Abstract
Background
Angular cheilitis, an infection mainly caused by Candida yeasts, is featured by the appearance of inflammatory lesions at the bilateral corners of the mouth, particularly in patients with poor oral hygiene, ill-fitting dentures and old age. The first isolation of an atypical yeast, Cystobasidium calyptogenae, from oral samples of a patient presenting with angular cheilitis is discussed in this study.
Case presentation
Angular cheilitis was diagnosed in a 60-year-old denture-wearing woman who presented with an irritation fibroma on her right lower buccal sulcus over the premolar region. Primary cultures of her oral swab and oral rinse samples grew a pure culture of an uncommon yeast strain resembling Rhodotorula sp. Sequence analysis of the yeast internal transcribed spacer (ITS) gene region and D1D2 domain showed highest similarity (99.6% and 100%, respectively) to C. calyptogenae CBS 9125 type strain. Following 2 weeks of treatment with miconazole/fusidic acid and mouthwash, the oral lesion showed improvement with less erythema. C. calyptogenae was not isolated from the patient’s oral samples upon repeat sampling.
Conclusion
This is the first report on the isolation of C. calyptogenae from human oral samples. The ability of C. calyptogenae to grow at 37 °C and the fact that it was the only yeast species isolated from the patient’s oral samples suggests its pathogenic potential and possible involvement in angular cheilitis. The ubiquitous nature of the Cystobasidium yeast is believed to increase the likelihood of opportunistic infections among immunocompromised individuals. As Cystobasidium is phenotypically indistinguishable from Rhodotorula, an emerging opportunistic pathogen, surveillance using molecular identification in clinical settings is essential in providing accurate diagnosis and treatment of uncommon yeast infections.
Similar content being viewed by others
Background
Angular cheilitis (angular stomatitis; cheilosis; perleche) is characterized by burning sensations, soreness, redness, bleeding and fissures at the bilateral corners of the oral cavity [1]. The disease has been linked to various underlying risk factors including poor oral hygiene, long-term and ill-fitting dentures, inadequate vitamin B consumption, protein and trace minerals (zinc or iron) deficiency, immunocompromised states (acquired immunodeficiency syndrome, diabetes, chemotherapy), prolonged antibiotic use and old age [1, 2]. The inflammatory erythematous lesions of angular cheilitis are commonly associated with the Candida yeasts and occasionally, Staphylococcus aureus and β-hemolytic streptococci [3, 4]. Candida albicans and S. aureus are acknowledged as co-infectors in 60—75% cases of angular cheilitis [1].
Cystobasidium (Langerheim) Neuhoff (1924) is a mycoparasitic yeast associated with coprophilous ascomycetes. The yeast forms soft, smooth and light orange to pinkish colonies upon culturing on mycological agars [5,6,7,8,9,10,11,12]. To date, 21 Cystobasidium species have been described, i.e., C. fimetarium, C. minutum, C. slooffiae, C. calyptogenae, C. pinicola, C. laryngis, C. benthicum, C. pallidum, C. lysinophilum, C. portillonensis, C. oligophagum, C. alpinum, C. psychroaquaticum, C. rietchieii, C. tubakii, C. ongulense, C. keelungensis [6,7,8,9,10], C. halotolerans [11], C. iriomotense C. raffinophilum and C. terricola [13]. Various habitats and diverse ecologies, i.e., supraglacial sediments [9], aquatic environments [10, 14,15,16,17,18], soil [8, 19,20,21] and phylloplanes [7, 22,23,24,25,26] have been associated with Cystobasidium yeasts. Previously classified in the Rhodotorula minuta clade [7], members of this genus have not been described as commensal organisms or human pathogens.
C. calyptogenae is ovoidal to elongate with polar budding appearance [7, 9]. It was first isolated from Calyptogena sp., a species of giant white clam, at a depth of 1156 m from Sagami Bay, Japan, and was initially named Rhodotorula calyptogenae [5]. The detection of C. calyptogenae from food sources [27], human household items [28], pets [29], soil, plant materials [30,31,32] and marine ecosystems [5, 33,34,35] suggests its high adaptability to various environments.
The emerging agents of opportunistic fungal infections may impact the treatment and management of oral diseases in elderly and immunocompromised individuals. This study reports the isolation and characterization of an uncommon yeast species, C. calyptogenae, from the oral swab and rinse samples of an elderly patient presenting with angular cheilitis.
Case presentation
A 60-year-old denture-wearing Malaysian woman was referred to the Oral Medicine Clinic, Universiti Malaya, in February 2020, with the complaint of a painless swelling on her lower right cheek for a month. She also had moderate xerostomia, with a Clinical Oral Dryness Score of five [36, 37]. Upon examination, the patient was partially edentulous with no obvious facial swelling or palpable lymph nodes. Slight erythema was noted at the bilateral angle of her mouth, suggestive of angular cheilitis. A non-tender, non-indurated lump measuring 0.5 × 0.5 cm resembling an irritation fibroma, most likely caused by an ill-fitting denture, was observed on her lower right buccal sulcus over the premolar region.
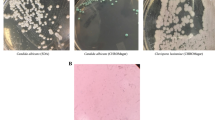
An oral swab was collected from the bilateral angle of her mouth for Gram staining and cultured for Candida yeasts on Brilliance Candida agar™ (BCA) (Oxoid, UK). An expectorated oral rinse sample collected from gargling 20 ml of saline was also obtained and centrifuged at 9600 revolutions per minute at 4 ºC for 10 min. The pellet (100 μl) was cultured for isolation of Candida yeasts. Gram-stained smear of the oral swab showed presence of scanty Gram-positive and Gram-negative bacteria and epithelial cells. Blastoconidia or hyphal filaments were not observed. Primary cultures from the oral swab and oral rinse samples grew dark blue colonies along the initial streak lines on BCA upon incubation at 37 °C after 48 h of incubation. The colonies were subcultured onto fresh BCA and Sabouraud’s dextrose agar (SDA) plates for incubation at room temperature and 37 °C. Smooth, mucoidal, and light orange or beige colonies were observed on SDA at both temperatures. Similar colonies were observed on BCA at room temperature; however, dark blue colonies were observed on BCA at 37 °C. The yeast growth was generally faster at room temperature compared to 37 ºC on both agar media. Gram stain examination of a pure culture showed round-to-oval yeast morphology. The yeast isolated was assigned as C. calyptogenae strain D1.
The yeast was identified to the species level through polymerase chain reaction (PCR) amplification and sequence determination of the internal transcribed spacer (ITS) gene and D1/D2 domain of the large subunit (LSU) ribosomal DNA (rDNA) of the yeast. The freeze–thaw method as described by Silva et al. [38] was used to extract yeast DNA. Two sets of primers, ITS1 (5ʹ-TCC GTA GGT GAA CCT GCG G-3ʹ) and ITS4 (5ʹ-TCC TCC GCT TAT TGA TAT GC-3ʹ) [39], and primer pair NL1 (5ʹ- GCA TAT CAA TAA GCG GAG GAA AAG-3ʹ) and NL4 (5ʹ-GGT CCG TGT TTC AAG ACG G-3ʹ) were used for species identification. The PCR reaction mixture (25 μl) contained 2 μl (16 ng) of DNA extract, 1 μM of each primer, 12.5 μl of exTen 2 × PCR Master Mix (1st Base, Singapore). The PCR amplification conditions included 5 min denaturation at 95 ºC for 1 cycle, followed by 35 cycles of 95 ºC for 1 min, 52 ºC for 1 min, 72 ºC for 1 min and one final extension step of 72 ºC for 10 min. The nucleotide sequences of the amplified products were determined by a commercial sequencing provider (Apical Scientific, Malaysia), using forward and reverse PCR primers. The sequences were assembled on the Biological Sequence Alignment Editing (Bioedit) software (RRID: SCR_007361) and searched for the highest sequence similarity using the GenBank Basic Local Alignment Search Tool (BLASTn) (RRID: SCR_004870) against the National Centre for Biotechnology Information (NCBI) nucleotide database (RRID: SCR_004860). A phylogenetic tree was constructed on the Molecular Evolutionary Genetics Analysis (MEGA) software (RRID: SCR_000667) using ITS gene sequences of C. calyptogenae strains retrieved from the GenBank database. Erythrobasidium hasegawianum strain CBS 8253 (AF444522) was used as an outgroup.
The yeast D1/D2 domain sequence (GenBank accession no. OK147747) was 100% (558/558 nucleotides) similar to the C. calyptogenae CBS 9125 type strain, which was first isolated from a giant white clam (AB025996) [5]. Other strains demonstrating 100% sequence similarity include C. calyptogenae strain 4107 (EU669877) [33], which was isolated from seawater in Taiwan, and strain CBS 11058 from a culture collection [40]. Meanwhile, the yeast ITS sequence (GenBank accession no. OK147742) exhibited 100% (463/463 nucleotides) similarity to C. calyptogenae strain 4107 (EU669877) [33], and CBS 11134 (KY103129) [40] but 99.5% (2 nucleotide difference) to C. calyptogenae CBS 9125 type strain. Figure 1 shows the phylogenetic tree constructed based on ITS sequences of various Cystobasidium reference strains. Strain D1 was clustered with C. calyptogenae CBS 9125 type strain, and other known C. calyptogenae strains in the same branch with high bootstrap value (100%). Based on phylogenetic analysis, the identity of strain D1 was thus confirmed as C. calyptogenae.
Neighbour-joining phylogenetic tree (Jukes–Cantor model) constructed based on ITS gene sequences of Cystobasidium species (refer to Table 1 for source and details of C. calyptogenae strains; black square = phylogenetic position of C. calyptogenae strain D1). Bootstrap values generated from 1000 replications are represented on the node of every branch
The patient was given topical treatment with 2% miconazole and fusidic acid, once daily, for the management of angular cheilitis. Mouthwash (Oral-7) was provided to improve oral dryness. The patient was also scheduled for the construction of a new lower denture to increase the vertical dimension of the mouth to prevent recurrence of angular cheilitis. The clinical condition of the patient improved during a follow-up visit 2 weeks later, with mild erythema observed on the affected area. Repeat oral swab and oral rinse cultures were negative for C. calyptogenae. However, a mixed growth of yeasts (Trichosporon asahii, Candida dubliniensis, and Candida parapsilosis) was noted in the oral rinse sample. Similar treatment was thus continued for another 2 weeks and the patient was placed on an open appointment that required her to come to the clinic only if the symptoms persisted.
Discussion and conclusions
C. calyptogenae was the only yeast isolated from both primary cultures of oral swab and rinse samples of the patient investigated in this study. The ability of C. calyptogenae to multiply at 37 °C suggests its virulence potential to infect human hosts [41], and further strengthens our speculation on its pathogenic potential. Currently, there are 12 published studies describing the isolation of C. calyptogenae from a variety of environmental sources [5, 27,28,29,30,31,32,33,34,35, 42, 43] (Table 1). The presence of C. calyptogenae in homemade fermented rice water [27], dishwashers [28], plant and soil samples [30,31,32], solidified radioactive waste disposal sites [43], marine ecosystems (including sea surface microlayer, underlying seawater, corals, clams and crabs) [29, 34, 42], as well as the external ear canal of a cat [35], may indicate possible human exposure through various points of environmental sources [44]. Given the ubiquitous nature of this Cystobasidium yeast, the likelihood of opportunistic infections may increase among immunocompromised individuals.
The observation of dark blue colonies of C. calyptogenae strain D1 on BCA at 37 ºC has not been described before in literature. Rhodotorula mucilaginosa, a close relative of Cystobasidium yeast, has been reported to grow salmon-pink colonies on BCA at 37 ºC [45]. Generally, pink to orange colonies of Cystobasidium spp. on SDA at 20–22 ºC [7, 8, 10,11,12] are due to the production of carotenoid pigment, torularhodin [46, 47]. Higher temperatures have been reported to reduce carotenoid production [12]. Hence, the development of dark blue yeast colonies on BCA at 37 ºC could be due to accumulated effects of reduced carotenoid production and enzymatic reactions with chromogens in BCA.
Among members of the genus Cystobasidium, C. fimetarium has been reported to be mycoparasitic towards ascomycetes such as Lasiobolus equinus, Saccobolus violaceus and Thelebolus crustaceus [48]. Mycoparasitism was similarly reported through the biocontrol ability of C. calyptogenae (MF438284) from a native umbú (Spondias tuberosa) fruit [30]. In vitro and in vivo antagonistic effects of the yeast were demonstrated against multiple fungal pathogens (i.e., Lasiodiplodia theobromae, Fusicoccum aesculli, Neofusicoccum parvum and Colletotrichum dianesei) affecting postharvest of mangoes. Biotechnological potentials have also been recognized in closely related species such as C. psychroaquaticum, for its extracellular enzyme and carotenoid production [12]. Therefore, further exploration of C. calyptogenae strain D1 as a mycoparasitic biocontrol agent may help provide an alternative approach for the treatment of oral fungal diseases.
An increase in the incidence of oral candidiasis has been recently reported in elderly patients [49, 50]. This study documents the first isolation and identification of C. calyptogenae, an environmental yeast, from the oral samples of an elderly patient presenting with angular cheilitis. In recent years, several clinically relevant red-color pigmented Rhodotorula species have been reported to cause opportunistic infections such as meningitis, endocarditis, fungemia, central venous catheter infections, keratitis [51], as well as non-healing oral ulcers [52]. As Cystobasidium and Rhodotorula yeasts are difficult to differentiate phenotypically [53], molecular screening is necessary in providing accurate identification for the surveillance of these opportunistic fungal infections. Further research is essential to unravel the mechanisms of infection and mycoparasitism of C. calyptogenae in the human oral cavity, in addition to various underlying factors that may increase the risk of angular cheilitis [1].
Availability of data and materials
The sequences generated and analyzed in this study were deposited into GenBank database, under the accession numbers OK147742 and OK147747.
Change history
06 December 2022
A Correction to this paper has been published: https://doi.org/10.1186/s40001-022-00884-9
Abbreviations
- Bioedit:
-
Biological Sequence Alignment Editing
- BCA:
-
Brilliance Candida agar™
- C. calyptogenae :
-
Cystobasidium calyptogenae
- C. albicans :
-
Candida albicans
- BLASTn:
-
GenBank Basic Local Alignment Search Tool
- ITS:
-
Internal transcribed spacer
- LSU:
-
Large subunit
- Min:
-
Minute
- MEGA:
-
Molecular Evolutionary Genetics Analysis
- PCR:
-
Polymerase chain reaction
- rDNA:
-
Ribosomal DNA
- SDA:
-
Sabouraud’s dextrose agar
- S. aureus :
-
Staphylococcus aureus
References
Federico JR, Basehore BM, Zito PM. Angular cheilitis. Treasure Island: StatPearls Publishing; 2021.
Cabras M, Gambino A, Broccoletti R, Lodi G, Arduino PG. Treatment of angular cheilitis: a narrative review and authors’ clinical experience. Oral Dis. 2019;26(6):1107–15. https://doi.org/10.1111/odi.13183.
Oza N, Doshi JJ. Angular cheilitis: a clinical and microbial study. Indian J Dent Res. 2017;28(6):661. https://doi.org/10.4103/ijdr.IJDR_668_16.
Pandarathodiyil AK, Anil S, Vijayan SP. Angular cheilitis—an updated overview of the etiology, diagnosis, and management. Int J Dent Oral Sci. 2021;8(2):1433–8.
Nagahama T, Hamamoto M, Nakase T, Horikoshi K. Rhodotorula benthica sp. nov. and Rhodotorula calyptogenae sp. nov., novel yeast species from animals collected from the deep-sea floor, and Rhodotorula lysiniphila sp. nov., which is related phylogenetically. Int J Syst Evol Microbiol. 2003;53(3):897–903. https://doi.org/10.1099/ijs.0.02395-0.
Laich F, Vaca I, Chavez R. Rhodotorula portillonensis sp. nov., a basidiomycetous yeast isolated from Antarctic shallow-water marine sediment. Int J Syst Evol Microbiol. 2013;63(10):3884–91. https://doi.org/10.1099/ijs.0.052753-0.
Yurkov AM, Kachalkin A, Daniel H, Groenewald M, Libkind D, de Garcia V, et al. Two yeast species Cystobasidium psychroaquaticum fa sp. nov. and Cystobasidium rietchieii fa sp. nov. isolated from natural environments, and the transfer of Rhodotorula minuta clade members to the genus Cystobasidium. Antonie Leeuwenhoek. 2015;107(1):173–85. https://doi.org/10.1007/s10482-014-0315-0.
Tsuji M, Tsujimoto M, Imura S. Cystobasidium tubakii and Cystobasidium ongulense, new basidiomycetous yeast species isolated from East Ongul Island, East Antarctica. Mycoscience. 2017;58(2):103–10. https://doi.org/10.1016/j.myc.2016.11.002.
Turchetti B, Selbmann L, Gunde-Cimerman N, Buzzini P, Sampaio JP, Zalar P. Cystobasidium alpinum sp. nov. and Rhodosporidiobolus oreadorum sp. nov. from European cold environments and Arctic region. Life. 2018;8(2):9. https://doi.org/10.3390/life8020009.
Chang C-F, Lee C-F, Liu S-M. Cystobasidium keelungensis sp. nov. a novel mycosporine producing carotenogenic yeast isolated from the sea surface microlayer in Taiwan. Arch Microbiol. 2019;201(1):27–33. https://doi.org/10.1007/s00203-018-1570-7.
Fotedar R, Fell JW, Boekhout T, Kolecka A, Zeyara A, Kaul R, et al. Cystobasidium halotolerans sp. nov., a novel basidiomycetous yeast species isolated from the Arabian Gulf. Int J Syst Evol Microbiol. 2019;69(3):839–45. https://doi.org/10.1099/ijsem.0.003250.
Chreptowicz K, Marlicka K, Milner-Krawczyk M, Korzeniowska E, Poterała M, Mierzejewska J. Cystobasidium psychroaquaticum as a new promising source of valuable bioactive molecules. Biocatal Agric Biotechnol. 2021;33: 101985. https://doi.org/10.1016/j.bcab.2021.101985.
Li A-H, Yuan F-X, Groenewald M, Bensch K, Yurkov AM, Li K, et al. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud Mycol. 2020;96:17–140. https://doi.org/10.1016/j.simyco.2020.01.002.
Slavikova E, Vadkertiova R. Seasonal occurrence of yeasts and yeast-like organisms in the river Danube. Antonie Leeuwenhoek. 1997;72(2):77–80. https://doi.org/10.1023/a:1000287005253.
Libkind D, Brizzio S, Ruffini A, Gadanho M, van Broock M, Paulo SJ. Molecular characterization of carotenogenic yeasts from aquatic environments in Patagonia, Argentina. Antonie Leeuwenhoek. 2003;84(4):313–22. https://doi.org/10.1023/a:1026058116545.
Nagahama T. Yeast biodiversity in freshwater, marine and deep-sea environments. In: Gabor Peter CR, editor. Biodiversity and ecophysiology of yeasts. The yeast handbook. Berlin: Springer; 2006. p. 241–62.
de Garcia V, Brizzio S, Libkind D, Buzzini P, van Broock M. Biodiversity of cold-adapted yeasts from glacial meltwater rivers in Patagonia, Argent. FEMS Microbiol Ecol. 2007;59(2):331–41. https://doi.org/10.1111/j.1574-6941.2006.00239.x.
Kachalkin AV. Yeasts of the White Sea intertidal zone and description of Glaciozyma litorale sp. nov. Antonie Leeuwenhoek. 2014;105(6):1073–83. https://doi.org/10.1007/s10482-014-0165-9.
de Azeredo LA, Gomes EA, Mendonca-Hagler LC, Hagler AN. Yeast communities associated with sugarcane in Campos, Rio de Janeiro, Brazil. Int Microbiol. 1998;1(3):205–8.
Golubtsova YV, Glushakova A, Chernov IY. The seasonal dynamics of yeast communities in the rhizosphere of soddy-podzolic soils. Eurasian Soil Sci. 2007;40(8):875–9. https://doi.org/10.1134/S1064229307080108.
Connell L, Redman R, Craig S, Scorzetti G, Iszard M, Rodriguez R. Diversity of soil yeasts isolated from South Victoria Land, Antarctica. Microb Ecol. 2008;56(3):448–59. https://doi.org/10.1007/s00248-008-9363-1.
Fonseca A, Inácio J. Phylloplane yeasts. In: Gabor Peter CR, editor. Biodiversity and ecophysiology of yeasts. The yeast handbook. Berlin: Springer; 2006. p. 263–301.
Yurkov AM, Chernov I, Tiunov AV. Influence of Lumbricus terrestris earthworms on the structure of the yeast community of forest litter. Mikrobiologiia. 2008;77(1):121–5.
Yurkov A, Vustin M, Tyaglov B, Maksimova I, Sineokiy S. Pigmented basidiomycetous yeasts are a promising source of carotenoids and ubiquinone Q10. Microbiology. 2008;77(1):1–6. https://doi.org/10.1134/S0026261708010013.
Glushakova A, Chernov IY. Seasonal dynamics of the structure of epiphytic yeast communities. Microbiology. 2010;79(6):830–9. https://doi.org/10.1134/S0026261710060160.
Kachalkin AV, Yurkov AM. Yeast communities in Sphagnum phyllosphere along the temperature-moisture ecocline in the boreal forest-swamp ecosystem and description of Candida sphagnicola sp. nov. Antonie Leeuwenhoek. 2012;102(1):29–43. https://doi.org/10.1007/s10482-012-9710-6.
Wongwigkarn J, Saemi K, Seeweera W, Konunta K, Fakthong W, Tasanapak K, et al. Detection and identification of naturally-occurring yeasts in homemade fermented rice water. Eco Env & Cons. 2020;26(2):871–8.
Zalar P, Novak M, de Hoog GS, Gunde-Cimerman N. Dishwashers–a man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 2011;115(10):997–1007. https://doi.org/10.1016/j.funbio.2011.04.007.
Niae S, Yurayart C, Thengchaisri N, Sattasathuchana P. Prevalence and in vitro antifungal susceptibility of commensal yeasts in the external ear canal of cats. BMC Vet Res. 2021;17(1):1–8. https://doi.org/10.1186/s12917-021-02995-7.
Gava CAT, de Castro APC, Pereira CA, Fernandes-Júnior PI. Isolation of fruit colonizer yeasts and screening against mango decay caused by multiple pathogens. Biol Control. 2018;117:137–46. https://doi.org/10.1016/j.biocontrol.2017.11.005.
Into P, Pontes A, Sampaio JP, Limtong S. Yeast diversity associated with the phylloplane of corn plants cultivated in Thailand. Microorganisms. 2020;8(1):80. https://doi.org/10.3390/microorganisms8010080.
Takashima M, Sugita T, Van BH, Nakamura M, Endoh R, Ohkuma M. Taxonomic richness of yeasts in Japan within subtropical and cool temperate areas. PLoS ONE. 2012;7(11): e50784. https://doi.org/10.1371/journal.pone.0050784.
Tien C-J, Chang C-W, Wang P-H. Rhodotorula calyptogenae, a new record yeast for Taiwan. Fung Sci. 2008;23:55–6.
Kaewkrajay C, Chanmethakul T, Limtong S. Assessment of diversity of culturable marine yeasts associated with corals and zoanthids in the Gulf of Thailand, South China Sea. Microorganisms. 2020;8(4):474. https://doi.org/10.3390/microorganisms8040474.
Shaumi A, Cheang U-C, Yang C-Y, Chang C-W, Guo S-Y, Yang C-H, et al. Culturable fungi associated with the marine shallow-water hydrothermal vent crab Xenograpsus testudinatus at Kueishan Island, Taiwan. Bot Mar. 2021;64(4):289–300. https://doi.org/10.1515/bot-2021-0034.
Osailan S, Pramanik R, Shirlaw P, Proctor G, Challacombe S. Clinical assessment of oral dryness: development of a scoring system related to salivary flow and mucosal wetness. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114(5):597–603. https://doi.org/10.1016/j.oooo.2012.05.009.
Jager DHJ, Bots CP, Forouzanfar T, Brand HS. Clinical oral dryness score: evaluation of a new screening method for oral dryness. Odontology. 2018;106(4):439–44. https://doi.org/10.1007/s10266-018-0339-4.
Silva GAd, Bernardi TL, Schaker PDC, Menegotto M, Valente P. Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Braz Arch Biol Technol. 2012;55(2):319–27. https://doi.org/10.1590/S1516-89132012000200020.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990;18(1):315–22. https://doi.org/10.1016/b978-0-12-372180-8.50042-1.
Vu D, Groenewald M, Szöke S, Cardinali G, Eberhardt U, Stielow B, et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol. 2016;85:91–105. https://doi.org/10.1016/j.simyco.2016.11.007.
Hogan LH, Klein BS, Levitz SM. Virulence factors of medically important fungi. Clin Microbiol Rev. 1996;9(4):469–88. https://doi.org/10.1128/CMR.9.4.469.
Nagahama T, Hamamoto M, Nakase T, Takami H, Horikoshi K. Distribution and identification of red yeasts in deep-sea environments around the northwest Pacific Ocean. Antonie Leeuwenhoek. 2001;80(2):101–10. https://doi.org/10.1023/a:1012270503751.
Li C-C, Chung H-P, Wen H-W, Chang C-T, Wang Y-T, Chou F-I. The radiation resistance and cobalt biosorption activity of yeast strains isolated from the Lanyu low-level radioactive waste repository in Taiwan. J Environ Radioact. 2015;146:80–7. https://doi.org/10.1016/j.jenvrad.2015.04.010.
Diaz PI, Dongari-Bagtzoglou A. Critically appraising the significance of the oral mycobiome. J Dent Res. 2021;100(2):133–40. https://doi.org/10.1177/0022034520956975.
Ghelardi E, Pichierri G, Castagna B, Barnini S, Tavanti A, Campa M. Efficacy of chromogenic candida agar for isolation and presumptive identification of pathogenic yeast species. Clin Microbiol Infect. 2008;14(2):141–7. https://doi.org/10.1111/j.1469-0691.2007.01872.x.
Re VL III, Fishman N, Nachamkin I. Recurrent catheter-related Rhodotorula rubra infection. Clin Microbiol Infect. 2003;9(8):897–900. https://doi.org/10.1046/j.1469-0691.2003.00641.x.
Deepa A, Nair BJ, Sivakumar T, Joseph AP. Uncommon opportunistic fungal infections of oral cavity: a review. J Oral Maxillofac Pathol. 2014;18(2):235. https://doi.org/10.4103/0973-029X.140765.
Sampaio JP, Oberwinkler F. Cystobasidium (Lagerheim) Neuhoff. In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts, vol. 3. Amsterdam: Elsevier; 2011. p. 1419–22. https://doi.org/10.1016/B978-0-444-52149-1.00110-5.
Sakaguchi H. Treatment and prevention of oral candidiasis in elderly patients. Med Mycol. 2017;58(2):J43–9. https://doi.org/10.3314/mmj.17.004.
Bessa ERL, de Oliveira LD, Muniz AB, da Silva GDG, Fernandes OCC, Herkrath FJ. Epidemiology of oral candidiasis: a household-based population survey in a medium-sized city in Amazonas. Res Soc Dev. 2021;10(10): e127101018664. https://doi.org/10.33448/rsd-v10i10.18664.
Wirth F, Goldani LZ. Epidemiology of Rhodotorula: an emerging pathogen. Interdiscip Perspect Infect Dis. 2012;2012: 465717. https://doi.org/10.1155/2012/465717.
Kaur R, Wadhwa A, Agarwal SK. Rhodotorula mucilaginosa: an unusual cause of oral ulcers in AIDS patients. AIDS. 2007;21(8):1068–9. https://doi.org/10.1097/QAD.0b013e328108f41c.
Libkind D, Sampaio J. Rhodotorula. In: Liu D, editor. Molecular detection of foodborne pathogens. Boca Raton: CRC Press; 2010. p. 603–18.
Acknowledgements
The authors would like to thank all project collaborators, clinicians, nurses and research students from Universiti Malaya for their contribution in this study.
Funding
The work is supported financially by the Ministry of Higher Education Malaysia via Fundamental Research Grant Scheme (FRGS/1/2019/SKK11/UM/01/1).
Author information
Authors and Affiliations
Contributions
All authors were involved in manuscript writing and approved the final draft of the case report. Student supervision was provided by STT and TGK. Clinical diagnosis was performed by JG and TGK, while laboratory investigations were conducted by ASK and STT. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Medical Research Ethics Committee, University Malaya Medical Centre (MRECID. no: 2019103–7894). Written informed consent was obtained from the patient in the study. A copy of the consent form is available for review by the Editor of this journal.
Consent for publication
The participant has provided informed consent for the publication of clinical details.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The statement in the Funding information section which was incorrectly published in the original version has been corrected.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Karajacob, A.S., Goh, J.P.E., Kallarakkal, T.G. et al. First isolation and identification of Cystobasidium calyptogenae from the oral samples of an elderly patient presenting with angular cheilitis. Eur J Med Res 27, 48 (2022). https://doi.org/10.1186/s40001-022-00671-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-022-00671-6