Abstract
Background
SLC10A3, a gene upregulated in pan-cancer, lacks full understanding regarding its prognostic implications and association with immune infiltration in colorectal cancer (CRC). This study comprehensively analyzed SLC10A3 in CRC, evaluating its prognostic significance and influence on the tumor's immune microenvironment.
Methods
Transcriptomic data from TCGA were obtained to compare SLC10A3 expression in both colorectal cancer (CRC) and normal tissues. Prognostic value was assessed for overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI). DNA methylation patterns of SLC10A3 and correlation with DNA mismatch repair (MMR) were explored. Genetic alterations in SLC10A3 were scrutinized. The study also delved into the influence of SLC10A3 on the immune microenvironment of CRC, including immune cell infiltration and chemokines. Involvement of cancer-associated fibroblasts (CAFs) was explored. Methylation status of specific CpG islands in the SLC10A3 gene correlated with CRC patient prognosis. CRC tissue microarray was performed to verify the expression of SLC10A3 and its relationship with prognosis.
Results
The research revealed that SLC10A3 is significantly upregulated in CRC and holds promise as a potential diagnostic marker. Elevated SLC10A3 expression was linked to poorer OS, DSS, and PFI. Methylation patterns of SLC10A3 displayed prognostic relevance, and genetic alterations in the gene were identified. SLC10A3 was shown to impact the immune microenvironment, with significant correlations observed between its expression and various immune cell types, chemokines, and markers associated with CAFs. Furthermore, an inverse relationship between SLC10A3 and MMR molecules was established. Methylation status of specific CpG islands within the SLC10A3 gene was associated with CRC patient prognosis. Tissue microarray showed that SLC10A3 was highly expressed in CRC and significantly correlated with poor prognosis.
Conclusion
The study underscores the importance of elevated SLC10A3 in CRC, associating it with decreased survival and immune infiltration, proposing it as a diagnostic biomarker and appealing immunotherapy target, given its significant overexpression and influence on the immune microenvironment and prognosis through methylation patterns.
Graphical Abstract

Similar content being viewed by others
Introduction
Colon cancer is a major human cancer, ranking the top three cancers in the United States in terms of incidence as well as mortality in 2022 [1]. According to the data from 2014, it accounts for about 10% of all cancer cases in China [2, 3]. Colorectal cancer (CRC), including colon and rectum cancer, is often grouped due to their common characteristics, primarily manifesting as adenocarcinomas [4]. CRC can result from a combination of age, lifestyle, and genetic factors [5]. The progression from polyps to CRC is multifactorial, involving gene mutations, epigenetic modifications, and local inflammatory changes. Over the past 30 years, extensive research has identified numerous molecular alterations that contribute to the development and advancement of colon polyps. This research has unveiled multiple pathways leading to the heterogeneous presentation of colon polyps and CRC, characterized by global DNA abnormalities, including aneuploidy and microsatellite instability, as well as specific patterns of epigenetic changes (e.g., CpG Island Methylator Phenotype or CIMP), mutations in specific genes, and aberrantly methylated genes [6]. Comprehensive transcriptomics confer a precise characterization of individual cancers that should help to improve clinical strategies for neoplastic diseases through the development of novel drugs. In the past decade, numerous gene expression studies on CRC have been reported, aided by advanced technologies such as microarray and RNA-sequencing, as well as public databases such as Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA). For instance, Liang, Li, and Zhao employed GEO datasets to identify a total of 3500 differentially expressed genes (DEGs) in CRC, consisting of 1370 upregulated and 2130 downregulated genes [7]. Pathway enrichment analysis revealed that upregulated DEGs were enriched in the cell cycle and DNA replication, while downregulated DEGs were enriched in drug metabolism. Additionally, Y. Guo, Bao, Ma, and Yang aimed to identify potential key candidate genes and pathways in CRC using four GEO cohorts and discovered 292 shared DEGs (165 upregulated and 127 downregulated) from the four datasets [8]. In another study (Wu, Wu, & Jiang, 2017) [9], gene and microRNA (miRNA) expression profiles from GEO datasets were utilized to identify 600 upregulated DEGs, 283 downregulated DEGs, 13 upregulated miRNAs, and 7 downregulated miRNAs [10].
The Solute Carrier (SLC) Family 10 (SLC10) is involved in the influx transport of bile acids, steroidal hormones, specific drugs, and a variety of other substrates, and comprises seven family members, including SLC10A1, SLC10A2, SLC10A3, SLC10A4, SLC10A5, SLC10A6 and SLC10A7. The SLC10 family is identified as the Bile Acid Sodium Symporter (BASS) family in the transporter classification system and belongs to the bile/arsenite/riboflavin transporter (BART) superfamily [11]. Bile acids (BA) are complex signaling molecules that modulate glucose, lipid and energy metabolism and are also implicated in the etiology of certain types of cancer [11]. Recent studies suggest that SLC10A3 may influence chemotherapy resistance and serve as a promising immune biomarker in liver cancer. Moreover, the expression of SLC10A3 protein is correlated with stromal CD4 T cells, CD20 B cells, macrophages, PDCD1 (PD-1) and CD274 (PD-L1) expression, indicating a potential regulatory role in liver cancer immunotherapy [12]. However, whether SLC10A3 plays a similar role in colorectal adenocarcinoma remains unknown.
Currently, updated public databases based on integrative bioinformatics analysis of the Cancer Genome Atlas (TCGA) have significantly enhanced the efficiency of identification of biomarkers and functional genes in cancerous diseases. Therefore, this study evaluates the transcriptional profiles and potential prognostic value of the SLC10A3 by systematical bioinformatics analysis and provides a novel role of SLC10A3 in the prognostic value and immune microenvironment in colorectal adenocarcinoma.
Material and methods
Transcriptome data sources
The mRNA expression data of SLC10A3 were downloaded from the TCGA database (http://cancergenome.nih.gov/abouttcga) for 33 different types of cancer [13]. The transcriptome data were then transformed into Log2 form and analyzed using the Wilcoxon rank-sum test to compare the expression levels between tumor and normal tissues. A p-value < 0.05 was considered statistically significant. To evaluate the predictive value of SLC10A3 for COAD and READ, AUC and ROC analysis were conducted.
Survival analysis
The hazard ratio (HR) and 95% confidence intervals were calculated using univariate survival analysis. To assess the survival outcomes of SLC10A3 in COAD and READ, Kaplan–Meier survival analysis was conducted for OS (overall survival), DSS (disease-specific survival), and PFI (progression-free interval). The survival differences were compared using ‘‘survminer’’ and ‘‘surviva’’ packages. The concordance index (C-index) was used to evaluate the discrimination of the nomograms.
To explore the DNA methylation of SLC10A3 in TCGA, MethSurv7 was utilized. The analysis involved assessing the methylation levels and prognostic values of each CpG site within SLC10A3. Furthermore, the relationship between the CpG methylation status of SLC10A3 and the overall survival (OS) of CRC was evaluated.
Analysis of mutation
For the genetic alteration analysis of SLC10A3, we utilized the cBioPortal (https://www.cbioportal.org/), a comprehensive website offering visualization, analysis, and download options for large-scale cancer genomics datasets [14]. Specifically, we examined the alteration frequency, mutation type, and copy number alterations (CNA) of SLC10A3 in COAD and READ.
Immune cells infiltration
We utilized ssGSEA [15] to investigate the relationship between SLC10A3 and 24 types of immune cell infiltration in the immune microenvironment of CRC. Furthermore, we examined the differences in immune cell abundance between the high and low SLC10A3 expression groups. Using the Tumor Immune Estimation Resource (TIMER) [16], we assessed the association between the expression levels of tumor-infiltrating immune cells, including B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, dendritic cells, and cancer-associated fibroblasts (CAFs), and SLC10A3 expression. The Pearson method was used to display the results. Finally, we explored the correlation between somatic copy number alterations (SCNA) of SLC10A3 and the abundance of 6 leukocytes using the ‘SCNA module’.
Functional enrichment analysis
We employed the Cluster Profiler R package to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the DEGs between the high and low SLC10A3 expression groups.
Immunohistochemical (IHC) staining
To assess the expression of SLC10A3 in human CRC tissues, we obtained the ZL-COC sur1602 CRC tissue microarrays from Shanghai Zhuolibiotech Company Co., Ltd. (Shanghai, China). Immunohistochemistry (IHC) was performed directly on the tissue microarrays, orderly dewaxed in xylene baths for 3 times, then rehydrated with graded alcohol series and retrieved in a pressure cooker with sodium citrate buffer (pH 6.0) heating for 15 min. The SLC10A3 (Proteintech, 19909-1-AP) antibody was utilized for IHC according to their protocols. The IHC steps followed our previous research [17]. We took two field shots on each slide. The IHC staining scores (IS) were classified into four score ranks: 0, negative; 1, weak; 2, moderate; and 3, strong. The percentage of positively stained cells (PS): 0 (< 5%), 1 (5–25%), 2 (25–50%), 3 (50–75%) and 4 (75–100%). The score of each slide: IS x PS (0–12) [18]. Additionally, clinical and pathological information regarding the tissue samples was obtained from the array manufacturer. Based on the cut-off value (n = 7), we classified those with > 7 as the SLC10A3-high group and those with ≤ 7 as the SLC10A3-low group.
Statistical analysis
We performed log2 transformation to normalize all data obtained from TCGA. Spearman's or Pearson's test was used to analyze the correlation between two variables. TCGA (https://portal.gdc.cancer.gov/) COAD and READ data were utilized for these analyses. For survival analysis, we calculated the hazard ratios (HRs) and p-value using either the univariate Cox regression analysis or log-rank test. A p-value < 0.05 was considered statistically significant for all analyses.
Results
High expression of SLC10A3 in colorectal cancer samples
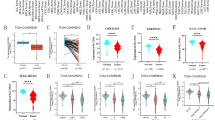
To explore the biological function of SLC10A3 in pan-cancer, especially CRC, we performed a series of bioinformatics analyses (Fig. 1A). Firstly, we compared the transcriptional levels of SLC10A3 in pan-cancer between tumor and normal samples by applying the TCGA databases. In TCGA data, differential SLC10A3 expression was significant among 16 of the 33 cancer types analyzed, including 12 upregulation and 4 downregulation (Fig. 1B). Moreover, SLC10A3 expression was higher in tumors than in normal tissues in CRC (Fig. 2A). We evaluated the accuracy of a colon cancer risk prediction model based on SLC10A3 using ROC curve analysis and found that the sensitivity was greater than the specificity, with an area under curve (AUC) of 0.745 (Fig. 2B). This indicates good predictive accuracy for the model. Based on the TCGA database and UALCAN, we found SLC10A3 was highly expressed in COAD and READ (Fig. 2CD). Based on the TCGA database, we found the methylation levels of SLC10A3 did not significantly change in colon and rectum cancer tissues than that in the corresponding normal tissues (Fig. 2EF).
Comparative analysis of SLC10A3 expression and methylation levels in colorectal cancer (CRC) and normal tissues based on the TCGA database. A SLC10A3 expression was higher in CRC than in normal tissues based on the TCGA database. B The ROC and AUC of CRC is based on the expression of SLC10A3. C–D SLC10A3 expression was higher in COAD and READ than in normal tissues based on the TCGA database. E–F the methylation levels of SLC10A3 in COAD, READ and normal tissues. *indicates p-value < 0.05; ** indicates p-value < 0.01; *** indicates p-value < 0.001. CRC colorectal cancer, READ rectal adenocarcinoma; COAD colon adenocarcinoma
Mutation of SLC10A3 in CRC
Gene mutations play a crucial role in the development and progression of cancer. To gain insights into the mutations associated with CRC, we conducted a study on SLC10A3 gene mutations. Our analysis of the cBioPortal database revealed that SLC10A3 gene alterations were detected in CRC patients with amplification (> 0.5%) (Fig. 3A). The genetic alterations in SLC10A3 were primarily categorized into four types: amplification, truncating mutation, inframe mutation, and missense mutation. We further investigated the types, sites, and case numbers of SLC10A3 gene modification using data from DFCI, Cell Reports 2016, and TCGA Firehose Legacy (Fig. 3B–D).
Association of SLC10A3 with clinical stage and prognosis
We examined the association between SLC10A3 expression and clinical stage in CRC, using high and low expression groups. Our analysis revealed positive correlations between SLC10A3 and lymph node invasion (p-value = 0.006), distant metastasis (p-value < 0.001), and patient age (p-value = 0.049) (Table 1). Moreover, we found that the mRNA level of SLC10A3 was higher in M1 stage than in M0 stage (Fig. 4F). In order to investigate the relationship between SLC10A3 expression and patient survival, we employed the Kaplan–Meier (KM) Plotter. Our results demonstrated that higher mRNA levels of SLC10A3 were significantly associated with shorter OS (p-value = 0.037), DSS (p-value = 0.034), and PFI (p-value = 0.008) in COAD patients (Fig. 4A–C). However, we also observed that high expression of SLC10A3 was linked to shorter OS (p-value = 0.022) and DFS (p-value = 0.026) (Fig. 4D–E). To assess survival rates in COAD patients, we constructed a nomogram based on age, gender, TNM stage, tumor stage, and risk score, and found good consistency between predicted and actual survival rates (Fig. 4G–H).
High-expressional SLC10A3 affects the prognosis of CRC patients. A The OS of CRC patients between low and high-expressional groups. B The DSS of CRC patients between low and high-expressional groups. C The PFI of CRC patients between low and high-expressional groups. D–E The survival analysis of SLC10A3 high expression and low expression in COAD and READ based on the TIMER database. F SLC10A3 expression in CRC patients was analyzed based on the tumor metastasis. G–H Nomogram to predict the COAD survival possibility among 1 year, 3 years and 5 years, including age, gender, TNM stage, tumor stage, and risk score. CRC, colorectal cancer; READ, rectal adenocarcinoma; COAD, colon adenocarcinoma. HR hazard ratio, OS overall survival, DSS disease-special survival, PFI progress-free interval
To confirm the prognostic significance of SLC10A3 in CRC patients, we conducted univariate and multivariate survival analyses (Tables 2, 3). Univariate analysis identified four prognostic factors in CRC: age (≤ 65 vs. > 65), N stage (N1–2 vs. N0), M stage (M1 vs. M0), and T stage (T1 vs. T4). Further multivariate analysis revealed that age (p-value = 0.019), BMI (p-value = 0.035), and N stage (p-value < 0.001) were independent predictors of OS in CRC patients (Table 2).
Role of SLC10A3 in CRC immune microenvironment
The tumor microenvironment plays a crucial role in tumor genesis and development. Therefore, investigating the relationship between SLC10A3 and immune cell infiltration is significant. Immune cell infiltration in CRC was determined using ssGSEA for 24 immune cell types, and the relationship between SLC10A3 expression and immune cell infiltration was analyzed using Pearson's correlation coefficient (Table 3). NK cells, Eosinophils, and TReg showed a positive correlation with SLC10A3 expression, while Tcm, Th2 cells, and T helper cells showed a negative association with SLC10A3 (Fig. 5A). The tumor infiltration levels of TReg cells, CD8 + cells, NK cells, and T cells were consistent with the Pearson's correlation analysis results (Fig. 5B–E). Furthermore, we analyzed the relationship between somatic copy number aberrations (CNA) and tumor immune infiltration using TIMER. Firstly, the ‘‘SCNA’’ module analysis showed that altered SLC10A3 gene copy numbers in COAD were associated with immune cell infiltration levels of B cells, macrophages, neutrophils, and dendritic cells (Fig. S1). In comparison, SLC10A3 gene copy numbers were only associated with dendritic cell infiltration levels in READ (Fig. S1). Secondly, the ‘‘Gene’’ module analysis confirmed that immune infiltration levels of B cells, CD8 + T cells, CD4 + T cells and macrophages were associated with SLC10A3 expression in READ and COAD (Fig. 5FG). Besides, the results of CIBERSORT, EPIC, QUANTISEQ, XCELL, and MCPCOUNTER about the relationship between SLC10A3 and immune infiltration are displayed in Table S1. In conclusion, high expression of SLC10A3 could affect the immune microenvironment of READ and COAD by regulating the proportions of immune cell infiltration.
Correlation of immune cell infiltration, chemokines and SLC10A3 expression in CRC patients. A Relationships among infiltration levels of 24 immune cell types and SLC10A3 expression profiles by Spearman’s analysis in CRC. B–E The infiltration levels of Treg cells, CD8 + cells, NK cells, and T cells in the high- and low-SLC10A3 expression groups in CRC. F–G SLC10A3 expression is related to immune infiltration levels in COAD and READ through TIMER. H The relationship between chemokines and SLC10A3 expression in COAD and READ. CRC colorectal cancer; READ rectal adenocarcinoma, COAD colon adenocarcinoma. *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001
Chemokine networks affect tumor immunity and tumorigenesis by regulating the tumor microenvironment. Therefore, we next examined the correlations between chemokines and SLC10A3 in CRC. The results showed significant (p-value < 0.05) and strong positive correlations between SLC10A3 expression and the following chemokines: CCL15, CCL24, and CXCL14. Additionally, strong negative correlations were observed between SLC10A3 expression and chemokines including CCL2-5/7–8, CXCL5-6 and CXCL8-13 in COAD (Fig. 5H). These results further confirmed that SLC10A3 was significantly correlated with immune infiltrating cells and chemokines in READ and COAD, indicating that SLC10A3 may play a crucial role in the microenvironment of READ and COAD.
Correlation analysis of SLC10A3 and cancer-associated fibroblasts
Cancer-associated fibroblasts (CAFs) create a conducive environment for tumor growth, as they are known to inhibit immune cell function through secretion of cytokines and metabolites, thereby promoting tumorigenesis, invasion, metastasis, and drug resistance. Our analysis of the correlation between SLC10A3 and CAFs using TIMER revealed a significant association in CRC (Fig. 6AC). Specifically, high expression of SLC10A3 was positively correlated with CAF markers such as ACAT2 and VIM in COAD and READ (Fig. 6BD).
Correlation analysis of SLC10A3 and cancer-associated fibroblasts in COAD and READ. A, C The association of SLC10A3 and cancer-associated fibroblast in COAD (p-value = 1.02e−04) and READ (p-value = 5.07e-03) by EPIC. B, D The correlation analysis between the SLC10A3 expression and the expression levels of VIM, ACAT2 in the TCGA dataset. READ rectal adenocarcinoma, COAD colon adenocarcinoma
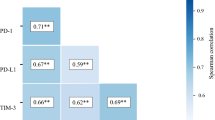
Association of SLC10A3 and DNA mismatch repair (MMR)
We analyzed the correlations between SLC10A3 expression and mismatch repair molecules using Spearman’s correlation coefficient. Obviously, SCL10A3 expression showed a negative association with PMS1, RAD21, DNA2, MLH3, and MSH4 (Fig. 7A–F). In summary, our findings suggest a negative link between SLC10A3 and MMR in CRC, with implications for error repair of nucleotide sequence and microsatellite instability.
The methylation status of the SLC10A3 is associated with the prognosis of COAD and READ patients
DNA methylation levels in SLC10A3 were analyzed, and the prognostic value of CpG islands in SLC10A3 was assessed using the MetSurv tool. The findings revealed the presence of 10 methylated CpG islands in COAD and 11 methylated CpG islands in READ. Notably, methylation levels of two specific CpG islands, namely cg06266461 and cg09418475, showed significant association with prognosis (p-value < 0.05) in COAD. In contrast, no CpG islands in READ were found to be associated with prognosis (Table 4).
Screening DEGs and functional enrichment analyses based on SLC10A3 single gene analysis
Based on the TCGA database, we conducted SLC10A3 single gene analysis, divided the tumor samples into SLC10A3 low expression group and SLC10A3 high expression group by R (version 4.1.1), and screened differentially expressed genes in volcano plot (Fig. 8A). DEGs, including 127 upregulated and 2492 downregulated genes, were identified between low and high SLC10A3 groups in CRC (Fig. 8A). The potential bio-function of DEGs was enriched in nucleosome, nucleosome assembly and small nuclear ribonucleoprotein complex via GO and KEGG enrichment analysis in CRC (Fig. 8B). SLC10A3 may promote CRC progression by increasing chromatin synthesis. GSEA showed that the SLC10A3-associated DEGs were significantly enriched in nucleosome organization, protein–DNA complex and nucleosome assembly to regulate the progression of CRC (Fig. 8C).
Screening DEGs and constructing functional analysis between SLC10A3 high-expressional group and SLC10A3 low-expressional group. A Volcano plots of DEGs between the expression of SLC10A3-high and SLC10A3-low in CRC samples. B GO and KEGG analysis among DEGs in CRC as a circle graph. C GSEA analysis of DEGs between SLC10A3-high and SLC10A3-low in CRC samples. CRC colorectal cancer
High SLC10A3 expression is strongly associated with CRC prognosis
We investigated the functions of SLC10A3 in CRC by examining the relative levels of SLC10A3 in patient tissues and corresponding normal tissues. 72 patient specimens were analyzed, and SLC10A3 was found to be expressed differently, with some specimens having low levels and others having high levels (Fig. 9A–B). Using IHC scores (cut-off = 7), we divided the specimens into high and low expression groups, with a higher proportion of high SLC10A3 expression in tumor tissues (Fig. 9C). Additionally, the amount of SLC10A3 was significantly increased in CRC tissues compared to normal tissues (Fig. 9C). To predict the diagnostic value of SLC10A3 in distinguishing CRC from the mucosa, we used ROC and AUC, which resulted in an AUC value of 0.879 (Fig. 9D). Our analysis indicated that the level of SLC10A3 is correlated with CRC malignancy in tumor relapse and metastasis (Fig. 9E–F), but not in lymph node invasion status and tumor stage (Fig. S2AB). Besides, high SLC10A3 expression predicted low OS (HR = 2.23, p-value = 0.022) and DFS (HR = 2.24, p-value = 0.026) (Fig. 9G–H). Our data suggest that SLC10A3 may serve as a useful diagnostic and prognostic indicator in CRC patients.
High expression of SLC10A3 predicts poor prognosis in CRC patients. A IHC analysis of SLC10A3 expression in a CRC patient tissue array (n = 72). SLC10A3 levels in cancer tissues and corresponding normal tissues. Representative images are shown. Scale bars = 100 μm. B IHC scores of SLC10A3 in human CRC tissues and adjacent tissues. C The percent of high and low SLC10A3 expression in CRC patients (normal and cancer tissues). D ROC curve with corresponding AUC value for SLC10A3 when classifying cancer from the mucosa. E–F SLC10A3 expression was positively associated with tumor relapse (R) and metastasis (M). G–H High SLC10A3 expression predicted low OS (HR = 2.23, p-value = 0.022) and DFS (HR = 2.24, p-value = 0.026). Data were shown as mean and standard deviation from three independent experiments. Data are means ± SDs. *, p-value < 0.05; **, p-value < 0.01; ***, p-value < 0.001. CRC colorectal cancer
Discussion
Colorectal cancer is the third most common cancer worldwide in terms of incidence and ranks fourth in terms of cancer-related mortality [19]. Although significant clinical advancements have been made in the treatment of colorectal cancer through medical techniques, surgical interventions, and chemotherapy [20], the current lack of an effective diagnostic method to monitor CRC progression or recurrence remains a challenge. Recent studies have highlighted the pivotal role of SLC10A3 in various cancers, with its mutation being implicated in malignant growth and cancer development. Specifically, SLC10A3 plays a critical role in regulating the immune response in liver cancer, related with stromal CD4 T cells, CD20 B cells, and macrophage [9]. Upregulation of SLC10A3 mRNA is statistically related with unfavorable survival outcomes and immune infiltration among low-grade gliomas [21]. Nonetheless, there is currently no literature on SLC10A3 in colorectal cancer. Thus, this study aims to elucidate the expression of SLC10A3, immunocorrelation and its significant role in predicting the prognosis of colorectal cancer, ultimately establishing a genetic risk-scoring model.
In our study, through TCGA database we found that SLC10A3 is significantly higher expressed in colorectal cancer, so we further investigated the role of SLC10A3 in colorectal cancer and unveiled its prognostic value, functional enrichment pathways, methylation patterns, tumor immune infiltration, and variance analysis between high and low SLC10A3 expression groups using bioinformatic analysis. Higher SLC10A3 expression was notably associated with worse prognosis, including shorter overall survival (OS, p-value = 0.037, HR = 1.45(1.02–2.05)), disease-free survival (DFS, p-value = 0.034, HR = 1.63(1.04–2.56)), and progression-free survival (PFI, p-value = 0.008, HR = 1.52(1.12–2.06)). Moreover, high SLC10A3 expression may be linked to metastasis in CRC patients. Abnormal methylation of specific genes in cells poses a high risk of cancer, and this aberrant methylation occurs during cell oncogenesis [22, 23]. The methylation levels of four CpG islands, including cg06266461 and cg09418475, were found to be associated with prognosis (p < 0.05) in COAD. Tissue microarray of CRC confirmed the high expression of SLC10A3 in tumor tissues, which was indicative of poor prognosis, including OS (p-value = 0.022, HR = 2.23(1.16–4.29) and DFS (p-value = 0.026, HR = 2.24(1.10–4.56)). Our results demonstrate that CRC patients with high SLC10A3 expression experienced shorter survival times and a higher likelihood of disease progression, highlighting SLC10A3 as an independent risk factor for survival.
Mounting evidence indicates the pivotal role of the immune microenvironment in tumorigenesis [24]. Tumor immunity closely intertwines with cell growth, and any disruption in the immune microenvironment can expedite cancer progression [25]. Computer algorithms analyzing transcriptomic data have gained widespread adoption in assessing the dynamic immune components within the tumor microenvironment (TME) of cancer patients. In this study, we employed the ssGSEA method to determine the extent of immune cell infiltration and the ratio of immune/mesenchymal components in colorectal cancer (CRC) samples sourced from the TCGA database. Our findings confirmed the prognostic biomarker, SLC10A3, as an influential factor in immune regulation within the CRC TME. CD8 + T cell can effectively kill various pathogens and tumor cell, while Treg cell plays a crucial role in inhibitory tumor immune microenvironment. These findings affirmed the substantial correlation between SLC10A3 and CD8 + T cell (negatively) and Treg cell (positively) in CRC, suggesting a crucial role of SLC10A3 within the microenvironment of these malignancies.
Subsequently, we conducted correlation analyses between chemokines and SLC10A3 in CRC. Spearman's method was utilized to examine the associations between SLC10A3 expression and immune checkpoint molecules. Additionally, we investigated the relationships between SLC10A3 and various chemokines in both COAD and READ. The results revealed statistically significant (p-value < 0.05) and robust positive correlations with the several chemokines: CCL15, CCL24, and CXCL14. Moreover, strong negative correlations were observed between SLC10A3 and chemokines such as CCL2-5/7–8, CXCL5-6 and CXCL8-13 in COAD. Cancer-associated fibroblasts (CAFs) represent a critical component within the tumor microenvironment, exerting a pivotal role in tumor initiation and progression [26, 27]. Rather than existing in isolation, CAFs interact with neighboring tumor cells, actively promoting their growth, survival, and sustaining their malignant behavior [28]. By secreting diverse cytokines, CAFs establish communication with other stromal cells and tumor cells, resulting in immune cell function inhibition and facilitation of tumor development [29]. Cytokines such as TGF-β are secreted by CAFs, leading to DC maturation suppression and Treg differentiation promotion [30,31,32]. Consequently, we assessed the correlation between SLC10A3 and CAFs in COAD and READ by utilizing R and TIMER, based on TCGA datasets. Notably, we identified a significant positive correlation between SLC10A3 and CAFs in both COAD (p-value = 1.02e−04) and READ (p-value = 5.07e−03). High expression levels of SLC10A3 were found to be positively associated with CAF markers, including ACAT2 (p-value = 0.008) and VIM (p-value < 0.001).
The mismatch repair system is a safety net in the body that maintains the integrity and stability of genetic material. In DNA mismatch repair (MMR), repair genes can ensure the ‘‘fidelity’’ of the replication process during DNA replication [33]. Our results showed SCL10A3 expression was negatively associated with PMS1 (p-value < 0.001), RAD21 (p-value = 0.012), DNA2 (p-value < 0.001), MLH3 (p-value < 0.001), and MSH4 (p-value < 0.001). Low expression in mismatch repair genes can lead to shorten or lengthen microsatellite segments. Microsatellite instability (MSI) will result in a high mutant phenotype of the genome, leading to an increased risk of tumor development [34].
However, it is important to acknowledge the limitations of our study. Our analysis heavily relied on tissue samples from the TCGA dataset, with only 72 pairs of clinical samples used for validation. Therefore, it would be beneficial to validate our findings using larger sample cohorts from different medical centers. Additionally, although we found an association between SLC10A3 and the immune microenvironment of colorectal cancer, confirmatory experiments were not conducted. Moving forward, our further plan includes verifying that SLC10A3 promotes CRC proliferation and metastasis through cytological and animal experiments, as well as exploring its mechanism. This aspect represents a potential future direction for our investigation.
Conclusion
Overall, the findings suggested that SLC10A3 high expression is an independent risk biomarker for worse prognosis in patients with CRC and is related to immune infiltration and distant metastasis. Besides, our study may provide new insights into the treatment of CRC and inform further studies of tumor immunity in CRC. However, studies with larger clinical samples and further functional experiments are required to verify our results.
Availability of data and materials
The datasets generated and analyzed during this study are available in the public database TCGA.
Abbreviations
- SLC10:
-
The solute carrier family 10
- AUC:
-
Area under the curve
- ROC:
-
Receiver operating characteristic
- DEGs:
-
Differentially expressed genes
- GSEA:
-
Gene set enrichment analysis
- OS:
-
Overall survival
- DSS:
-
Disease-specific survival
- PFI:
-
Progression-free interval
- DFS:
-
Disease-free survival
- GEO:
-
Gene expression omnibus
- TCGA:
-
The Cancer Genome Atlas
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- BASS:
-
Bile acid sodium symporter
- BART:
-
Bile/arsenite/riboflavin transporter
- BA:
-
Bile acids
- C-index:
-
The concordance index
- TIMER:
-
Tumor immune estimation resource
- IHC:
-
Immunohistochemistry
- IS:
-
The IHC staining scores
- PS:
-
Positively stained cells
- CNA:
-
Copy number aberrations
- MSI:
-
Microsatellite instability
- CRC:
-
Colorectal cancer
- READ:
-
Rectal adenocarcinoma
- COAD:
-
Colon adenocarcinoma
- TME:
-
Tumor micro-environment
- CAFs:
-
Cancer-associated fibroblasts
- SCNA:
-
Somatic copy number alterations
- HR:
-
Hazard ratio
- MMR:
-
Mismatch repair
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics. BMC Cancer. 2022;2022(72):7–33.
Xu R, Wang W, Zhu B, Lin X, Ma D, Zhu L, Zhao Q, Nie Y, Cai X, Li Q, Fang W, Li H, Wang N, Chen Y, Peng C, Fang H, Shen L. Disease characteristics and treatment patterns of Chinese patients with metastatic colorectal cancer: a retrospective study using medical records from China. BMC Cancer. 2020;20:131.
Yang Y, Han Z, Li X, Huang A, Shi J, Gu J. Epidemiology and risk factors of colorectal cancer in China. Chinese J Cancer Res. 2020;32:729–41.
Fleming M, Ravula S, Tatishchev SF. Colorectal carcinoma. Pathol Aspects. 2012;3:153–73.
Jia SN, Han YB, Yang R, Yang ZC. Chemokines in colon cancer progression. Semin Cancer Biol. 2022;86:400–7.
Grady WM, Markowitz SD. The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig Dis Sci. 2015;60:762–72.
Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016. https://doi.org/10.1007/s12032-016-0829-6.
Guo Y, Bao Y, Ma M, Yang W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci. 2017. https://doi.org/10.3390/ijms18040722.
Wu S, Wu F, Jiang Z. Identification of hub genes, key miRNAs and potential molecular mechanisms of colorectal cancer. Oncol Rep. 2017;38:2043–50.
Sun G, Li Y, Peng Y, Lu D, Zhang F, Cui X, Zhang Q, Li Z. Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis. J Cell Physiol. 2019;234:15215–24.
Claro da Silva T, Polli JE, Swaan PW. The solute carrier family 10 (SLC10): beyond bile acid transport. Mol Aspects Med. 2013;34:252–69.
Tian S, Li J, Xiang J, Peng P. The clinical relevance and immune correlation of SLC10 family genes in liver cancer. J Hepatocell Carcinoma. 2022;9:1415–31.
Gao W, Li Y, Zhang T, Lu J, Pan J, Qi Q, Dong S, Chen X, Su Z, Li J. Systematic analysis of chemokines reveals CCL18 is a prognostic biomarker in glioblastoma. J Inflamm Res. 2022;15:2731–43.
Liang K, Guo Z, Zhang S, Chen D, Zou R, Weng Y, Peng C, Xu Z, Zhang J, Liu X, Pang X, Ji Y, Liao D, Lai M, Peng H, Ke Y, Wang Z, Wang Y. GPR37 expression as a prognostic marker in gliomas: a bioinformatics-based analysis. Aging. 2023;15:10146–67.
Hao W, Zhang Y, Dou J, Cui P, Zhu J. S100P as a potential biomarker for immunosuppressive microenvironment in pancreatic cancer: a bioinformatics analysis and in vitro study. BMC Cancer. 2023;23:997.
Liu Y, Jiang C, Xu C, Gu L. Systematic analysis of integrated bioinformatics to identify upregulated THBS2 expression in colorectal cancer cells inhibiting tumour immunity through the HIF1A/Lactic Acid/GPR132 pathway. Cancer Cell Int. 2023;23:253.
Wang B, Zhang J, Wang X, Zhao L, Wang Y, Fan Z, Liu L, Gao W. Identification and clinical validation of key genes as the potential biomarkers in colorectal adenoma. BMC Cancer. 2023;23:39.
Liu X, Li X, Zheng P, Yang W. DAX1 promotes cervical cancer cell growth and tumorigenicity through activation of Wnt/β-catenin pathway via GSK3β. Cell Death Dis. 2018;9:339.
Thrumurthy SG, Thrumurthy SS, Gilbert CE, Ross P, Haji A. Colorectal adenocarcinoma: risks, prevention and diagnosis. BMJ. 2016;354: i3590.
Weng J, Li S, Zhu Z, Liu Q, Zhang R, Yang Y, Li X. Exploring immunotherapy in colorectal cancer. J Hematol Oncol. 2022;15:95.
He Y, Song J, Qin Y, Mao D, Ding D, Wu S, Wu H. The prognostic significance and immune correlation of SLC10A3 in low-grade gliomas revealed by bioinformatic analysis and multiple immunohistochemistry. Aging. 2023;15:3771–90.
Katsman E, Orlanski S, Martignano F, Fox-Fisher I, Shemer R, Dor Y, Zick A, Eden A, Petrini I, Conticello SG, Berman BP. Detecting cell-of-origin and cancer-specific methylation features of cell-free DNA from Nanopore sequencing. Genome Biol. 2022;23:158.
Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Can Res. 2016;76:3446–50.
Inamura K, Hamada T, Bullman S, Ugai T, Yachida S, Ogino S. Cancer as microenvironmental, systemic and environmental diseases: opportunity for transdisciplinary microbiomics science. Gut. 2022;71(10):2107.
Cismaru CA, Pirlog R, Calin GA, Berindan-Neagoe I. Stem Cells in the tumor immune microenvironment -part of the cure or part of the disease? Ontogeny Dichotomy Stem Immune Cells Led Better Understanding Stem Cell Rev Rep. 2022;18:2549–65.
Glabman RA, Choyke PL, Sato N. Cancer-associated fibroblasts: tumorigenicity and targeting for cancer therapy. Cancers. 2022;14:3906.
Jenkins BH, Buckingham JF, Hanley CJ, Thomas GJ. Targeting cancer-associated fibroblasts: challenges, opportunities and future directions. Pharmacol Ther. 2022;240: 108231.
Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discovery. 2019;18:99–115.
Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, Zhang B, Meng Q, Yu X, Shi S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20:131.
Chen B, Mu C, Zhang Z, He X, Liu X. The love-hate relationship between TGF-β signaling and the immune system during development and tumorigenesis. Front Immunol. 2022;13: 891268.
Moreau JM, Velegraki M, Bolyard C, Rosenblum MD, Li Z. Transforming growth factor-β1 in regulatory T cell biology. Sci Immunol. 2022;7:4613.
Xiang X, Niu YR, Wang ZH, Ye LL, Peng WB, Zhou Q. Cancer-associated fibroblasts: Vital suppressors of the immune response in the tumor microenvironment. Cytokine Growth Factor Rev. 2022;67:35–48.
Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–99.
Pećina-Šlaus N, Kafka A, Salamon I, Bukovac A. Mismatch repair pathway. Genome Stability Cancer Frontiers Mol Biosci. 2020;7:122.
Funding
This research was supported by grants from The National Natural Science Foundation of China (82202828), Ili Kazkh Autonomous Clinical Research Institute (yl2020ms10) and Six talent peaks of Jiangsu Province (WSW-006).
Author information
Authors and Affiliations
Contributions
BW, YW and YT designed the experiments; BW, JW and WF performed the experiments; BW and WF prepared all figures; JW and YT analyzed the data; ZF, YW and LL supervised the work; BW, JW, YT and WF wrote the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent participate
Ethical approval was acquired for this study from the Ethical Committee of Medical Research, Jiangsu province Hospital of Nanjing medical University (2018-SR-258).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Correlation between somatic copy number alterations (SCNA) and immune infiltration abundance in relation to SLC10A3 expression. Figure S2. Impact of age, gender, and SLC10A3 expression on the overall and disease-free survival of CRC patients. Table S1. The relationship between SLC10A3 and immune infiltration based on TIMER2.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, B., Fan, W., Tao, Y. et al. The impact of SLC10A3 on prognosis and immune microenvironment in colorectal adenocarcinoma. Eur J Med Res 29, 20 (2024). https://doi.org/10.1186/s40001-023-01526-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-023-01526-4