh P, Goretsky T, Managlia E, Grimm GR, Ryu H, Zadeh M, Dirisina R, Barrett TA (2011) Epithelial phosphatidylinositol-3-kinase signaling is required for β-catenin activation and host defense against Citrobacter rodentium infection. Infect Immun 79(5):1863–1872
Bunney TD, Katan M (2011) PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem Sci 36(2):88–96
Cenni B, Picard S (1999) Two compounds commonly used for phospholipase C inhibition activate the nuclear estrogen receptors. Biochem Biophys Res Commun 261:340–344
Charpentier TH, Waldo GL, Barrett MO, Huang W, Zhang Q, Harden TK, Sondek J (2014) Membrane-induced allosteric control of phospholipase C-β isozymes. J Biol Chem 289(43):29545–29557
Cheng H, Luo C, Wu X, Zhang Y, He Y, et al (2011) shRNA targeting PLCe1 inhibits bladder cancer cell growth in vitro and in vivo. Urology 78: 474.e477–474.e411
Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ Jr, Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB et al (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci 23(8):281–282
Cowley S, Paterson H, Kemp P, Marshall CJ (1994) Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77(6):841–852
Cremasco V, Benasciutti E, Cella M, Kisseleva M, Croke M, Faccio R (2010) Phospholipase C gamma 2 is critical for development of a murine model of inflammatory arthritis by affecting actin dynamics in dendritic cells. PLoS ONE 5(1):e8909
Dard N, Louvet-Vallee S, Santa-Maria A et al (2004) Phosphorylation of ezrin on threonine T567 plays a crucial role during compaction in the mouse early embryo. Dev Biol 271:87–97
Defacque H, Egeberg M, Habermann A, Diakonova M, Roy C, Mangeat P, Voelter W, Marriott G, Pfannstiel J, Faulstich H, Griffiths G (2000) Involvement of ezrin/moesin in de novo actin assembly on phagosomal membranes. EMBO J 19(2):199–212
Defacque H, Bos E, Garvalov B, Barret C, Roy C, Mangeat P, Shin HW, Rybin V, Griffiths G (2002) Phosphoinositides regulate membrane-dependent actin assembly by latex bead phagosomes. Mol Biol Cell 13(4):1190–1202
Divecha N, Irvine RF (1995) Phospholipid signaling. Cell 80(2):269–278
Dobos GJ, Norgauer J, Eberle M, Schollmeyer PJ, Traynor-Kaplan AE (1992) C5a reduces formyl peptide-induced actin polymerization and phosphatidylinositol(3,4,5)trisphosphate formation, but not phosphatidylinositol (4,5) bisphosphate hydrolysis and superoxide production, in human neutrophils. J Immunol 149(2):609–614
Dzhura I, Chepurny OG, Leech CA, Roe MW, Dzhura E, Xu X, Lu Y, Schwede F, Genieser HG, Smrcka AV, Holz GG (2011) Phospholipase C-ε links Epac2 activation to the potentiation of glucose-stimulated insulin secretion from mouse islets of Langerhans. Islets 3(3):121–128
Eberle M, Traynor-Kaplan AE, Sklar LA, Norgauer J (1990) Is there a relationship between phosphatidylinositol trisphosphate and F-actin polymerization in human neutrophils? J Biol Chem 265(28):16725–16728
Epple H, Cremasco V, Zhang K, Mao D, Longmore GD et al (2008) Phospholipase Cgamma2 modulates integrin signaling in the osteoclast by affecting the localization and activation of Src kinase. Mol Cell Biol 28:3610–3622
Faccio R, Cremasco V (2010) PLCgamma2: where bone and immune cells find their common ground. Ann N Y Acad Sci 1192:124–130
Faenza I, Bavelloni A, Fiume R, Santi P, Martelli AM, Billi AM, Lo Vasco VR, Manzoli L, Cocco L (2004) Expression of Phospholipase C beta family isoenzymes in C2C12 myoblasts during terminal differentiation. J Cell Physiol 200(2):291–296
Feisst C, Albert D, Steinhilber D, Werz O (2005) The aminosteroid phospholipase C antagonist U-73122 (1-[6-[[17-beta-3-methoxyestra-1,3,5(10)-trien-17-yl]amino]hexyl]-1Hpyrrole-2,5-dione) potently inhibits human 5-lipoxygenase in vivo and in vitro. Mol Pharmacol 67:1751–1757
Ferrari S, Zanella L, Alberghini M et al (2008) Prognostic significance of immunohistochemical expression of ezrin in non-metastatic high-grade osteosarcoma. Pediatr Blood Cancer 50:752–756
Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M (2004) Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol 164:653–659
Fievet B, Louvard D, Arpin M (2007) ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta 1773:653–660
Fukami K, Inanobe S, Kanemaru K, Nakamura Y (2010) Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res 49(4):429–437
Fukayama S, Tashjian AH Jr (1990) Stimulation by parathyroid hormone of 45Ca2+ uptake in osteoblast-like cells: possible involvement of alkaline phosphatase. Endocrinology 126(4):1941–1949
Gachet C, Payrastre B, Guinebault C, Trumel C, Ohlmann P, Mauco G, Cazenave JP, Plantavid M, Chap H (1997) Reversible translocation of phosphoinositide 3-kinase to the cytoskeleton of ADP-aggregated human platelets occurs independently of Rho A and without synthesis of phosphatidylinositol (3,4)-bisphosphate. J Biol Chem 272(8):4850–4854
Gatta G, Capocaccia R, Stiller C, Kaatsch P, Berrino F, Terenziani M (2005) EUROCARE Working Group. Childhood cancer survival trends in Europe: a EUROCARE Working Group study. J Clin Oncol 3(16):3742–3751
Gautreau A, Poullet P, Louvard D et al (1999) Ezrin, a plasma membrane-microfilament linker, signals cell survival through the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA 96:7300–7305
Gilmore AP, Burridge K (1996) Molecular mechanisms for focal adhesion assembly through regulation of protein–protein interactions. Structure 4(6):647–651
Glassford J, Soeiro I, Skarell SM, Banerji L, Holman M, Klaus GG, Kadowaki T, Koyasu S, Lam EW (2003) BCR targets cyclin D2 via Btk and the p85alpha subunit of PI3-K to induce cell cycle progression in primary mouse B cells. Oncogene 22(15):2248–2259
Gorlick R (2009) Current concepts on the molecular biology of osteosarcoma. Cancer Treat Res 152:467–478
Gorlick R, Toretsky J, Marina N (2003) Bone tumors. In: Kufe D, Pollock R, Weichselbaum R et al (eds) Cancer medicine, vol 2, 6th edn. BC Decker, Hamilton, pp 2383–2406
Gratacap MP, Payrastre B, Viala C, Mauco G, Plantavid M, Chap H (1998) Phosphatidylinositol 3,4,5-trisphosphate- dependent stimulation of phospholipase C-gamma2 is an early key event in FcgammaRIIA-mediated activation of human platelets. J Biol Chem 273(38):24314–24321
Hao W, Tan Z, Prasad K, Reddy KK, Chen J, Prestwich GD, Falck JR, Shears SB, Lafer EM (1997) Regulation of AP-3 function by inositides. Identification of phosphatidylinositol 3,4,5-trisphosphate as a potent ligand. J Biol Chem 272(10):6393–6398
Hao JJ, Liu Y, Kruhlak M, Debell KE, Rellahan BL, Shaw S (2009) Phospholipase C-mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J Cell Biol 184(3):451–462
Hao N-B, He Y-F, Zhang D, Luo G, Chen B-J, Zhang Y, Yang S-M (2013) PLCE1 polymorphism and upper gastrointestinal cancer risk: a meta-analysis. PLoS ONE 8(6):e67229
Hartwig JH, Bokoch GM, Carpenter CL, Janmey PA, Taylor LA, Toker A, Stossel TP (1995) Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell 82(4):643–653
Heemskerk JW, Farndale RW, Sage SO (1997) Effects of U73122 and U73343 on human platelet calcium signalling and protein tyrosine phosphorylation. Biochim Biophys Acta 1355(1):81–88
Heiska L, Alfthan K, Gronholm M, Vilja P, Vaheri A, Carpen O (1998) J Biol Chem 273:2189
Hellberg C, Molony L, Zheng L, Andersson T (1996) Ca21 signaling mechanisms of the b2 integrin on neutrophils: involvement of phospholipase Cg2 and Ins(1,4,5)P3. Biochem J 317(Pt 2):403–409
Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nurnberg G, Garg P, Verma R, Chaib H, Hoskins BE, Ashraf S, Becker C et al (2006) Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet 38:1397–1405
Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol 135(1):37–51
Hisatsune C, Nakamura K, Kuroda Y et al (2005) Amplification of Ca2+ signaling by diacylglycerolmediatedinositol 1,4,5-trisphosphate production. J Biol Chem 280(12):11723–11730
Hollywood MA, Sergeant GP, Thornbury KD, McHale NG (2010) The PLC inhibitor U-73122 is a potent inhibitor of the SERCA pump in smooth muscle. Br J Pharmacol 160(6):1293–1294
Hughes S, Gibson WJ, Young JM (2000) The interaction of U-73122 with the histamine H-1 receptor: implications for the use of U-73122 in defining H-1 receptor-coupled signalling pathways. Naunyn Schmiedebergs Arch Pharmacol 362:555–558
Hunter KW (2004) Ezrin, a key component in tumor metastasis. Trends Mol Med 10:201–204
Isenberg G, Niggli V (1998) Interaction of cytoskeletal proteins with membrane lipids. Int Rev Cytol 178:73–125
Ishida S, Matsu-Ura T, Fukami K, Michikawa T, Mikoshiba K (2014) Phospholipase C-β1 and β4 contribute to non-genetic cell-to-cell variability in histamine-induced calcium signals in HeLa cells. PLoS ONE 9(1):e86410
Jasmin Spray DC, Campos de Carvalho AC, Mendez-Otero R (2010) Chemical induction of cardiac differentiation in P19 embryonal carcinoma stem cells. Stem Cells Dev 19(3):403–411
Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC, Pazdur R, FDA (2005) Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 11(10):3604–3608
Katan M, Williams RL (1997) Phosphoinositide-specific phospholipase C: structural basis for catalysis and regulatory interactions. Semin Cell Dev Biol 8(3):287–296
Kelley GG, Reks SE, Ondrako JM, Smrcka AV (2001) Phospholipase C(epsilon): a novel Ras effector. EMBO J 20(4):743–754
Kertész Z, Gyori D, Körmendi S, Fekete T, Kis-Tóth K, Jakus Z, Schett G, Rajnavölgyi E, Dobó-Nagy C, Mócsai A (2012) Phospholipase Cγ2 is required for basal but not oestrogen deficiency-induced bone resorption. Eur J Clin Invest 42(1):49–60
Khanna C, Khan J, Nguyen P, Prehn J, Caylor J, Yeung C, Trepel J, Meltzer P, Helman L (2001) Metastasis-associated differences in gene expression in a murine model of osteosarcoma metastasis. Cancer Res 61(9):3750–3759
Khanna C, Wan X, Bose S et al (2004) The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med 10:182–186
Kim SD, Kim HJ, Shim JW, Lee HY, Lee SK, Kwon S, Jung YS, Baek SH, Park JS, Zabel BA, Bae YS (2012) Phospholipase C activator m-3M3FBS protects against morbidity and mortality associated with sepsis. J Immunol 189(4):2000–2005
Kouchi Z, Igarashi T, Shibayama N, Inanobe S, Sakurai K, Yamaguchi H, Fukuda T, Yanagi S, Nakamura Y, Fukami K (2011) Phospholipase Cdelta3 regulates RhoA/Rho kinase signaling and neurite outgrowth. J Biol Chem 286(10):8459–8471
Lattanzio R, Marchisio M, La Sorda R, Tinari N, Falasca M, Alberti S, Miscia S, Ercolani C, Di Benedetto A, Perracchio L, Melucci E, Iacobelli S, Mottolese M, Natali PG, Piantelli M (2013) CINBO (Consorzio Interuniversitario Nazionale per Bio-Oncologia). Overexpression of activated phospholipase Cγ1 is a risk factor for distant metastases in T1–T2, N0 breast cancer patients undergoing adjuvant chemotherapy. Int J Cancer 132(5):1022–1031
Lea JP, Jin SG, Roberts BR, Shuler MS, Marrero MB, Tumlin JA (2002) Angiotensin II stimulates calcineurin activity in proximal tubule epithelia through AT-1 receptor-mediated tyrosine phosphorylation of the PLC-gamma1 isoform. J Am Soc Nephrol 13(7):1750–1756
Legg JW, Isacke CM (1998) Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol 8(12):705–708
Li M, Edamatsu H, Kitazawa R, Kitazawa S, Kataoka T (2009) Phospholipase Cepsilon promotes intestinal tumorigenesis of Apc(Min/+) mice through augmentation of inflammation and angiogenesis. Carcinogenesis 30(8):1424–1432
Lo Vasco VR, Calabrese G, Manzoli L, Palka GD, Spadano A, Morizio E, Guanciali-Franchi P, Fantasia D, Cocco L (2004) Inositide-specific phospholipase C beta1 gene deletion in the progression of myelodisplastic syndrome to acute myeloid leukemia. Leukemia 18(6):1122–1126
Lo Vasco VR, Fabrizi C, Panetta B, Fumagalli L, Cocco L (2010a) Expression pattern and sub cellular distribution of phosphoinositide specific phospholipase C enzymes after treatment with U-73122 in rat astrocytoma cells. J Cell Biochem 110(4):1005–1012
Lo Vasco VR, Fabrizi C, Fumagalli L, Cocco L (2010b) Expression of phosphoinositide specific phospholipase C isoenzymes in cultured astrocytes activated after stimulation with lipopolysaccharide. J Cell Biochem 109(5):1006–1012
Lo Vasco VR, Pacini L, Di Raimo T, D’Arcangelo D, Businaro R (2011) Expression of Phosphoinositide-specific phospholipase C isoforms in HUVEC. J Clin Pathol 64(10):911–915
Lo Vasco VR, Leopizzi M, Chiappetta C, Businaro R, Polonia P, Della Rocca C, Litta P (2012) Expression of phosphoinositide-specific phospholipase C enzymes in normal endometrium and in endometriosis. Fertil Steril 98(2):410–414
Lo Vasco VR, Leopizzi M, Chiappetta C, Puggioni C, Di Cristofano C, Della Rocca C (2013) Expression of phosphoinositide-specific phospholipase C enzymes in human osteosarcoma cell lines. J Cell Commun Signal 7(2):141–150
Lo Vasco VR, Leopizzi M, Puggioni C, Della Rocca C (2014a) Ezrin silencing remodulates the expression of phosphoinositide-specific Phospholipase C enzymes in human osteosarcoma cell lines. J Cell Commun Signal 8(3):219–229. doi:10.1007/s12079-014-0235-9
Lo Vasco VR, Leopizzi M, Stoppoloni D, Della Rocca C (2014b) Silencing of phosphoinositide-specific phospholipase C ε remodulates the expression of the phosphoinositide signal transduction pathway in human osteosarcoma cell lines. Anticancer Res 34(8):4069–4075
Lo Vasco VR, Leopizzi M, Della Rocca C (2015) Ezrin-related phosphoinositide pathway modifies RhoA and Rac1 in human osteosarcoma cell lines. J Cell Commun Signal 9(1):55–62
Lopez I, Mak EC, Ding J, Hamm HE, Lomasney JW (2001) A novel bifunctional phospholipase c that is regulated by Galpha 12 and stimulates the Ras/mitogen-activated protein kinase pathway. J Biol Chem 276(4):2758–2765
Marina N, Gorlick R, Bielack S (2010) Pediatric osteosarcoma. In: CarrollW FinlayJ (ed) Cancer in children and adolecents. Jones and Bartlett, Sudbury, pp 383–394
Martin GS (2003) Cell signaling and cancer. Cancer Cell 4:167–174
Martins M, McCarthy A, Baxendale R, Guichard S, Magno L, Kessaris N, El-Bahrawy M, Yu P, Katan M (2014) Tumor suppressor role of phospholipase C epsilon in Ras-triggered cancers. Proc Natl Acad Sci USA 111(11):4239–4244
Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W, Gebhardt M, Goorin AM, Harris MB, Healey J, Huvos A, Link M, Montebello J, Nadel H, Nieder M, Sato J, Siegal G, Weiner M, Wells R, Wold L, Womer R, Grier H (2005) Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol 23(9):2004–2011
Mirabello L, Troisi RJ, Savage SA (2009a) International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer 125:229–234
Mirabello L, Troisi RJ, Savage SA (2009b) Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 115:1531–1543
Mogami H, Lloyd Mills C, Gallacher DV (1997) Phospholipase C inhibitor, U73122, releases intracellular Ca2+, potentiates Ins(1,4,5)P3-mediated Ca2+ release and directly activates ion channels in mouse pancreatic acinar cells. Biochem J 324:645–651
Naito Y, Okada M, Yagisawa H (2006) Phospholipase C isoforms are localized at the cleavage furrow during cytokinesis. J Biochem 140(6):785–791
Nakamura T, Takeuchi K, Muraoka S, Takezoe H, Takahashi N, Mori N (1999) A neurally enriched coronin-like protein, ClipinC, is a novel candidate for an actin cytoskeleton-cortical membrane-linking protein. J Biol Chem 274(19):13322–13327
Niggli V, Rossy J (2008) Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol 40:344–349
Noh DY, Shin SH, Rhee SG (1995) Phosphoinositide-specific phos- pholipase C and mitogenic signalling. Biochim Biophys Acta 1242:99–114
O’Carroll SJ, Mitchell MD, Faenza I, Cocco L, Gilmour RS (2009) Nuclear PLCbeta1 is required for 3T3-L1 adipocyte differentiation and regulates expression of the cyclin D3-cdk4 complex. Cell Signal 21(6):926–935
Oka M, Edamatsu H, Kunisada M, Hu L, Takenaka N, Dien S, Sakaguchi M, Kitazawa R, Norose K, Kataoka T, Nishigori C (2010) Enhancement of ultraviolet B-induced skin tumor development in phospholipase Cε-knockout mice is associated with decreased cell death. Carcinogenesis 31(10):1897–1902
Orchel J, Witek L, Kimsa M, Strzalka-Mrozik B, Kimsa M, Olejek A, Mazurek U (2012) Expression patterns of kinin-dependent genes in endometrial cancer. Int J Gynecol Cancer 22(6):937–944
Ou L, Guo Y, Luo C, Wu X, Zhao Y et al (2010) RNA interference suppressing PLCE1 gene expression decreases invasive power of human bladder cancer T24 cell line. Cancer Genet Cytogenet 200:110–119
Pujuguet P, Del Maestro L, Gautreau A et al (2003) Ezrin regulates E-cadherin-dependent adherens junction assembly through Rac1 activation. Mol Biol Cell 14:2181–2191
Pulcinelli FM, Gresele P, Bonuglia M, Gazzaniga PP (1998) Evidence for separate effects of U73122 on phospholipase C and calcium channels in human platelets. Biochem Pharmacol 56:1481–1484
Qiu MS, Green SH (1991) NGF and EGF rapidly activate p21ras in PC12 cells by distinct, convergent pathways involving tyrosine phosphorylation. Neuron 7(6):937–946
Rhee SG (2001) Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem 70:281–312
Rhee SG, Kim H, Suh PG, Choi WC (1991) Multiple forms of phosphoinositide-specific phospholipase C and different modes of activation. Biochem Soc Trans 19(2):337–341
Santos NC, Figueira-Coelho J, Martins-Silva J, Saldanha C (2003) Multidisciplinary utilization of dimethyl sulfoxide: pharmacological, cellular, and molecular aspects. Biochem Pharmacol 65(7):1035–1041
Schmidt M, Evellin S, Weernink PA, von Dorp F, Rehmann H, Lomasney JW, Jakobs KH (2001) A new phospholipase-C-calcium signalling pathway mediated by cyclic AMP and a Rap GTPase. Nat Cell Biol 3(11):1020–1024
Smallridge RC, Kiang JG, Gist ID, Fein HG, Galloway RJ (1992) U-73122, an aminosteroid phospholipase C antagonist, non-competitively inhibits thyrotropin-releasing hormone effects in GH3 rat pituitary cells. Endocrinology 131:1883–1888
Smrcka AV, Brown JH, Holz GG (2012) Role of phospholipase Cε in physiological phosphoinositide signaling networks. Cell Signal 24(6):1333–1343
Song C, Hu CD, Masago M, Kariyai K, Yamawaki-Kataoka Y, Shibatohge M, Wu D, Satoh T, Kataoka T (2001) Regulation of a novel human phospholipase C, PLCepsilon, through membrane targeting by Ras. J Biol Chem 276(4):2752–2757
Song C, Satoh T, Edamatsu H, Wu D, Tadano M, Gao X, Kataoka T (2002) Differential roles of Ras and Rap1 in growth factor-dependent activation of phospholipase C epsilon. Oncogene 21(53):8105–8113
Stålberg P, Granberg D, Carling T, Wilander E, Eriksson B, Gobl A, Akerström G, Rastad J, Modlin IM, Oberg K, Skogseid B (2003) In situ RNA–RNA hybridisation of phospholipase C beta 3 shows lack of expression in neuroendocrine tumours. Anticancer Res 23(3B):2227–2232
Suh PG, Park J, Manzoli L, Cocco L, Peak JC, Katan M, Fukami K, Kataoka T, Yuk S, Ryu SH (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep 41:415–434
Tadano M, Edamatsu H, Minamisawa S, Yokoyama U, Ishikawa Y, Suzuki N, Saito H, Wu D, Masago-Toda M, Yamawaki-Kataoka Y, Setsu T, Terashima T, Maeda S, Satoh T, Kataoka T (2005) Congenital semilunar valvulogenesis defect in mice deficient in phospholipase C epsilon. Mol Cell Biol 25(6):2191–2199
Takenouchi T, Ogihara K, Sato M, Kitani H (2005) Inhibitory effects of U73122 and U73343 on Ca2+ influx and pore formation induced by the activation of P2X7 nucleotide receptors in mouse microglial cell line. Biochim Biophys Acta 1726:177–186
Tan X, Chen M (2014) MYLK and MYL9 expression in non-small cell lung cancer identified by bioinformatics analysis of public expression data. Tumour Biol 35(12):12189–12200
Tan XG, Yang ZL (2010) Expression of Ezrin, HGF, C-met in pancreatic cancer and non-cancerous pancreatic tissues of rats. Hepatobiliary Pancreat Dis Int 9(6):639–644
Tang CH, Yang RS, Fu WM (2005) Prostaglandin E2 stimulates fibronectin expression through EP1 receptor, phospholipase C, protein kinase Calpha, and c-Src pathway in primary cultured rat osteoblasts. J Biol Chem 280(24):22907–22916
Thomas A, Allouche M, Basyn F, Brasseur R, Kerfelec B (2005) Role of the lid hydrophobicity pattern in pancreatic lipase activity. J Biol Chem 280:40074–40083
Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ, Hung YT (1999) Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat Neurosci 2:246–253
Vickers AE, Fisher RL (2004) Organ slices for the evaluation of human drug toxicity. Chem Biol Interact 150:87–96
Walker EM, Bispham JR, Hill SJ (1998) Nonselective effects of the putative phospholipase C inhibitor, U73122, on adenosine A(1) receptor-mediated signal transduction events in Chinese hamster ovary cells. Biochem Pharmacol 56:1455–1462
Wang H, Oestreich EA, Maekawa N, Bullard TA, Vikstrom KL, Dirksen RT, Kelley GG, Blaxall BC, Smrcka AV (2005) Phospholipase C epsilon modulates beta-adrenergic receptor-dependent cardiac contraction and inhibits cardiac hypertrophy. Circ Res 97(12):1305–1313
Waugh MG (2016) Chromosomal instability and phosphoinositide pathway gene signatures in glioblastoma multiforme. Mol Neurobiol 53(1):621–630
Xiao W, Ando T, Wang HY, Kawakami Y, Kawakami T (2010) Lyn- and PLC-beta3-dependent regulation of SHP-1 phosphorylation controls Stat5 activity and myelomonocytic leukemia-like disease. Blood 116(26):6003–6013
Yang L, Guo T, Jiang S, Yang Z (2012) Expression of ezrin, HGF and c-met and its clinicopathological significance in the benign and malignant lesions of the gallbladder. Hepatogastroenterology 59(118):1769–1775. doi:10.5754/hge11744
Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G (2004) Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulator. Nat Med 10:175–181
Zhang Y, Yan L, Zhao Y, Ou L, Wu X, Luo C (2013) Knockdown of phospholipase C-epsilon by short-hairpin RNA-mediated gene silencing induces apoptosis in human bladder cancer cell lines. Cancer Biother Radiopharm 28(3):233–239
Zhao H, Shiue H, Palkon S et al (2004) Ezrin regulates NHE3 translocation and activation after Na + -glucose cotransport. Proc Natl Acad Sci USA 101:9485–9490
Zhao J, Zhang X, Xin Y (2011) Up-regulated expression of Ezrin and c-Met proteins are related to the metastasis and prognosis of gastric carcinomas. Histol Histopathol 26(9):1111–1120
Zhu Y, Lee HC, Zhang L (2002) An examination of heme action in gene expression: heme and heme deficiency affect the expression of diverse genes in erythroid k562 and neuronal PC12 cells. DNA Cell Biol 21(4):333–346
Zhu L, Zhou R, Mettler S et al (2007) High turnover of ezrin T567 phosphorylation: conformation, activity, and cellular function. Am J Physiol Cell Physiol 293:C874–C884
Authors’ contributions
VRL experimental design, Data elaboration, article writing. ML, VDM experiments. CD critical discussion. All authors read and approved the final manuscript.
Acknowledgements
The authors wish to thank the “Serena Talarico Onlus” for encouragement.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lo Vasco, V.R., Leopizzi, M., Di Maio, V. et al. U-73122 reduces the cell growth in cultured MG-63 ostesarcoma cell line involving Phosphoinositide-specific Phospholipases C. SpringerPlus 5, 156 (2016). https://doi.org/10.1186/s40064-016-1768-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-1768-6