Abstract
Phytophthora capsici, which causes diseases in solanaceous crops, secretes necrosis and ethylene-inducing peptide 1-like proteins (NLPs) that induce plant defense responses and leaf necrosis. In this study, we used RNA interference (RNAi) technique, a proven strategy for crop protection and gene regulation in plants, to suppress P. capsici infection through the inhibition of PcNLPs. In the RNAi mechanism, Dicer processes double-stranded RNA (dsRNA) into smaller entities known as small interfering RNAs (siRNAs). These siRNAs subsequently integrate into the RNA-induced silencing complex to form sequence-specific base pairing with complementary regions of the target mRNA. This interaction effectively initiates the degradation process of the target mRNA. We designed and synthesized dsRNAs targeting the “AIMY” and “GHRHDWE” conserved motifs of PcNLP gene family, which are predicted to be key elements for the expression of NLPs and pathogen infection. After infiltration of dsRNAs targeting the motifs and inoculation with P. capsici, we confirmed a significant suppression of P. capsici infection and downregulation of the PcNLP gene family. These findings imply that the dsRNA-mediated RNAi technique holds potential for mitigating a wide range of pathogens, while simultaneously suppressing the expression of a particular gene family using dsRNA targeting functional conserved motifs in the gene family.
Similar content being viewed by others
Introduction
RNA interference (RNAi) has emerged as a promising tool for crop protection and gene regulation in plants. Following the exogenous introduction of double-stranded RNA (dsRNA) molecules, the RNAi pathway triggers a gene silencing mechanism, offering a potential means to control plant genes and pathogens [1]. In the cellular system of plants, dsRNA is cleaved by Dicer-like endonucleases into small interfering RNAs (siRNAs). These siRNAs are then incorporated into the Argonaute protein, forming the RNA-induced silencing complex, which can specifically target and silence complementary sequences in the target mRNA [1,2,3].
The oomycete plant pathogen Phytophthora capsici (P. capsici) targets solanaceous crops, including pepper, which causes root rot and results in substantial yield losses [4, 5]. The secretion of necrosis and ethylene-inducing peptide 1-like proteins (NLPs) by oomycetes induces both the plant defense response and leaf necrosis [6, 7]. In P. capsici, significant upregulation of PcNLP2 and PcNLP6 was observed during the infection stage. Furthermore, when PcNLP2 and PcNLP6 were introduced in agroinfection assays, Capsicum annuum and Nicotiana benthamiana leaves showed the most extensive necrotic areas, indicating the crucial role of these genes in promoting virulence throughout the infection phases [8]. Most of the NLPs contain the “AIMY” motif and highly sequence-conserved “GHRHDWE” motif. Mutations in the “AIMY” motif in NLP1 reduce the production of reactive oxygen species and suppress Colletotrichum orbiculare infection in cucumber [9]. Additionally, the “GHRHDWE” motif is a requirement for the activity of NLPs. In addition, this motif is critically involved in cavity formation on the protein surface, which is important for the necrosis-inducing activity of Pectobacterium carotovorum [10]. These results suggest that the “AIMY” and “GHRHDWE” motifs in PcNLP2 and PcNLP6 may play a particularly important role in P. capsici infection.
In our previous studies, we suppressed pathogens, including the pepper mottle virus and P. capsici, using dsRNAs varying position of their targets [11,12,13]. We confirmed that P. capsici infection was suppressed by dsRNAs targeting PcNLP2 and PcNLP6 of P. capsici. Significant differences in the suppression of P. capsici infection were observed depending on the regions of PcNLP2 and PcNLP6 that were targeted by the dsRNA [13]. Interestingly, the dsRNAs that effectively suppressed P. capsici infection targeted regions containing the “AIMY” and “GHRHDWE” motifs. Therefore, to investigate the role of the “AIMY” and “GHRHDWE” motifs in P. capsici infection, we used dsRNAs specifically targeting regions containing these motifs in PcNLP2 and PcNLP6, as well as dsRNAs targeting regions not containing the motifs. Furthermore, we examined whether dsRNAs targeting regions containing the motifs also suppressed the expression of other PcNLPs. Through this approach, we aimed to ascertain whether our designed multi-target dsRNA, which targets the functional conserved motifs of a particular gene family, can effectively regulate the expression of that gene family.
Materials and methods
Plant growth conditions and P. capsici maintenance
The wild-type Nicotiana benthamiana was grown at 25 ℃ and 50% humidity, exposed to a daily photoperiod of 16 h of light followed by 8 h of darkness, all within a growth chamber. For the experiments, N. benthamiana leaves were selected when plants were 3 weeks old. The KACC 40476 strain of P. capsici, generously supplied by Dr. Doil Choi’s Laboratory (Seoul National University, Seoul, Republic of Korea), was grown on V8 juice agar medium for 8 days, maintained in continuous darkness at 23 ℃.
Design and synthesis of dsRNAs
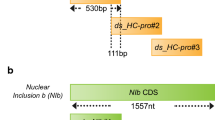
Two dsRNAs, each 200 bp in length, were designed to target the regions containing the “AIMY” and “GHRHDWE” motifs of PcNLP2 and PcNLP6, and regions not containing the motifs, respectively (Fig. 1a, b). The conservation of amino acids and RNA sequences was measured using Jalview software [14]. A 200 bp dsRNA, designed to target the Renilla luciferase gene, was employed as a mock in the experiments. As reported in our previous study [13], for the synthesis of dsRNAs, we first prepared the corresponding DNA templates, incorporating T7 promoter sequences (5′-TAATACGACTCACATATAAGAGAG-3′). This was achieved through a polymerase chain reaction (PCR) utilizing Phusion High-Fidelity DNA polymerase (Thermo Scientific, United States), in accordance with the manufacturer’s protocol. The resulting PCR products were subsequently employed in the dsRNA synthesis process utilizing MEGAscript RNAi Kit (Invitrogen, United States), in accordance with the manufacturer’s protocol. The purity of dsRNAs was shown in Additional file 1: Fig. S1. The primers used for PCR are listed in Additional file 1: Table S1.
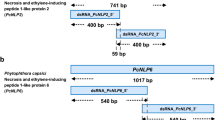
Design of double-stranded RNAs (dsRNAs) targeting the “AIMY” and “GHRHDWE” motifs of PcNLP2 and PcNLP6. a dsNLP2_MTF and dsNLP2_OReg were designed to target the regions containing the “AIMY” and “GHRHDWE” motifs of PcNLP2, and the outer region not containing the motifs, respectively. The length of each dsRNA was 200 bp. b dsNLP6_MTF and dsNLP6_OReg were designed to target the region containing the “AIMY” and “GHRHDWE” motifs of PcNLP6, and the outer region not containing the motifs, respectively. The length of each dsRNA was 200 bp. c Conservations of amino acids in PcNLPs in the regions targeted by dsNLP2_MTF and dsNLP6_MTF was measured using Jalview software. MTF: Motif, OReg: Outer region. Red boxes: “AIMY” and “GHRHDWE” motifs region
Administration of dsRNAs and P. capsici inoculation in N. benthamiana
As reported in our previous study [13], 2 days prior to P. capsici inoculation, we introduced 500 μL of dsRNAs (200 nM) into the abaxial side of N. benthamiana leaves using a needle-free syringe. A day before the P. capsici inoculation, the mycelium of P. capsici, cultured on V8 juice agar medium, was scrapped and left to incubate overnight under continuous exposure to light at 23 ℃ to facilitate sporangia formation. On the day of inoculation, the culture plate was flooded with 10 mL of distilled water, followed by a 1 h incubation at 4 ℃ to collect the zoospores. When the concentration of the zoospores reached at the 5 × 104 zoospores mL−1, 12 μL of collected zoospore suspension was inoculated in the abaxial side of the N. benthamiana leaves. Following inoculation, the leaves were incubated in darkness at 23 ℃ for approximately 24 h. The confirmation of P. capsici infection was conducted through phenotypic observation prior to sampling of leaves.
Assessment of chlorophyll fluorescence expression in N. benthamiana leaves
The infection of P. capsici on N. benthamiana leaves was confirmed by chlorophyll fluorescence expression using FOBI fluorescence in vivo imaging system (Neoscience, Republic of Korea), as previously described [13]. ImageJ was used to quantify the size of lesion [15].
Total RNA extraction and complementary DNA (cDNA) synthesis
Following assessment of chlorophyll fluorescence expression, N. benthamiana leaves were collected and ground in liquid nitrogen. Total RNA extraction was performed using RiboEx (GeneAll, republic of Korea), and the extracted total RNA was treated with recombinant DNase I (Takara, Japan) to remove both single-stranded and double-stranded DNAs, in accordance with the manufacturer’s instructions.
To synthesize cDNA, we utilized 1 μg of each RNA sample. This process was executed using PrimeScript Reverse Transcriptase (Takara, Japan) with oligo (dT) primers (Thermo Scientific, United States), in accordance with the manufacturer’s instructions.
Quantitative real-time PCR (qRT-PCR)
Light Cycler 480 (Roche, United States) with AccuPower 2X GreenStar qPCR Master Mix (Bioneer, Republic of Korea) was used for performing qRT-PCR, along with cDNA and gene-specific primers. The Ct values for the target genes were normalized to those of the housekeeping gene NbEF1a, as a control. To calculate relative expression levels, the ΔΔCt method was employed. Statistical analysis was performed using Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). Due to high homology among PcNLPs, we designed qRT-PCR primers considering potential off-target effects and the motif regions affected by dsRNAs. The primer sequences are listed in Additional file 1: Table S2. The qRT-PCR target regions within PcNLPs are depicted in Additional file 1: Fig. S2.
Results
Design of dsRNAs targeting “AIMY” and “GHRHDWE” motifs of PcNLP2 and PcNLP6
In our previous study [13], plants treated with dsRNAs targeting 5′ and 3′ regions of PcNLP2 and PcNLP6 sequence yielded different effects on the target gene and P. capsici infection. The dsRNAs that targeted the 5′-region of PcNLP2 and the 3′-region of PcNLP6 resulted in effective suppression of target gene expression and P. capsici infection. Interestingly, the target regions of the effective dsRNAs contained the “AIMY” and “GHRHDWE” motifs (Fig. 1a, b), which are predicted to be key elements in P. capsici infection and the expression of PcNLPs, based on previous studies [9, 10]. Therefore, we designed two dsRNAs for each gene: dsNLP2_MTF and dsNLP6_MTF (MTF; Motif) targeting both the “AIMY” and “GHRHDWE” motifs of PcNLP2 and PcNLP6, respectively, and dsNLP2_OReg and dsNLP6_OReg (OReg; Outer region) targeting regions that do not contain motifs, which were the outer regions of effective dsRNA in previous study [13], respectively (Fig. 1a, b). dsNLP2_MTF and dsNLP6_MTF are designed to target the “AIMY” motif on nine PcNLPs and the “GHRHDWE” motif on all PcNLPs (Fig. 1c). In addition, the RNA sequences of “AIMY” and “GHRHDWE” motifs, which are targets of dsRNA, are identical in most PcNLPs (Additional file 1: Fig. S3). In total four dsRNAs were synthesized to evaluate ability to suppress P. capsici infection and alter the expression of PcNLPs.
Suppression of P. capsici infection via dsRNAs targeting “AIMY” and “GHRHDWE” motifs of PcNLP2 and PcNLP6
Consistent with the previous study [13], we introduced dsRNAs targeting the motif regions and the outer region of PcNLP2, PcNLP6, or mock into the N. benthamiana leaves to assess the effect of dsRNAs on P. capsici infection (Figs. 2a, 3a). We measured the size of the infected lesions and the expression of PcNLP2 and PcNLP6 in P. capsica-inoculated leaves at 24 h post-inoculation (hpi) using FOBI and qRT-PCR, respectively. At 24 hpi, infected lesions were significantly suppressed in dsNLP2_MTF-treated leaves compared with mock-treated leaves (Fig. 2b, d), but there was less suppression in dsNLP2_OReg-treated leaves than the dsNLP2_MTF-treated leaves (Fig. 2c, d). Similarly, there was significant suppression of lesions in dsNLP6_MTF-treated leaves (Fig. 3b, d), whereas this did not occur in dsRNA6_OReg-treated leaves (Fig. 3c, d).
Regulation of Phytophthora capsici infection and PcNLP2 expression via dsNLP2_MTF and dsNLP2_OReg. a The experimental scheme for dsRNA treatment and P. capsici infection. b The phenotype of P. capsici infection lesion introduced with mock and dsNLP2_MTF, respectively, in 3-week-old Nicotiana benthamiana leaves, determined using a FOBI in vivo fluorescence imaging system. Scale bar = 1 cm. c The phenotype of P. capsici infection lesion introduced with mock and dsNLP2_OReg, respectively. Scale bar = 1 cm. d Quantification of P. capsici infection lesion size using ImageJ. e Relative expression level of PcNLP2 using quantitative real-time PCR (qRT-PCR). Mock: Treatment with dsRNA targeting Renilla luciferase; dsNLP2_MTF: Treatment with dsNLP2_MTF; dsNLP2_OReg: Treatment with dsNLP2_OReg. MTF: Motif, OReg: Outer region. Data represent the mean ± standard error of mean (SEM; N = 3). Statistical significance is determined by Student’s t-test (*P < 0.05 and ***P < 0.001)
Regulation of Phytophthora capsici infection and PcNLP6 expression via dsNLP6_MTF and dsNLP6_OReg. a The experimental scheme for dsRNA treatment and P. capsici infection. b The phenotype of P. capsici infection lesion introduced with mock and dsNLP6_MTF, respectively, in 3-week-old Nicotiana benthamiana leaves, determined using a FOBI in vivo fluorescence imaging system. Scale bar = 1 cm. c The phenotype of P. capsici infection lesion introduced with mock and dsNLP6_OReg, respectively. Scale bar = 1 cm. d Quantification of P. capsici infection lesion size using ImageJ. e Relative expression level of PcNLP6 using quantitative real-time PCR (qRT-PCR). Mock: Treatment with dsRNA targeting Renilla luciferase; dsNLP6_MTF: Treatment with dsNLP6_MTF; dsNLP6_OReg: Treatment with dsNLP6_OReg. MTF: Motif, OReg: Outer region. Data represent the mean ± standard error of mean (SEM; N = 3). Statistical significance is determined by Student’s t-test (*P < 0.05)
Additionally, we analyzed the levels of PcNLP2 and PcNLP6 transcript in leaves treated with dsRNAs, respectively. Compared with mock-treated leaves, PcNLP2 expression was 57-fold lower in dsNLP2_MTF-treated leaves, and PcNLP6 expression was 6,133-fold lower in dsNLP6_MTF-treated leaves (Figs. 2e, 3e). These results indicate that dsRNAs targeting the motifs suppress P. capsici infection and suppress the expression of PcNLP2 and PcNLP6. However, dsRNAs targeting the outer region did not significantly affect the expression of PcNLP2 and PcNLP6 (P > 0.05), causing only a slight reduction (Figs. 2e, 3e). In addition, they had no effect on P. capsici infection (Figs. 2d, 3d).
Expression of PcNLPs in P. capsici treated with dsRNAs targeting “AIMY” and “GHRHDWE” motifs of PcNLP2 and PcNLP6
We performed qRT-PCR on PcNLPs of P. capsici to investigate the multi-target effects of dsNLP2_MTF and dsNLP6_MTF. Interestingly, compared with mock-treated leaves, the expression of PcNLPs decreased from 2-fold to 362-fold in dsNLP2_MTF-treated leaves (Fig. 4a), and from 5-fold to 6,133-fold in dsNLP6_MTF-treated leaves (Fig. 4b). However, there was no significant decrease in the expression of PcNLPs in dsNLP2_OReg and dsNLP6_OReg-treated leaves (Additional file 1: Fig. S4). A Ct value for PcNLP8 was not detected in mock-treated and dsNLP6_MTF-treated leaves (Fig. 4b). This was attributed to the significantly lower expression level of PcNLP8 at the P. capsici infection stage compared with other PcNLPs, resulting in the non-detection of PcNLP8 using qRT-PCR. (Additional file 1: Fig. S5). Overall, these results suggest that multi-target dsRNAs targeting the “AIMY” and “GHRHDWE” motifs of PcNLPs not only suppress P. capsici infection, but also reduce the expression of PcNLPs.
Changes in the expression of the PcNLP gene family after treatment with dsRNAs targeting dsNLP2_MTF and dsNLP6_MTF. Quantification of the expression of the PcNLP gene family after treatment with mock, a dsNLP2_MTF, and b dsNLP6_MTF using qRT-PCR. Mock: Treatment with dsRNA targeting Renilla luciferase; dsNLP2_MTF: Treatment with dsNLP2_MTF; dsNLP6_MTF: Treatment with dsNLP6_MTF. MTF: Motif. N.D.: Not detected. Data represent the mean ± SEM (N = 3). Statistical significance is determined by Student’s t-test (*P < 0.05 and ***P < 0.001)
Discussion
Traditional fungicides have conventionally controlled P. capsici [5, 16], but their prolonged use and accumulation may induce fungicide-resistant pathogens and unknown mutations in the pathogen. dsRNA-mediated RNAi technology is now extensively employed to target plant genes, insects, viruses, and fungi [1]. This approach allows the fastest response to the mutation of plant pathogens as dsRNAs can be targeted to the specific mutated gene.
In a previous study [13], we showed the effective suppression of P. capsici by dsRNAs targeting specific regions of the P. capsici effector genes PcNLP2 and PcNLP6. In this study, we confirmed that the “AIMY” and “GHRHDWE” motifs were included in the target regions of the effective dsRNAs used in the previous study. The dsRNAs were designed and synthesized by dividing the target regions of the effective dsRNAs used in the previous study into regions containing motifs (dsNLP2_MTF, dsNLP6_MTF) and regions not containing motifs (dsNLP2_OReg, dsNLP6_OReg) (Fig. 1a, b). dsNLP2_MTF and dsNLP6_MTF, which targeted the motif regions, effectively suppressed P. capsici infection (Figs. 2, 3) and the expression of other PcNLPs (Fig. 4), including PcNLP2 and PcNLP6. However, dsNLP2_OReg and dsNLP6_OReg did not significantly affect the expression of PcNLP2, PcNLP6, and P. capsici infection, respectively (Figs. 2, 3). The region targeted by dsNLP2_OReg does not contain conserved motifs and the RNA sequences of PcNLPs compared with the region targeted by dsNLP2_MTF (Additional file 1: Figs. S6, S7). However, the region targeted by dsNLP6_OReg contains conserved amino acid residues and RNA sequences except for the “AIMY” and “GHRHDWE” motifs (Additional file 1: Figs. S8, S9). The regulation of the target region except for the motifs by dsNLP6_OReg suggests that it cannot control P. capsici infection (Fig. 3c, d) or the expression of other PcNLPs (Additional file 1: Fig. S4b), even if some conserved amino acid residues or RNA sequences are regulated by dsRNA. These results showed that using only dsNLP2_MTF and dsNLP6_MTF to target the “AIMY” and “GHRHDWE” motifs can suppress P. capsici infection and the expression of PcNLPs (Figs. 2, 3, 4). In addition, when dsNLP2_OReg and dsNLP6_OReg were used, the suppression of PcNLP2 and PcNLP6 expression was weaker than when dsNLP2_MTF and dsNLP6_MTF were used (Figs. 2e, 3e), suggesting that “AIMY” and “GHRHDWE” motifs are key elements for P. capsici infection and PcNLPs expression, similar to previous studies [9, 10]. In future research, it will be necessary to separate the dsRNA targeting both the “AIMY” and “GHRHDWE” motifs into dsRNAs targeting only one of these motifs to compare the degree of suppression of P. capsici infection and PcNLPs expression. This approach will help to uncover the roles of the “AIMY” motif and “GHRHDWE” motif, respectively, in regulating P. capsici infection and PcNLPs expression.
In conclusion, this study demonstrated that multi-target dsRNAs targeting conserved motifs in the PcNLP gene family could suppress P. capsici infection and the expression of PcNLPs. In addition, we provided further evidence that the “AIMY” and “GHRHDWE” motifs could be key elements regulating P. capsici infection and the expression of PcNLPs. These results can contribute to the development of RNAi technology that can control the entire gene family using one specific multi-target dsRNA.
Availability of data and materials
Not applicable.
Abbreviations
- NLP:
-
Necrosis and ethylene-inducing peptide 1-like protein
- RNAi:
-
RNA interference
- dsRNA:
-
Double-stranded RNA
- siRNA:
-
Small interfering RNA
- hpi:
-
Hours post-inoculation
- cDNA:
-
Complementary DNA
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
References
Dalakouras A, Wassenegger M, Dadami E, Ganopoulos I, Pappas ML, Papadopoulou K (2020) Genetically modified organism-free RNA interference: exogenous application of RNA molecules in plants. Plant Physiol 182:38–50
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci 6:208
Das PR, Sherif SM (2020) Application of exogenous dsRNAs-induced RNAi in agriculture: challenges and triumphs. Front Plant Sci 11:946
Rehrig WZ, Ashrafi H, Hill T, Prince J, Van Deynze A (2014) CaDMR1 cosegregates with QTL Pc51 for resistance to Phytophthora capsici in pepper (Capsicum annuum). Plant Genome. https://doi.org/10.3835/plantgenome2014.03.0011
Barchenger DW, Lamour KH, Bosland PW (2018) Challenges and strategies for breeding resistance in Capsicum annuum to the multifarious pathogen, Phytophthora capsici. Front Plant Sci 9:628
Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nurnberger T (2002) NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J 32:375–390
Qutob D, Kemmerling B, Brunner F, Kufner I, Engelhardt S, Gust AA, Luberacki B, Seitz HU, Stahl D, Rauhut T et al (2006) Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18:3721–3744
Feng BZ, Zhu XP, Fu L, Lv RF, Storey D, Tooley P, Zhang XG (2014) Characterization of necrosis-inducing NLPproteins in Phytophthora capsici. BMC Plant Biol. https://doi.org/10.1186/1471-2229-14-126
Azmi NSA, Singkaravanit-Ogawa S, Ikeda K, Kitakura S, Inoue Y, Narusaka Y, Shirasu K, Kaido M, Mise K, Takano Y (2018) Inappropriate expression of an NLP effector in Colletotrichum orbiculare impairs infection on Cucurbitaceae cultivars via plant recognition of the C-terminal region. Mol Plant Microbe In 31:101–111
Ottmann C, Luberacki B, Kufner I, Koch W, Brunner F, Weyand M, Mattinen L, Pirhonen M, Anderluh G, Seitz HU et al (2009) A common toxin fold mediates microbial attack and plant defense. PNAS 106:10359
Yoon J, Fang M, Lee D, Park M, Kim K-H, Shin C (2021) Double-stranded RNA confers resistance to pepper mottle virus in Nicotiana benthamiana. Appl Biol Chem 64:1–8
Kweon Y, Fang M, Shin S-Y, Lee D, Kim K-H, Shin C (2022) Sequence optimization and multiple gene-targeting improve the inhibitory efficacy of exogenous double-stranded RNA against pepper mottle virus in Nicotiana benthamiana. Appl Biol Chem. https://doi.org/10.1186/s13765-022-00756-0
Park M, Kweon Y, Lee D, Shin C (2023) Suppression of Phytophthora capsici using double-stranded RNAs targeting NLP effector genes in Nicotiana benthamiana. Appl Biol Chem. https://doi.org/10.1186/s13765-023-00768-4
Procter JB, Carstairs GM, Soares B, Mourao K, Ofoegbu TC, Barton D, Lui L, Menard A, Sherstnev N, Roldan-Martinez D et al (2021) Alignment of biological sequences with jalview. Methods Mol Biol 2231:203–224
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Sanogo S, Ji P (2012) Integrated management of Phytophthora capsicion solanaceous and cucurbitaceous crops: current status, gaps in knowledge and research needs. Can J Plant Pathol 34:479–492
Acknowledgements
We are grateful for helpful discussions with members of the Shin laboratory. We also thank Dr. Doil Choi’s laboratory for their materials and helpful comments.
Funding
This study was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (No. 2021R1A5A1032428). This was also supported by a grant from the New breeding technologies development Program (Project No. RS-2022-RD009544), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
CS conceived the project. MP, YK, JE, MO performed experiments. MP, YK, and CS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Primer sequences used in dsRNA synthesis. Table S2. Primer sequences used in qRT-PCR. Figure S1. The purity of Mock, dsNLP2_MTF, dsNLP2_OReg, dsNLP6_MTF, and dsNLP6_OReg. M: 100 bp marker, 1: After transcription, 2: After nuclease treatment, 3: After purification. MTF: Motif, OReg: Outer region. Figure S2. Schematic diagram of target regions in PcNLPs using qRT-PCR. Figure S3. RNA sequences conservation of the region containing the “AIMY” and “GHRHDWE” motifs of PcNLPs targeted by dsNLP2_MTF and dsNLP6_MTF. MTF: Motif. Figure S4. Quantification of the expression of PcNLP family genes after treatment with mock, (a) dsNLP2_OReg, and (b) dsNLP6_OReg using qRT-PCR. Mock: Treated with dsRNA targeting Renilla luciferase; dsNLP2_OReg: Treated with dsNLP2_OReg; dsNLP6_OReg: Treated with dsNLP6_OReg. OReg: Outer region. Data represent mean ± SEM (N = 3). Figure S5. Dot plot representation of the delta Ct values of PcNLPs in P. capsica-infected wild-type leaves using qRT-PCR. The dot represents outliers of replicated samples, and whiskers represent standard deviation.of mean. Figure S6. Amino acid sequences conservation of the region targeted by dsNLP2_OReg. OReg: Outer region. Figure S7. RNA sequences conservation of the region targeted by dsNLP2_OReg. OReg: Outer region. Figure S8. Amino acid sequences conservation of the region targeted by dsNLP6_OReg. OReg: Outer region. Figure S9. RNA sequences conservation of the region targeted by dsNLP6_OReg. OReg: Outer region.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, M., Kweon, Y., Eom, J. et al. Development of multi-target dsRNAs targeting PcNLP gene family to suppress Phytophthora capsici infection in Nicotiana benthamiana. Appl Biol Chem 66, 69 (2023). https://doi.org/10.1186/s13765-023-00828-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-023-00828-9