Abstract
Plant exosomes are nanosized (30–150 nm) membrane vesicles that contain biomolecules and influence the development of a plant and protect the plant from pathogens. Recently, plant exosomes are in the spotlight as a new biologically active substance. However, whether plant exosomes have similar efficacy to conventional secondary metabolites of plants is unknown. In this study, the difference in efficacy between plant exosomes and conventional secondary metabolites was analyzed with three or four types of plant extracts, including ginseng (Panax ginseng) and green tea (Camellia sinensis). After 6 h of treatment, the analysis of gene expression pattern of each sample showed that the exosome treatment group and the extract treatment group were clearly distinguished. After selecting the genes that showed differential expression of > twofold change, the number of genes that were up- or downregulated appeared to be 398 or 438 for the extract and 861 or 648 for the exosome, on average. This suggests that the change in transcriptome is more expressed in the exosome treatment group than in the extract treatment group. In addition, in the comparative analysis of expression of genes that are known to affect aging, regeneration, skin barrier, and moisturization—MMP12, MMP13, NOTCH3, FGF12, HS3ST3A1, LOX, VIM, ELOVL3, and KRTI—the exosome treatment group was predicted to more effectively contribute to maintaining a healthy skin when compared to the extract treatment group. The number of genes that were identified to specifically react to the Panax ginseng or Camellia sinensis treatment group during the transcriptome change phase was 11 and 8, respectively. This suggests that exosomes bear its specific effect according to the plant it is derived from. In conclusion, the results of this study indicate that plant exosomes, as natural biologically active substances, have different effects from conventional plant extracts, and have the potential to be commercialized as a cosmeceutical product.
Similar content being viewed by others
Introduction
Plants survive by defending against pathogens or adapting to the surrounding environment with various secondary metabolites, including alkaloid, flavonoid, polyphenol, terpenoid, and quinone, when exposed to biological and non-biological stress. These secondary metabolites are commonly used by consumers in ecosystems, including humans, to obtain nutrition or to expect specific efficacy from bioactive substances [1, 2]. In particular, plant-derived natural substances are widely used as cosmeceutical materials because they exert beneficial effects on the human skin, such as antiaging, moisturizing, whitening, regeneration, and nutritional supply. Typical examples are ginseng (Panax ginseng) and green tea (Camellia sinensis).
Ginseng has several ginsenosides as representative active ingredients and exerts antiaging, anti-inflammatory, and antioxidative effects [3,4,5]. Green tea has reportedly been effective in antioxidation, photoprotection, and improvement in skin-related conditions owing to the active component of flavonoids including catechin and polyphenols [6,7,8].
The plant-derived materials that are being studied recently, other than secondary metabolites that plants produce for the defense, are exosome-like nanoparticles (ELNs) or exosomes that are called extracellular vesicles (EVs) [9, 10]. Exosomes are nanosized (30–150 nm) membrane vesicles that interacts between cells from producer to recipient cells. Mammalian exosomes contain various biomolecules, including proteins, metabolites, and nucleic acids; possess characteristics similar to that of the cells from which they are derived; and play a major role in cell metabolism, including mediating cell–cell communication, immune response, and signal delivery [11,12,13]. The facts known about plant exosomes are that they contain DNA, mRNA, miRNA, proteins, lipids and substances with protective function, and they are produced in response to various biological and non-biological environmental stresses, including pathogen infection [14,15,16].
Exosome is garnering attention as a natural drug delivery nanoplatform due to its biocompatibility characteristic of evading decomposition by the immune system, and its function as a therapeutic material is actively investigated [17, 18]. In particular, plant exosomes have the advantage in that they could be produced in a large scale. Cabbage exosomes encapsulated with therapeutic drugs have been demonstrated to be efficiently delivered to human cells [19]. Plant exosomes are reported to have various bioactive functions according to the plant they are derived from. Blueberries-derived exosomes increase the survival rate of cells by regulating the expression of genes that affect inflammation and oxidation stress [20]. Exosomes derived from strawberry, along with vitamin C, were reported to prevent oxidative stress in human cells [21]. Other plant exosomes (grape, grapefruit, ginger, and carrot) were reported to help maintain intestinal homeostasis owing to their anti-inflammatory functions [22]. It has been reported that this effect of plant exosomes is due to the structural properties that can be absorbed in the gastrointestinal tract and miRNAs loaded in plant exosomes can cross kingdom regulation [19, 23,24,25]. Hence, it is anticipated that plant exosomes will be widely used in various fields, including pharmaceutics and foods as a major biologically active substance.
To date, the research of plant exosomes was restricted to the individual efficacy or the utility as a drug delivery system, there is almost no research that has reported the difference between the function of plant exosomes and conventional secondary metabolites.
This study treated the extracts and exosomes from ginseng and green tea with human keratinocytes, analyzed the change of transcriptomes, and conducted a comparative analysis on how extracts and exosomes differ in the manner they affect skin cells. The results of this study provide the evidence that plant exosomes affect human body in a different manner as compared to the extracts, and have the potential to be newly applied as a cosmeceutical product.
Materials and methods
Separation of plant extract and exosomes
Plants for analysis were purchased from a farmhouse. The leaf part of Cantella asiatica (Cica), Portulaca oleracea (Purslane), and C. Sinensis (Green tea) was used (1 kg for each plant) and the root part of 10 individual P. ginseng (Ginseng) plants was used. After hot-air drying the plants at 45 °C for 24 h, the tissues were finely grinded using a blender.
To obtain the extract, 25 g of the preprocessed sample was soaked in 25 L of distilled water (DW) at 80 °C for 3 h and was filtered through a 0.45 µm mesh filter.
To separate the exosomes, 50 g of the preprocessed sample was sealed in a plastic bag with 1 L of DW, which was passed through high pressure processing of 200 MPa at 25 °C for 30 s. This was then squeezed at 30 rpm with slow-speed screws of a juice extractor, and floating particles were removed using a mesh filter. Then it was divided into two parts; one part was used for ultracentrifugation at 150,000 ×g, 4 °C for 70 min, the other part was used for the previously reported procedure [26] of aqueous two-phase extraction system (ATPS) to obtain the exosomes. In the ATPS method, PEG (Polyethylene glycol) with molecular weight of 10,000–35,000 and dextran with molecular weight of 300,000–650,000 were added to the juice so that the concentrations reach 3.3% and 1.7%, respectively, and the centrifugation was conducted at 1000 ×g, 4 °C for 10 min to obtain the supernatant exosomes.
Analysis of exosome
To conduct the morphological analysis of exosomes, Cryo-TEM was performed using the exosomes purified with the ATPS method. 5 µL of exosome samples was put on to the lacey carbon grid, subjected to 30 s of wait time in Vitrobot Mark IV (FEI, USA), and treated with 5 s of bolt time, followed by which Cryo-TEM image was analyzed as previously described [27].
To determine the size and concentration of exosomes used for Cryo-TEM analysis, blue laser (45 mW at 488 nm) and C11440-5B camera-attached nanoparticle tracking analysis (NTA) NanoSight LM10 (Malvern Instruments) were used to observe the exosome suspension and NTA was performed as previously described [28]. The size and concentration of the nanoparticle was analyzed using NTA software 2.3.
Treatment of keratinocytes with exosomes and extract
The HaCaT (Human Epidermal Keratinocytes) cell line was cultured in Dulbecco Modified Eagle Medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin, and 1% streptomycin at 37 °C and 5% CO2. The experiments were conducted when the cell density reached 70–80% confluency. Keratinocytes were seeded at a density of 1 × 105 in a 35 mm cell culture plate and incubated for 24 h. After incubation, cells were treated with fresh media containing 2% plant extracts or 1 × 108 mL−1 concentration of plant exosomes and mock-treated with DW as control for 6 h. The exosomes used for analysis were separated by ultra-centrifugation. The groups were treated with the process three times, categorized as one set, and total two sets were each used for RNA sequencing and qRT-PCR analysis.
RNA sequencing and qRT-PCR
For the RNA sequencing and qRT-PCR analysis of keratinocytes, TransZol (TransGen, China) was used to separate RNA. Total RNA integrity was assessed using 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) with an RNA Integrity Number value of ≥ 7. RNA sequencing was performed with TruSeq Stranded mRNA LT Sample Prep Kit following the user manual. Illumina platform (San Diego, CA, USA) was used for sequencing. The raw reads data obtained from sequencing were used to create trimmed data using the Trimmomatic program. The reads that went through preprocessing were mapped to a genomic reference (GRCh38) using the Bowtie2 aligner. Gene/transcript was assembled using the String Tie program, and the abundance and normalized values of the transcript were estimated using read count and Transcript Per Kilobase Million, respectively.
A semi-quantitative real-time PCR (qRT-PCR) was performed using the CFX96TM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). GAPDH was used as the internal quantitative control and relative expression level was calculated by the 2 − ∆∆Ct method [29]. Primer sequences were designed using Primer3 software (http://fokker.wi.mit.edu). The sequences are provided in Additional file 1: Table S1.
Statistical analysis
Statistical analysis was performed using SigmaPlot v12.0. The significance of differences between groups was calculated using Student’s t-test and significance level is indicated by asterisks (*P < 0.05; **P < 0.01; and ***P < 0.001).
Results
Characterization of exosome
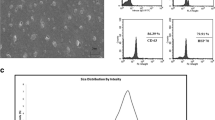
The shape of exosomes separated from P. ginseng and C. sinensis was analyzed through Cryo-TEM. Exosomes were round, with the diameter of approximately 150 nm, and the surface was surrounded by a lipid bilayer (Fig. 1A). To investigate the characteristics of plant-derived exosomes in detail, the distribution of size and concentration of exosomes obtained from ginseng (P. ginseng), green tea (C. sinensis), cica (C. asiatica), and purslane (P. oleracea) was calculated. Exosomes within the separated and purified suspension showed a Gaussian distribution of nanoparticles within the range of 87–256 nm for ginseng, 108–234 nm for green tea, 93–277 nm for cica, and 97–240 nm for purslane (Fig. 1B). The average exosome size ranged from ≤ 140 nm to < 170 nm, and the concentration of exosomes was 2.2 × 108 mL−1–4.3 × 109 mL−1 (Fig. 1C).
Characterization of exosomes isolated from plants. A Cryo-TEM images of exosomes derived from ginseng and green tea. The yellow arrow head indicates exosomes. The white arrows depict a lipid bilayer membrane of vesicle. Scale bars are 100 nm. B Size distributions of two types of plant exosomes as analyzed by Software ZataView (version 8.05.14 SP7). C Nanoparticle tracking analysis (NTA) of particle sizes and original concentrations of exosomes derived from ginseng, green tea, cica, and purslane
Transcriptome assembly of keratinocytes treated with plant extract and exosome
To confirm whether exosomes exert effects on human cell, keratinocytes were treated with the exosomes and extracts separated from ginseng, green tea, cica, purslane, and the control group (mock) for 6 h and the effects were compared. From 8 treatment groups, an average of 71,500 thousand reads with an average length of 7.22G bp were created. The trimmed data from the reads were average 70,750, with the total average length of 7.11 bp. More than 99% of cleaned reads were mapped with genomic reference (GRCh38) and < 1% of reads were unmapped.
Analysis of differentially expressed genes (DEGs) of keratinocytes according to the treatment with exosomes and extract
To investigate the correlation between exosome treatment samples and extract treatment samples, hierarchical clustering analysis of genes was conducted that showed significant changes compared with the control group. By visualizing the clustered information with heat map using the degree of similarity of gene expression pattern of each sample, exosome treatment group and extract treatment group were distinctly differentiated, and the control was clustered with extract treatment group (Fig. 2A). By selecting genes that are differentially expressed with > twofold change compared with the control group, the following results were obtained: in the extract treatment group, 385, 386, and 420 genes were upregulated and 465, 390, and 460 genes were downregulated for cica, ginseng, and green tea, respectively. In the exosome treatment group, 794, 877, 843, and 930 genes were upregulated and 747, 658, 620, and 568 were downregulated for cica, ginseng, green tea, and purslane, respectively. The average total number of regulated genes was 835 in the extract treatment group and 1509 in the exosome treatment group, with the latter possessing 1.8 times more number of differentially expressed genes (DEGs).
Transcriptome analysis regarding the keratinocytes treated with exosomes extracted from plants (cica, ginseng, green tea, and purslane) and extracts separated from plants (cica, ginseng, and green tea). A Samples and genes, demonstrated using Heatmap, that have a gene expression pattern that is similar to that of differentially expressed genes (DEGs) significant in at least one comparison group and calculated by the sample gene’s normalized expression value. Differentially expressed genes in each treatment compared with the control group were calculated by log twofold change value. B The number of up- or downregulated DEGs compared with the control group with the standard fold change value
Analysis of expression tendency of genes associated with skin benefits
The expression tendency of genes associated with skin aging, regeneration, barrier, and moisture in keratinocytes treated with exosomes and extract obtained from green tea and ginseng was analyzed. The levels of expressions of MMP12, MMP13, and NOTCH3 genes which are associated with skin aging showed a decrease in the number of ginseng exosomes compared with the control group. In exosomes originated from green tea, MMP13 and NOTCH3 expression showed a decreasing trend. For ginseng and green tea extracts, all three genes did not show any significant changes (Fig. 3A–C). Regarding the genes associated with skin regeneration, FGF12 showed an increase in only ginseng exosome treatment group compared with the control group, HS3ST3A1 showed an increase in all treatment groups, and LOX showed an increase in all treatment groups except the ginseng extract treatment group (Fig. 3D–F). Regarding the genes associated with skin barrier and moisture, VIM showed an increase in only the exosome treatment group, whereas ELOVL3 and KRT1 showed an increase in the extracts or exosomes treatment group (Fig. 3G–I).
Transcriptome analysis regarding the genes of keratinocytes treated with exosomes and extracts separated from ginseng and green tea that are expected to affect the skin function. Expression of each indicated gene is plotted. In each plot, black bars (right Y-axis) indicate TPM (Transcripts Per Kilobase Million) values from RNA-seq data while red bars (left Y-axis) indicate qRT-PCR results. A–C Expression of genes related to skin aging. D–F Expression of genes related to skin regeneration. G–I Expression of genes related to skin barrier and moisturizing.The ID of a gene name was written in parenthesis as follows: MMP12 (4321); MMP13 (4322); NOTCH3 (4854); FGF12 (2257); HS3ST3A1 (9955); LOX (4015); VIM (7431); ELOVL3 (83401), and KRT1 (3848). GAPDH (2597) was used as a reference gene for qRT-PCR. Error bars indicate S.E. (*P-value < 0.1, **P-value < 0.01, ***P-value < 0.001, ****P-value < 0.0001, unpaired t-test). gene-specific primer used in this analysis are shown in Additional file 1: Table S1
To further validate RNA-seq data, qRT-PCR analysis was conducted on nine genes from cells of different sets, all obtained under the same experimental conditions. This result showed a trend consistent with the gene expression patterns identified above (Fig. 3).
Identification of genes that have differential expression tendency toward specific treatment
To determine whether it is possible to incur a specific response in keratinocytes according to the species of plants from which exosomes and extracts are derived from, genes that showed increase or decrease of > 2 times in the gene expression in a specific treatment group while showing no significant change (< 2 times) in the gene expression were analyzed. Consequently, the number of genes with specific response to ginseng extract, ginseng exosomes, green tea extract, and green tea exosomes was found to be 3–13 for each treatment group. Additionally, genes that showed a specific response to certain plant treatment group, such as green tea exosome and extract group as well as ginseng exosome and extract group, were selected. Moreover, genes that showed a specific response to the extract treatment group or the exosome treat group, regardless of the plant, were identified. The number of genes that showed a specific response to exosome treatment group of various plant types in the experiment was 35, the highest value (Fig. 4). The number of genes that changed its expression significantly in all treatment group compared with the control group was found to be 28 (Additional file 1: Table S2).
The identification of genes that are specifically expressed in certain treatment group and the analysis of their gene expression tendency. The genes that showed a specific response to nine groups were selected. The nine groups are as follows: (ginseng [extract, exosome, extract and exosome]), (Green tea [extract, exosome, extract and exosome]), (ginseng and green tea [extract]), (ginseng and green tea [exosome]), and (ginseng and green tea [extract, exosome, extract and exosome]). The unit of the expression value is transcripts per million (TPM), and the value is normalized. Genes that showed fold change of > 3 compared with the control group while the other groups showed fold change of < 2 were selected. Fold change was calculated based on TPM values and is depicted using a color gradient. The list of genes with fold change value of ≥ 3 are presented in Additional file 1: Table S2
Discussion
In this study, the extracts and exosomes from ginseng and green tea were treated keratinocytes, and the change in transcriptomes was compared to analyze the difference in their effects on cells. We first focused on the fact that exosomes are nanosized particles and possess biocompatibility characteristics [17, 18]. Exosomes were assumed to affect keratinocytes more rapidly than those of the extract. To confirm this, the extracts and exosomes were treated with cells at a concentration of 2%, 1 × 108 mL−1, respectively. It is still not known how much bioactive substances are contained in the exosomes and there is no research that compared the extracts and exosomes, it was difficult making the decision of appropriate concentration. To solve this, the concentration that was deemed to be most appropriate was selected from the results of cell viability, cell efficacy, and wound healing tests according to the diverse concentration of the extracts and exosomes from various plants including cica, tea tree, cabbage, etc. (not published). While the treatment period for cells when analyzing the effect of ginseng or green tea extracts on human skin cells is generally 24–72 h for the transcriptome analysis, in this study, cells were treated for 6 h, which is quite a short period of time [30,31,32]. As expected, exosome treatment group had significantly higher numbers of expressed genes than the extract treatment or control groups. Hence, the cells were mediated more quickly or effectively (Fig. 2B). Data reliability was confirmed by increasing the types of plant exosomes. All four types of the exosome treatment group showed such tendency, and the DEG expression distinctly differed among the three types of extract treatment group and four types of exosome treatment group (Fig. 2A). This result implies that exosomes were effectively absorbed to Keratinocytes owing to their biocompatibility, taking a fast time to affect cells than the extracts. In addition, a previous study has also reported that cabbage-derived exosomes were effectively absorbed to human cells by penetrating into them [19].
To investigate whether exosomes can affect human skin cells, expression patterns of genes related to skin aging, regenerating, barriers, and moisturizing were analyzed. Gene expression level of MMP12, MMP13, and NOTCH3 reportedly increases when exposed to light stress such as UVB and cause decomposition of collagen and skin aging [33,34,35]. Expressions of these genes have significantly decreased in exosome treatment groups of ginseng and green tea, with no notable changes in the extract treatment groups (Fig. 3A–C). Analyzing the expression of genes related to skin regeneration revealed that FGF12 which suppresses radiation damage to skin tissue increased specifically in ginseng exosome treatment group [36]. HS3ST3A1 which facilitates skin regeneration by affecting skin regeneration signal delivery and LOX which considerably influences healing of skin wounds by facilitating stabilization showed an increase in the exosome treatment group (Fig. 3D–F) [37, 38]. VIM, which affects the differentiation and migration of keratinocytes; ELOVL3, which affects lipid acid biosynthesis; and KRT1, a fibrillary protein that form skin, hair, and nail, are all recognized as genes that construct skin barrier and prevent water loss [39,40,41,42]. These genes responded particularly to the exosome treatment group and showed the highest increase in ginseng exosomes.
The analysis results of the expression mode of genes associated with skin benefits demonstrated that exosomes separated from plants may help form healthy skin. Although, in the extract treatment group, the expression of these genes did not change significantly or the degree of change was inadequate compared with the exosome treatment group. This does not mean that extracts have less effect compared with exosomes. As aforementioned, extracts are normally treated for > 24 h. Therefore, if the extracts and exosomes are treated for more than 6 h, the result of the transcriptome of keratinocytes is expected to be different. This is because although ginseng treatment group did not show significant gene expression of MMP12 and MMP13 in this study, a previous study reported that the gene expression was decreased by > twofold change when compound K, obtained from ginseng extract, was treated with keratinocytes for 24 h [30].
This study also aimed to confirm whether exosomes have different effect according to the plant species. Currently, plant exosomes are known to contain DNA, mRNA, miRNA, secondary cell wall biosynthesis enzyme, and protection substance [14,15,16], while detailed research on the constitutive elements of exosomes according to the plant species are rarely conducted. We assumed that if constitutive elements in exosomes vary by plant species, there should be a gene that specifically reacts to only exosomes of certain species. To find this out, genes that are expressed significantly only in certain treatment groups were identified. Consequently, the number of genes that were identified to show no significant change in gene expression in other treatment groups but specifically reacts to ginseng or green tea exosome treatment group were 11 and 8 respectively. Therefore, it was cautiously anticipated that a specific substance that depends on the plant species is likely to be contained within exosomes.
Additionally, profiling small RNAs within exosomes of ginseng or green tea revealed that 75% or 41% of RNA composition is unknown (data not shown), implying that exosome is an uncharted territory that has infinite potential as a new resource of plant-driven natural substances.
In conclusion, this study revealed the potential of plant exosomes as a new cosmeceutical material, with different effect and influence on human keratinocytes from the conventional secondary metabolites.
Availability of data and materials
The datasets used in this study and any additional information are available from the corresponding author upon reasonable request.
References
Pagare S, Bhatia M, Tripathi N, Pagare S, Bansal YK (2015) Secondary metabolites of plants and their role: overview. Curr Trends Biotechnol Pharm 9:293–304
Karuppusamy S (2009) A review on trends in production of secondary metabolites from higher plants by in vitro tissue, organ and cell cultures. J Med Plant Res 3:1222–1239
Kang TH, Park HM, Kim YB, Kim H, Kim N, Do JH, Kang C, Cho Y, Kim SY (2009) Effects of red ginseng extract on UVB irradiation-induced skin aging in hairless mice. J Ethnopharmacol 123:446–451
Hong CE, Lyu SY (2011) Anti-inflammatory and antioxidative effects of Korean red ginseng extract in human keratinocytes. Immune Netw 11:42–49
Park HJ, Kim DH, Park SJ, Kim JM, Ryu JH (2012) Ginseng in traditional herbal prescriptions. J Ginseng Res 36:225–241
Yarnell E, Abascal K (2012) Herbal sunscreens and ultraviolet protectants. Altern Compl Ther 18:141–144
Meetham P, Kanlayavattanakul M, Lourith N (2018) Development and clinical efficacy evaluation of anti-greasy green tea tonner on facial skin. Rev Bras Farmacogn 28:214–217
Koch W, Zagórska J, Marzec Z, Kukula-Koch W (2019) Applications of tea (Camellia sinensis) and its active constituents in cosmetics. Molecules 24:4277
Hessvik NP, Llorente A (2018) Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 75:193–208
Schorey JS, Bhatnagar S (2008) Exosome function: from tumor immunology to pathogen biology. Traffic 9:871–881
Colombo M, Raposo G, Théry C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289
Wortzel I, Dror S, Kenific CM, Lyden D (2019) Exosome-mediated metastasis: communication from a distance. Dev Cell 49:347–360
Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H (2018) New technologies for analysis of extracellular vesicles. Chem Rev 118:1917–1950
Hansen LL, Nielsen ME (2017) Plant exosomes: using an unconventional exit to prevent pathogen entry? J Exp Bot 69:59–68
Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, De La Canal L (2017) Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J Exp Bot 68:5485–5495
Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360:1126–1129
Peng LH, Wang MZ, Chu Y, Zhang L, Niu J, Shao HT, Yuan TJ, Jiang ZH, Gao JQ, Ning XH (2020) Engineering bacterial outer membrane vesicles as transdermal nanoplatforms for photo-TRAIL-programmed therapy against melanoma. Sci Adv 6:2735
Dad HA, Gu TW, Zhu AQ, Huang LQ, Peng LH (2021) Plant exosome-like nanovesicles: emerging therapeutics and drug delivery nanoplatforms. Mol Ther 29:13–31
You JY, Kang SJ, Rhee WJ (2021) Isolation of cabbage exosome-like nanovesicles and investigation of their biological activities in human cells. Bioact Mat 6:4321–4332
De Robertis M, Sarra A, D’Oria V, Mura F, Bordi F, Postorino P, Fratantonio D (2020) Blueberry-derived exosome-like nanoparticles counter the response to TNF-α-induced change on gene expression in EA.hy926 cells. Biomolecules 10:742
Perut F, Roncuzzi L, Avnet S, Massa A, Zini N, Sabbadini S, Giampieri F, Mezzetti B, Baldini N (2021) Strawberry-derived exosome-like nanoparticles prevent oxidative stress in human mesenchymal stromal cells. Biomolecules 11:87
Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, Zhang L, Kakar S, Jun Y, Miller D, Zhang HG (2014) Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res 58:1561–1573
Wang QL, Ren Y, Mu JY, Egilmez NK, Zhuang XY, Deng ZB, Zhang LF, Yan J, Miller D, Zhang HG (2015) Grapefruit derived nanovectors use an activated leukocyte trafficking pathway to deliver therapeutic agents to inflammatory tumor site. Cancer Res 75:2520–2529
Pérez-Bermúdez P, Blesa J, Soriano JM, Marcilla A (2017) Extracellular vesicles in food: experimental evidence of their secretion in grapefruits. Eur J Pharm Sci 98:40–50
Woith E, Fuhrmann G, Melzig MF (2019) Extracellular vesicles—connecting kingdoms. Int J Mol Sci 20:569
Kırbaş OK, Bozkurt BT, Asutay AB, Mat B, Ozdemir B, Öztürkoğlu D, Ölmez H, İşlek Z, Şahin F, Taşlı PN (2019) Optimized isolation of extracellular vesicles from various organic sources using aqueous two-phase system. Sci Rep 9:19159
Emelyanov A, Shtam T, Kamyshinsky R, Garaeva L, Verlov N, Miliukhina I, Kudrevatykh A, Gavrilov G, Zabrodskaya Y, Pchelina S, Konevega A (2020) Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid. PLoS ONE 15:e0227949
Chan MY, Dowling QM, Sivananthan SJ, Kramer RM (2016) Particle sizing nanoparticle adjuvant formulations by dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA), vaccine adjuvants. Methods Mol Biol 17:239–252
Pfaffl MW (2001) Development and validation of an externally standardized quantitative insulin like growth factor-1 (IGF-1) RT-PCR using Light Cyclear SYBR Green I technology. In: Meuer S, Witter C, Nakagawara K (eds) Rapid cycle real-time PCR. Springer, Heidelberg, pp 281–291
Kim S, Kang BY, Cho SY, Sung DS, Chang HK, Yeom MH, Kim DH, Sim YC, Lee YS (2004) Compound K induces expression of hyaluronan synthase 2 gene in transformed human keratinocytes and increases hyaluronan in hairless mouse skin. Biochem Biophys Res Commun 316:348–355
Yoo HS, Kim JM, Jo E, Cho CK, Lee SY, Kang HS, Lee MG, Yang PY, Jang IS (2017) Modified Panax ginseng extract regulates autophagy by AMPK signaling in A549 human lung cancer cells. Oncol Rep 37:3287–3296
Esghaei M, Ghaffari H, Esboei BR, Ebrahimi TZ, Bokharaei SF, Motevalian M (2018) Evaluation of anticancer activity of Camellia Sinensis in the Caco-2 colorectal cancer cell line. Asian Pac J Cancer Prev 19:1697–1701
Tewari A, Grys K, Kollet J, Sarkany R, Young AR (2014) Upregulation of MMP12 and its activity by UVA1 in human skin: potential implications for photoaging. J Invest Dermatol 134:2598–2609
Pittayapruek P, Meephansan J, Prapapan O, Komine M, Ohtsuki M (2016) Role of matrix metalloproteinases in photoaging and photocarcinogenesis. Int J Mol Sci 17:868
Rossi M, Abdelmohsen K (2021) The emergence of senescent surface biomarkers as senotherapeutic targets. Cells 10:1740
Nakayama F, Yasuda T, Umeda S, Asada M, Imamura T, Meineke V, Akashi M (2011) Fibroblast growth factor-12 (FGF12) translocation into intestinal epithelial cells is dependent on a novel cell-penetrating peptide domain. J Biol Chem 286:25823–25834
Patel VN, Lombaert IM, Cowherd SN, Shworak NW, Xu Y, Liu J, Hoffman MP (2014) Hs3st3-modified heparan sulfate controls KIT+ progenitor expansion by regulating 3-O-sulfotransferases. Dev Cell 29:662–673
Cai L, Xiong X, Kong X, Xie J (2017) The role of the lysyl oxidases in tissue repair and remodeling: a concise review. Tissue Eng Regen Med 14:15–30
Velez-delValle C, Marsch-Moreno M, Castro-Muñozledo F, Galván-Mendoza IJ, Kuri-Harcuch W (2016) Epithelial cell migration requires the interaction between the vimentin and keratin intermediate filaments. Sci Rep 6:24389
Cheng F, Shen Y, Mohanasundaram P, Lindström M, Ivaska J, Ny T, Eriksson JE (2016) Vimentin coordinates fibroplast proliferation and keratinocyte differentiation in wound healing via TGF—slug signaling. PNAS E4320-E4327
Westerberg R, Tvrdik P, Undén AB, Månsson JE, Norlén L, Jakobsson A, Holleran WH, Elias PM, Asadi A, Flodby P, Toftgård R (2004) Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem 279:5621–5629
Roth W, Kumar V, Beer HD, Richter M, Wohlenberg C, Reuter U, Thiering S, Staratschek-Jox A, Hofmann A, Kreusch F, Schultze JL (2012) Keratin 1 maintains skin integrity and participates in an inflammatory network in skin through interleukin-18. J Cell Sci 125:5269–5279
Acknowledgements
The authors are grateful the funding resources for this work, the Small and Medium Business Administration (SMBA, Korea).
Funding
This work was supported by the Starting Growth Technological R&D Program (S3073598) funded by the Small and Medium Business Administration (SMBA, Korea).
Author information
Authors and Affiliations
Contributions
J-SC, JHC, SJP: conceived and designed the analysis; DK, YDH: data collection; H-MK: contributed data/analysis tools; EJY, JSK: performed the analysis; J-SC, JHC: wrote the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests or conflicts in relation to the work described.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Primers used in this study. Table S2. List of genes with fold change value of ≥3 in all treatment groups.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cho, J.H., Hong, Y.D., Kim, D. et al. Confirmation of plant-derived exosomes as bioactive substances for skin application through comparative analysis of keratinocyte transcriptome. Appl Biol Chem 65, 8 (2022). https://doi.org/10.1186/s13765-022-00676-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-022-00676-z