Abstract
Introduction
Vancomycin-resistant Staphylococcus aureus, identified as a “high priority antibiotic-resistant pathogen” by the World Health Organization, poses a significant threat to human health. This systematic review and meta-analysis aimed to estimate the pooled prevalence of vancomycin-resistant Staphylococcus aureus in Ethiopia.
Methods
This systematic review and meta-analysis was reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. Studies that reported VRSA prevalence due to infection or carriage from human clinical specimens were extensively searched in bibliographic databases and grey literatures using entry terms and combination key words. Electronic databases like PubMed, Google Scholar, Wiley Online Library, African Journal Online, Scopus, Science Direct, Embase, and ResearchGate were used to find relevant articles. In addition, the Joanna Briggs Institute quality appraisal tool was used to assess the quality of the included studies. Stata version 14 software was used for statistical analysis. Forest plots using the random-effect model were used to compute the overall pooled prevalence of VRSA and for the subgroup analysis. Heterogeneity was assessed using Cochrane chi-square (I2) statistics. After publication bias was assessed using a funnel plot and Egger’s test, trim & fill analysis was carried out. Furthermore, sensitivity analysis was done to assess the impact of a single study on pooled effect size.
Results
Of the 735 studies identified, 31 studies that fulfilled the eligibility criteria were included for meta-analysis consisted of 14,966 study participants and 2,348 S. aureus isolates. The overall pooled prevalence of VRSA was 14.52% (95% CI: 11.59, 17.44). Significantly high level of heterogeneity was observed among studies (I2 = 93.0%, p < 0.001). The region-based subgroup analysis depicted highest pooled prevalence of 47.74% (95% CI: 17.79, 77.69) in Sidama region, followed by 14.82% (95% CI: 8.68, 19.88) in Amhara region, while Oromia region had the least pooled prevalence 8.07% (95% CI: 4.09, 12.06). The subgroup analysis based on AST methods depicted a significant variation in pooled prevalence of VRSA (6.3% (95% CI: 3.14, 9.43) for MIC-based methods, and 18.4% (95% CI: 14.03, 22.79) for disk diffusion AST method) which clearly showed that disk diffusion AST method overestimates the pooled VRSA prevalence. The total number of S. aureus isolates was found to be the responsible variable for the existence of heterogeneity among studies (p = 0.033).
Conclusion
This study showed an alarmingly high pooled prevalence of VRSA necessitating routine screening, appropriate antibiotic usage, and robust infection prevention measures to manage MRSA infections and control the emergence of drug resistance. Furthermore, mainly attributable to the overestimation of VRSA burden while using disk diffusion method, there is an urgent need to improve the methods to determine vancomycin resistance in Ethiopia and incorporate MIC-based VRSA detection methods in routine clinical laboratory tests, and efforts should be directed at improving it nationally.
Trial Registration
PROSPERO registration identification number: CRD42023422043.
Similar content being viewed by others
Introduction
Bacterial multidrug resistance has emerged as a global threat, and continues to pose a significant challenge to medicine and healthcare systems worldwide [1]. There has been a devastating report of about 5 million deaths globally associated with bacterial antimicrobial resistance (AMR) only in the year 2019, of which sub-Saharan Africa bear the highest burden, with 27.3 deaths per 100,000 attributable to AMR. Surprisingly, it is also predicted that AMR will possibly kill 10 million people annually by 2050, while tumbling the global economy by $100 trillion [2].
Staphylococcus aureus (S. aureus), which is a Gram-positive coccus responsible for various human infections, ranging from skin and soft tissue infections to life-threatening systemic diseases as an opportunistic, nosocomial and community-acquired pathogen [3]. Over the years, S. aureus has developed various drug resistance mechanisms, which make it difficult to treat with conventional antibiotics, including βeta-lactamase production, methicillin resistance (MRSA), vancomycin resistance (VRSA), macrolide, aminoglycoside and quinolone resistances, and biofilm formation [4]. Highly drug resistant S. aureus including MRSA have been effectively treated with vancomycin as a first line drug since 1980s [5, 6], and vancomycin has been used as a last resort antibiotic for the management of severe infections due to MRSA and other MDR Gram-positive pathogens [7]. However, S. aureus isolates resistant to vancomycin have emerged in the past two decades, and are now becoming a major cause of morbidity and mortality worldwide [7, 8], with the first VRSA being reported in 1997 from Japan [9].
The World Health Organization has recently listed VRSA as a “high priority antibiotic-resistant pathogens” [10] due to its significant impact on public health. Vancomycin resistance in S. aureus (MIC ≥ 16 µg/ml) is mainly conferred by vanA operon encoded on transposon Tn1546, and other van gene clusters including vanB, vanC, vanD, vanF, vanE, vanG vanI, vanL, vanM and vanN phenotypes [11, 12]. These genetic elements alter the cell wall structure, preventing vancomycin from effectively inhibiting cell wall synthesis [13, 14]. Primarily due to their evidently decreased permeability and altered cell wall, VRSA strains are immensely multidrug resistant against various antibacterial agents currently in use [6].
Recently, published systematic review and meta-analysis articles assessed the epidemiology of VRSA globally and revealed the prevalence based on diverse years and regions [15, 16]. Despite the reported morbidity rates of VRSA were relatively low in developed countries, the burden is still high in developing countries such as Africa. Thus, comprehensive countrywide studies are critical in low-income countries to reflect the real burden of VRSA nationally and devise control strategies.
In Ethiopia, there is a rapidly increasing bacterial antimicrobial resistance to the routinely used antibacterial drugs as depicted by a recent systematic review [17]. Thus, epidemiological studies and evidence-based practices are of paramount significance for developing effective prevention and control strategies and improving healthcare services. Although upsurging rates of VRSA are nowadays being reported in different parts of the world, there is no national pooled data in Ethiopia. This study is the first systematic review and meta-analysis to report the national burden of VRSA in Ethiopia; and it aimed to summarize the findings of local studies reporting VRSA infection or colonization, and estimate the pooled prevalence of VRSA in Ethiopia.
Methods
Guidelines and protocol registration
This systematic review and meta-analysis was reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (PRISMA) [18]. The protocol for this review was originally registered in the International Prospective Register of Systematic Reviews (PROSPERO) database with registration identification number of CRD42023422043.
Search strategy and selection of studies
A comprehensive and systematic literature searches were carried out to retrieve studies reporting the prevalence of vancomycin-resistant Staphylococcus aureus (VRSA) in Ethiopia from different electronic bibliographic databases including PubMed/ Medline, Google Scholar, Wiley Online Library, African Journal Online, Scopus, Science Direct, Embase, and ResearchGate. Furthermore, grey literatures and university repositories were screened, and a direct Google search was carried out using the reference lists of the included studies to incorporate further relevant studies that was missed during electronic database searches. The search was conducted from May 1 to 20, 2023. Studies that were published/reported until April 30, 2023 and fulfilled the eligibility criteria were included.
A thorough searching strategy was deployed using the condition, context, population, and outcome of interest (CoCoPop) formulating questions, and all potentially eligible studies were accessed by using the following Medical Subject Headings (MeSH) terms and combination key words: “Prevalence”, “epidemiology”, “burden”, “Staphylococcus aureus”, “S. aureus”, “vancomycin-resistant Staphylococcus aureus”, “vancomycin-resistant S. aureus”, “VRSA” and “Ethiopia”. In the advanced searching databases, the abovementioned search terms were linked using Boolean operators (“OR” and “AND”) as necessary. Moreover, the bibliographies of all included studies were checked for additional articles and authors were contacted to receive any missing papers. Search results were consolidated into Endnote 20 software (Clarivate Analytics USA) and duplicates were removed. Three independent reviewers (MAB, AG and EA) identified the articles from databases and other sources. Duplicates were removed and four independent reviewers (HD, MT, OM, HE) screened the titles and abstracts of all retrieved studies, and were double-checked by a third reviewer (AG). The full texts of potentially eligible studies were then evaluated in detail against the inclusion criteria by two reviewers (MAB and EA), double-checked by a third reviewer (AG), and added to the extraction collection. Any disagreements among reviewers throughout each stage of screening were unraveled through discussion or with the intruding of a third reviewer (AG). Detailed article search strategies and search lines were indicated in Supplementary file 1.
Eligibility criteria
Original studies published in peer-reviewed journals or grey literature, articles published in English language, studies that reported prevalence of vancomycin-resistant Staphylococcus aureus among clinical specimens recovered from any human study participants which encompassed infection or carriage, studies that detected vancomycin-resistance using phenotypic or genotypic methods, laboratory-based observational (e.g. cross-sectional) studies conducted in Ethiopia from January 1, 2000 to April 30, 2023, addressing the research question, and studies involving human (infected individuals or asymptomatic carriers) were included.
Studies were excluded if they were done from non-human sources. Qualitative studies, reviews, commentaries, letters to the editor, author replies, and studies that did not include quantitative data on the prevalence of VRSA were excluded. Furthermore, studies with duplicate data or overlapping articles, studies with outcomes of interest were missing or vague, and studies with a small number of S. aureus isolates (less than 10) were excluded.
Outcome variables
The outcome variable for this study is the pooled prevalence of VRSA (infection and colonization) among Ethiopian populations. We included studies that reported the prevalence of VRSA among clinical specimens recovered from any human study participants, which encompassed both infection or carriage. In this study, an outcome of “infection” is defined as a form of diseases with suspected S. aureus aetiology by clinicians, while an outcome of “carriage” is defined as colonization of human with S. aureus as asymptomatic carrier, both of which are explained with detection of VRSA from human clinical specimens using phenotypic or genotypic methods.
Quality assessment
Three authors (MAB, DG and EA) critically assessed the methodological and finding quality of the eligible studies using the Joanna Briggs Institute (JBI) quality appraisal tool for prevalence studies [19]. Using the critical appraisal checklist, studies with an average quality score of 50% or higher were deemed to be of good quality and hence included for analysis (Supplementary file 2). Studies were assessed using title, abstract and full text screening.
Data extraction
Essential data from the eligible studies were extracted onto an excel spreadsheet by three reviewers (MAB, EA and AG). The extracted data include author (s) name, publication year, region, study area, study period, study design, study population, specimen types, antimicrobial susceptibility testing (AST) method, sample size, number of S. aureus isolates, number of VRSA isolates and prevalence of VRSA. The three reviewers thoroughly cross-checked their extraction outputs, and disagreements were resolved by discussion, data cross-checking and validation.
Statistical data analysis
Data analysis was conducted using Stata version 14.0 software (Stata Corp., College Station, TX). We used logit transformation in our analysis to pool proportions. A random-effect model of DerSimonian and Laird analysis was used to estimate the pooled prevalence of VRSA [20]. The Cochran’s Q test and I2 statistics were used to quantify and assess the presence of heterogeneity between studies [21]. The p-value of < 0.05 for I2 statistics was used to determine the presence of heterogeneity. A predefined subgroup analysis was performed based on publication year, region, city, study design and AST method. Moreover, sensitivity analysis was carried out to assess the effect of a single study on the overall pooled estimate using a leave-one-out approach. Meta-regression was also used to further explore the potential sources of heterogeneity among the included studies by examining the relationship between study characteristics (such as publication year, sample size, or number of S. aureus) and the observed variations in the prevalence of VRSA, allowing for a more comprehensive understanding of the factors contributing to the heterogeneity. Publication bias was evaluated using inspection of funnel plot symmetry and Egger’s test statistics [22, 23]. The Trim-and-Fill analysis was then used in asymmetrical funnel plots to incorporate missing studies and provide an indication of the reliability of the estimate in relation to publication bias. The findings were presented using a pooled prevalence with a 95% CI, corresponding p-value and forest plots.
Results
Selection of studies
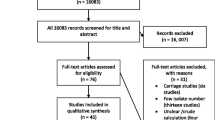
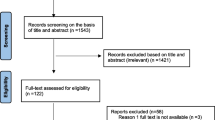
A total of 735 studies were retrieved from database searches and other sources, from which 367 were removed due to duplication. The remaining 368 articles were screened based on title and abstract review, and 281 were removed. Finally, a total of 87 articles were thoroughly evaluated against the eligibility criteria, and only 31 were found to be potentially eligible for inclusion in the systematic review and meta-analysis (Fig. 1).
Study characteristics
This systematic review and meta-analysis included a total of 31 original articles from different regions of Ethiopia. All the included studies had a quality score of greater than 75%. The overall number of participants in all studies included in the analysis was 14,966, with 315 VRSA isolates investigated from a total of 2,348 S. aureus isolates. Majority of the included (83.9%) deployed cross-sectional study design while the rest employed retrospective study design (Table 1).
Prevalence of VRSA in Ethiopia
In this systematic review and meta-analysis, the overall pooled prevalence of VRSA in Ethiopia was 14.52% (95% CI: 11.59, 17.44). A huge discrepancy in the prevalence of VRSA was revealed among the included studies, ranging from 1.2% (95% CI: 0.02, 2.38) reported in Jimma to 75.8% (95% CI: 61.19, 90.41) reported in Hawassa. Significantly high level of heterogeneity was observed among studies (I2 = 93.0%, p < 0.001) (Fig. 2).
Subgroup analysis of VRSA prevalence in Ethiopia
Subgroup analysis was carried out based on region, city, year of publication, study design and AST method. The region-based subgroup analysis depicted highest pooled prevalence of 47.74% (95% CI: 17.79, 77.69) in Sidama region, followed by 14.82% (95% CI: 8.68, 19.88) in Amhara region, while Oromia region had the least pooled prevalence 8.07% (95% CI: 4.09, 12.06). High heterogeneity was demonstrated in all included regions of the country. The pooled prevalence of VRSA was highest in Hawassa 47.74% (95% CI: 17.79, 77.69), followed by 36.71% (95% CI: 24.99, 48.43) in Dessie. Relatively low level of heterogeneity was observed from studies conducted in Dessie (I2 = 37.8%, p = 0.205) and Bahir Dar (I2 = 44.7%, p = 0.179), whereas no heterogeneity (I2 = 0.0%, p = 0.721) was seen among studies in Gondar. Nevertheless, there was high heterogeneity in Addis Ababa, Debre Markos, Hawassa and Jimma. Likewise, highest pooled prevalence of VRSA 21.01% (95% CI: 11.58, 30.45) was observed in the years 2015–2017, and low level of heterogeneity was seen among studies in the period 2018–2020. The prevalence of VRSA pooled from studies showed increment from the period ≤ 2014 to 2015–2017, then declined in the later publication years. The subgroup analysis based on AST methods depicted a significant variation in pooled prevalence of VRSA (6.3% (95% CI: 3.14, 9.43) for MIC-based methods, and 18.4% (95% CI: 14.03, 22.79) for disk diffusion AST method). On the other hand, the prevalence of VRSA in terms of study design was 15.07% (95% CI: 11.82, 18.31) in cross-sectional studies and 12.54% (95% CI: 4.66, 20.42) in studies with retrospective design (Table 2).
Meta-regression
Meta-regression was carried out to further explore the potential sources of heterogeneity or variability among studies included in the meta-analysis. We included continuous study characteristics as covariates including publication year, sample size and total number of S. aureus isolates in the meta-regression model and assess their potential influence on the overall effect size (pooled prevalence of VRSA) (Fig. 3). In this study, total number of S. aureus isolates was found to be the responsible variable for the existence of heterogeneity among studies (p = 0.033) (Table 3).
Publication bias
In this study, the symmetry of the funnel plot illustrated the presence of publication bias, with over 67% of the studies skewed to the left side of the triangular zone (Fig. 4). This finding was further supported by the Egger’s test, which revealed the presence of substantial publication bias (p < 0.001) (Table 4) (Fig. 5).
Trim and fill analysis of pooled prevalence of VRSA in Ethiopia
Attributable to the presence of marginally significant publication bias, we performed a trim and fill analysis. After incorporating 16 additional studies, the trim and fill analysis revealed a pooled prevalence of 3.56% (95% CI: 0.39, 6.73) VRSA in Ethiopia (Table 5).
Sensitivity analysis
Based on the results of the sensitivity analysis, which was conducted using a random effect model, the pooled effect size fell within the 95% CI of the overall pooled affect size when the individual studies were omitted. This demonstrated that no single study had an impact on the overall pooled prevalence of VRSA infection in Ethiopia (Table 6).
Discussion
Nowadays, frequent use of vancomycin as the drug of choice for treatment of infections caused by MRSA and other Gram-positive MDR pathogens has led to the emergence of S. aureus isolates with high resistance to vancomycin [13, 55, 56]. According to our evidence so far, we carried out the first large-scale systematic review and meta-analysis of available data on the epidemiology of VRSA in Ethiopia. The main aim of this study was to determine the national pooled prevalence of VRSA in Ethiopia by pooling data from various studies and assess the distribution patterns of VRSA across the country. The overall pooled prevalence estimate of VRSA in Ethiopia was found to be 14.52% (95% CI: 11.59, 17.44), with high level of heterogeneity (I2 = 93.0%, p < 0.001). This finding is comparable with a previous review reporting the pooled prevalence of VRSA in Africa 16% (95% CI: 3, 35) [15]. On the contrary, the finding of the present systematic review and meta-analysis is massively higher than global studies that reported an overall pooled prevalence of VRSA as 1.5% (95% CI: 1.0, 2.0] [16] and 6% (95% CI: (0.04, 0.09) [15]. In addition, our overall pooled prevalence finding is higher than a systematic review and meta-analysis studies conducted in Iran which only reported 24 VRSA isolates from the included thirteen studies with a pooled prevalence of 2.4% [57] and in the Middle east which reported a total of only 19 VRSA isolates with a pooled prevalence of 2.1% [58]. This higher finding indicates the huge burden of VRSA in Africa including Ethiopia than other continents as evidenced by lower findings reported from Asia 5% (95% CI: 0.03, 0.08), South America 3% (95% CI: 0.00, 0.17), North America 4% (95% CI: 0.02, 0.07), and Europe 1% (95% CI: 0.00, 0.05) [15]. The possible reasons for the higher rate of VRSA could possibly be poor hygiene standards [59], inadequate monitoring of nosocomial infections, and improper use of available antibacterial drugs in Africa in comparison to developed countries [60]. Furthermore, the problem will probably get worse as a result of the irrational use of antibiotics in health facilities and the accessibility of antibacterial drugs over the counter in many developing countries [61]. Nevertheless, our findings indicated a higher prevalence of VRSA strains within the country, revealing a more concerning level of S. aureus resistance to vancomycin than initially estimated or anticipated. The discrepancy in these estimations could be attributed to several factors. Firstly, the absence of a molecular approach for vancomycin resistance detection in almost all studies conducted in Ethiopia has contributed to an inadequate global report. Additionally, the absence of a national genomic repository in the country further complicates the situation. Moreover, a significant number of these studies did not adhere to specific guidelines, such as the recommendations provided by the Centers for Disease Control and Prevention, resulting in incomplete adherence to standardized protocols [62].
This significantly high pooled prevalence of VRSA in Ethiopia is indicative of the alarming widespread of multidrug-resistant S. aureus throughout all regions of the country. This finding, compounded with an escalated reports of MRSA in the country 10.94% [63], 32.5% [64], 47% [17] and 50.0% [65], necessitate urgent improvements to the national treatment guidelines to incorporate alternative, highly effective antimicrobial agents targeting MRSA. Simultaneously, the implementation of comprehensive antimicrobial stewardship strategies, accompanied by robust systemic surveillance, is imperative. Additionally, to curb the transmission of VRSA, it is essential to prioritize infection control measures such as contact precautions, meticulous screening, proper sterilization of healthcare equipment, and ensuring a sanitized environment [66].
Besides, this study revealed high level of heterogeneity (I2 = 93.0%, p < 0.001) depicting the presence of variations among included studies. The likely reason for this immense heterogeneity could be variations in methodology, study participants, study design and sample size all of which exert an influence on the prevalence of VRSA. A key contributor to this heterogeneity is the diversity of the target population, encompassing a range of individuals such as healthy food handlers, wound patients, children, healthcare professionals, burn patients, and individuals with diverse underlying medical conditions. Notably, surgical wound and burn patients are particularly prone to staphylococcal infections due to the loss of their skin’s protective barrier and the immunosuppression resulting from the systemic inflammatory response induced by the damaged tissue. This variety stemming from the diverse target population undoubtedly contributes to the elevated level of heterogeneity observed in this study.
Due to the diverse nature of the included studies, we anticipated heterogeneity and considered subgroup analysis in terms of region, city, publication year, study design, specimen type and AST method. In the subgroup analysis, we reported a moderate increment in the pooled prevalence of VRSA from the period ≤ 2014 (20.30%) to 2015–2017 (21.01%). This finding is in line with previous report of global meta-analysis, which reported a rise in the pooled prevalence of VRSA from the period < 2006 (2%), 2006–2014 (5%), 2015–2020 (7%) [15]. In addition, similar finding was revealed in a global study depicting a twofold upsurge in pooled prevalence of VRSA from 1.2% in studies conducted before 2010 to 2.4% in studies conducted after 2010 [16]. Nevertheless, our finding revealed a decline in pooled prevalence in the latest publication years. The reason for such discrepancy could be due to variations in the number of included studies across the categorized years. The recent subgroup periods 2018–2020 and ≥ 2021 comprised of fewer number of studies, which could be due to a shift in healthcare priorities to SARS-CoV-2 (COVID-19) pandemic response, and thus the number of studies may have decreased in these periods, causing the findings in these periods to be underestimated.
Region-based pooled prevalence was also estimated. The highest pooled prevalence of 47.74% (95% CI: 17.79, 77.69) was depicted in Sidama region, which is about three-times higher than Amhara region 14.82% (95% CI: 8.68, 19.88), four-times higher than the central (Addis Ababa) region 11.25% (955CI: 5.78, 16.72), and six-times higher than the least pooled prevalence from Oromia region 8.07% (95% CI: 4.09, 12.06). This regional variation could be attributable to differences in the study population, study period and antimicrobial susceptibility testing method and type of clinical sample used to isolate VRSA. Although such highest pooled prevalence in some regions and cities were mainly attributed to the use of disk diffusion technique of VRSA detection, the magnitude is still high and need further evaluation and genomic confirmation.
In the accurate diagnosis of VRSA, the role of clinical laboratory is critical for detecting, isolating and determining the antimicrobial susceptibility pattern [67]. In this regard, various techniques can be used to determine the resistance or susceptibility of S. aureus against vancomycin. In this study, the VRSA rates were significantly different based on AST methods. The pooled prevalence of VRSA using disk diffusion AST method (18.41%) is higher than the MIC-based methods (6.3%). This finding clearly showed that disk diffusion AST method overestimates the VRSA prevalence. Disk diffusion technique is not a reliable method as it showed poor sensitivity in differentiating the wild type isolates from isolates with non-vanA-inferred glycopeptide resistance [68, 69]. The MIC test technique of detecting vancomycin resistance, which include E-test and broth dilution tests, is considered a gold standard technique. However, these methods are not commonly being used in clinical laboratories of developing countries due to the fact that they are time-consuming, costly, labor intensive, and technically difficult. Consequently, clinical laboratories in developing countries are still using disk diffusion method to detect VRSA and this might result in overestimation of VRSA. Despite the incredibly high overall prevalence of 14.52% from all pooled studies, the pooled prevalence from studies using correct VRSA detection methods (MIC-based methods) was 6.3%, which is still high and a cause of national concern. This finding showed that there is an urgent need to improve the methods to determine vancomycin resistance in Ethiopia, and efforts should be directed at improving this nationally. In this sense, the incorporation of MIC-based methods for VRSA detection in routine clinical laboratory tests in Ethiopia is of paramount significance to show the real burden, and should be given due attention.
The methodology employed in a study plays a crucial role in accurately assessing the burden of a pathogen. Our meta-regression analysis identified the total number of S. aureus isolates as a significant factor contributing to heterogeneity among studies (p = 0.033) while publication year was not found to be a significant cause. It is common for prevalence studies conducted in developing countries, including Ethiopia, to involve a limited number of study participants, primarily due to financial and funding constraints. Consequently, this leads to a small number of bacterial isolates and may contribute to the observed heterogeneity among studies [70]. However, this finding contradicts a global report [16] that identified publication year as a source of heterogeneity.
One of the notable strengths of this study is its comprehensive nature, being the first of its kind to conduct a thorough analysis of VRSA within Ethiopia. It encompasses a wide range of studies conducted across multiple regions and cities of the country, providing a robust overview. Furthermore, the study included various studies done in different target populations using diversified clinical specimens in order to show the clear picture of VRSA in the country. However, the results should be interpreted with caution as the reviewed studies were highly heterogeneous in terms of VRSA magnitude, study setups, study participants, outcomes, diseases conditions, clinical specimens, sample sizes and AST methods, which collectively might introduce bias and have effect on result interpretation. Therefore, to account for this heterogeneity, the random-effects model of DerSimonian and Laird analysis was implemented in the meta-analysis. However, it should be taken into consideration that the DerSimonian-Laird (DSL) estimation method may have limitations when applied to estimate prevalence in studies with small sample sizes, and have shortcomings of being influenced by the number of included studies for meta-analysis and heavily biased when it is applied to proportions [71]. Moreover, subgroup analyses, sensitivity analysis, and meta-regression were conducted to further address and mitigate the impact of heterogeneity on the findings.
Conclusions
This systematic review and meta-analysis showed an alarmingly high pooled prevalence of VRSA which raises significant concerns for public health. The high burden of VRSA emphasizes the urgency of implementing routine screening practices and ensuring the appropriate utilization of antibiotics for effective management of MRSA infections. Mainly attributable to the overestimation of VRSA burden while using disk diffusion method, there is an urgent need to improve the methods to determine vancomycin resistance in Ethiopia and incorporate MIC-based VRSA detection methods in routine clinical laboratory tests, and efforts should be directed at improving it nationally. Furthermore, it serves as a clear call to action for the development and implementation of robust infection prevention measures and antimicrobial stewardship programs aimed at curbing the emergence and spread of drug resistance in Staphylococcal infections.
Data Availability
All relevant data are included in the manuscript and its supplementary data.
Abbreviations
- AMR:
-
Antimicrobial resistance
- CI:
-
Confidence interval
- CLSI:
-
Clinical Laboratory Standards Institute
- CSF:
-
Cerebrospinal fluid
- MDR:
-
Multidrug resistance
- MIC:
-
Minimum inhibitory concentration
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- NICU:
-
Neonatal and intensive care unit
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- Stata:
-
Statistics and data
- VRSA:
-
Vancomycin-resistant Staphylococcus aureus
- WHO:
-
World Health Organization
References
Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D, et al. Ready for a world without antibiotics? The Pensieres Antibiotic Resistance Call to Action. Antimicrob Resist Infect Control. 2012;1(1):11.
Naghav M, Collaborators AR. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55.
Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547–69.
Mlynarczyk-Bonikowska B, Kowalewski C, Krolak-Ulinska A, Marusza W. Molecular mechanisms of drug resistance in Staphylococcus aureus. Int J Mol Sci. 2022;23(15).
Sorrell TC, Packham DR, Shanker S. Vancomycin Therapy for Methicillin-Resistant Staphylococcus aureus. Ann Intern Med. 1982;97(3):344–50.
Cong Y, Yang S, Rao X. Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J Adv Res. 2020;21:169–76.
McGuinness WA, Malachowa N, DeLeo FR. Vancomycin Resistance in Staphylococcus aureus. YALE J BIOLOGY Med. 2017;90:269–81.
Choo EJ, Chambers HF. Treatment of Methicillin-Resistant Staphylococcus aureus Bacteremia. Infect Chemother. 2016;48(4):267–73.
Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, et al. Dissemination in japanese hospitals of strains of < em > Staphylococcus aureus heterogeneously resistant to vancomycin. The Lancet. 1997;350(9092):1670–3.
WHO. World Health Organization WHO published list of bacteria for which new antibiotics are urgently needed. 2017.
Perichon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(11):4580–7.
Zeng D, Debabov D, Hartsell TL, Cano RJ, Adams S, Schuyler JA, et al. Approved glycopeptide antibacterial drugs: mechanism of action and resistance. Cold Spring Harb Perspect Med. 2016;6(12).
Gardete S, Tomasz A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J Clin Invest. 2014;124(7):2836–40.
Tenover FC, Biddle BW, Lancaster MV. Increasing resistance to Vancomycin and other glycopeptides in Staphylococcus aureus. Emerg Infect Dis. 2001;7(2).
Wu Q, Sabokroo N, Wang Y, Hashemian M, Karamollahi S, Kouhsari E. Systematic review and meta-analysis of the epidemiology of vancomycin-resistance Staphylococcus aureus isolates. Antimicrob Resist Infect Control. 2021;10(1):101.
Shariati A, Dadashi M, Moghadam MT, van Belkum A, Yaslianifard S, Darban-Sarokhalil D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10(1):12689.
Berhe DF, Beyene GT, Seyoum B, Gebre M, Haile K, Tsegaye M, et al. Prevalence of antimicrobial resistance and its clinical implications in Ethiopia: a systematic review. Antimicrob Resist Infect Control. 2021;10(1):168.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71.
Munn Z, Moola S, Riitano D, Lisy K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int J Health Policy Manag. 2014;3(3):123–8.
Higgins JPTS, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Duval STR. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–63.
Egger MSG, Phillips AN. Meta-analysis: principles and procedures. Br Med J. 1997;315:1533–7.
Alebachew TY, Derabe G, Sisay A. Staphylococcus aureus burn wound infection among patients attending Yekatit 12 Hospital burn unit, Addis Ababa, Ethiopia. Ethiop J Health Sci. 2012;22(3):209–13.
Tadesse S. Antimicrobial resistance profile of Staphylococcus aureus isolated from clinical specimens and nasal swabs of patients at Tikur Anbessa Specialized Hospital. Addis Ababa University Research Repository; 2014.
Negussie AM, Bedrub G, Alia A, Shimelesc I, Lemab D, T. and, Aseffa A. Bacteriological Profile and Antimicrobial susceptibility pattern of Blood Culture isolates among Septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709–17.
Dilnessa T, Bitew A. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus isolated from clinical samples at Yekatit 12 Hospital Medical College, Addis Ababa, Ethiopia. BMC Infect Dis. 2016;16:398.
Dilnessa T, Biew A. Antimicrobial susceptibility pattern of Staphylococcus aureus with emphasize on Methicilin Resistance with Patients postoperative and wound infections at Yekatit 12 Hospital Medical College in Ethiopia. Am J Clin Experimental Med. 2016;4(1).
Atlaw A, Kebede HB, Abdela AA, Woldeamanuel Y. Bacterial isolates from diabetic foot ulcers and their antimicrobial resistance profile from selected hospitals in Addis Ababa, Ethiopia. Front Endocrinol (Lausanne). 2022;13:987487.
Gebremariam NM, Bitew A, Tsige E, Woldesenbet D, Tola MA. A high level of Antimicrobial Resistance in Gram-Positive Cocci isolates from different clinical samples among patients referred to Arsho Advanced Medical Laboratory, Addis Ababa, Ethiopia. Infect Drug Resist. 2022;15:4203–12.
Kahsay AM, Abebe A, T.; and, Andualem T. Isolation and antimicrobial susceptibility pattern of Staphylococcus aureus in patients with surgical site infection at Debre Markos Referral Hospital, Amhara Region, Ethiopia. Archives of Public Health. 2014;72(16):1–7.
Shibabaw A, Abebe T, Mihret A. Antimicrobial susceptibility pattern of nasal Staphylococcus aureus among Dessie Referral Hospital health care workers, Dessie, Northeast Ethiopia. Int J Infect Dis. 2014;25:22–5.
Denboba AA, Abejew AA. Antibiotic-resistant Bacteria are major threats of Otitis Media in Wollo Area, northeastern Ethiopia: a ten-year retrospective analysis. Int J Microbiol. 2016;2016:8724671.
Abebe M, Tadesse S, Meseret G, Derbie A. Type of bacterial isolates and antimicrobial resistance profile from different clinical samples at a Referral Hospital, Northwest Ethiopia: five years data analysis. BMC Res Notes. 2019;12(1):568.
Gobena A. Proportion of bloodstream infection, bacterial profile and their antimicrobial susceptibility patterns among pediatric patients at Felege Hiwot Referral Hospital, Bahir Dar, North west Ethiopia. Bahir Dar University Research Repository; 2019.
Abosse S, Genet C, Derbie A. Antimicrobial Resistance Profile of Bacterial isolates identified from Surgical Site infections at a Referral Hospital, Northwest Ethiopia. Ethiop J Health Sci. 2021;31(3):635–44.
Tefera S, Awoke T, Mekonnen D. Methicillin and Vancomycin Resistant Staphylococcus aureus and Associated factors from Surgical Ward Inpatients at Debre Markos Referral Hospital, Northwest Ethiopia. Infect Drug Resist. 2021;14:3053–62.
Jemal M, Tinshku F, Nigussie Y, Kefyalew B, Alemu C, Belay M, et al. Trend Analysis of Multidrug-Resistant bacterial pathogens causing neonatal Sepsis at University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia: a retrospective study. Int J Microbiol. 2021;2021:9992994.
Getaneh A, Ayalew G, Belete D, Jemal M, Biset S. Bacterial etiologies of ear infection and their Antimicrobial susceptibility pattern at the University of Gondar Comprehensive Specialized Hospital, Gondar, Northwest Ethiopia: a six-year retrospective study. Infect Drug Resist. 2021;14:4313–22.
Abebe W, Tegene B, Feleke T, Sharew B. Bacterial bloodstream infections and their antimicrobial susceptibility patterns in children and adults in Ethiopia: a 6-year retrospective study. Clin Lab. 2021;67(11).
Abrha A, Abdissa A, Beyene G, Getahun G, Girma T. Bacteraemia among severely malnourished children in Jimma University Hospital, Ethiopia. Ethiop J Health Sci. 2011;21(3):175–82.
Wubishet BL, Sabe ZS, Alemu HM, Z/Mariam FD. Antibiotic susceptibility pattern of Staphylococcus aureus strains from patients in Ethiopia. IJPSR. 2012;3(12):4889–94.
Kejela T, Bacha K. Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus aureus (MRSA) among primary school children and prisoners in Jimma Town, Southwest Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12(11):1–11.
Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12(17):1–7.
Tesfaye T, Beyene G, Gelaw Y, Bekele S, Saravanan M. Bacterial Profile and Antimicrobial susceptibility pattern of external ocular infections in Jimma University Specialized Hospital, Southwest Ethiopia. Am J Infect Dis Microbiol. 2013;1(1):13–20.
Beyene G, Mamo G, Kassa T, Tasew G, Mereta ST. Nasal and Hand Carriage Rate of Staphylococcus aureus among Food Handlers Working in Jimma Town, Southwest Ethiopia. Ethiop J Health Sci. 2019;29(5):605–12.
Sorsa A, Fruh J, Stotter L, Abdissa S. Blood culture result profile and antimicrobial resistance pattern: a report from neonatal intensive care unit (NICU), Asella teaching and referral hospital, Asella, south East Ethiopia. Antimicrob Resist Infect Control. 2019;8:42.
Kejela T, Dekosa F. High prevalence of MRSA and VRSA among inpatients of Mettu Karl Referral Hospital, Southwest Ethiopia. Trop Med Int Health. 2022;27(8):735–41.
Deresse D. Antibiotic-resistant Staphylococcus aureus isolated from mobile phone and hands of Health care workers in the Hawassa referral hospital, South Ethiopia. J Microbiol Antimicrobials. 2014;6(4):72–8.
Meseret G, Kassaye A, Yared M. Bacteria from infected surgical wounds and their antimicrobial resistance in Hawassa University Referral Teaching Hospital, Southern Ethiopia. Afr J Microbiol Res. 2014;8(11):1118–24.
Deyno S, Toma A, Worku M, Bekele M. Antimicrobial resistance profile of staphylococcus aureus isolates isolated from ear discharges of patients at University of Hawassa comprehensive specialized hospital. BMC Pharmacol Toxicol. 2017;18(1):35.
Mechal T, Hussen S, Desta M. Bacterial Profile, Antibiotic Susceptibility Pattern and Associated factors among patients attending adult OPD at Hawassa University Comprehensive Specialized Hospital, Hawassa, Ethiopia. Infect Drug Resist. 2021;14:99–110.
Abebe T, Teklemariam Z, Shume T, Mekuria S, Urgesa K, Weldegebreal F. Bacterial Profile of External Ocular Infections, its Associated factors, and Antimicrobial susceptibility pattern among patients attending Karamara Hospital, Jigjiga, Eastern Ethiopia. Int J Microbiol. 2023;2023:8961755.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. Springerplus. 2015;4:314.
Zhang S, Sun X, Chang W, Dai Y, Ma X. Systematic review and meta-analysis of the epidemiology of Vancomycin-intermediate and heterogeneous vancomycin intermediate Staphylococcus aureus isolates. PLoS One. 2015;10(8).
Kest H, Kaushik A. Vancomycin-resistant Staphylococcus aureus: formidable threat or silence before the storm? J Infect Dis Epidemiol. 2019;5(5).
Askari E, Zarifian A, Pourmand MR, Naderi-Nasab M. High-level vancomycin-resistant Staphylococcus aureus (VRSA) in Iran: a systematic review. JMed Bacteriol. 2012;3(4):53–61.
Rahimipour F, Ghazvini K, Youssefi M. Reports of vancomycin-resistant Staphylococcus aureus from Middle East Countries. Arch Clin Infect Dis. 2018;13(2).
Supa P, Karl P. Hygiene behaviour and health attitudes in african countries. Curr Opin Psychiatry. 2012;25(2):149–54.
Fraser JL, Mwatondo A, Alimi YH, Varma JK. Healthcare-associated outbreaks of bacterial infections in Africa, 2009–2018: a review. Int J Infect Dis. 2021;103:469–77.
Unni S, Siddiqui TJ, Bidaisee S. Reduced susceptibility and resistance to Vancomycin of Staphylococcus aureus: a review of global incidence patterns and related genetic mechanisms. Cureus. 2021;13(10):1–11.
CDC. Investigation and Control of Vancomycin- Resistant Staphylococcus aureus (VRSA): 2015 update. Division of Healthcare Quality Promotion Centers for Disease Control and Prevention Update; 2015.
Reta A, Mengist A, Tesfahun A. Nasal colonization of methicillin resistant Staphylococcus aureus in Ethiopia: a systematic review and meta-analysis. Ann Clin Microbiol Antimicrob. 2019;18:25.
Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. BMC Infect Dis. 2016;16(689):1–9.
Chelkeba L, Melaku T. Epidemiology of staphylococci species and their antimicrobial-resistance among patients with wound infection in Ethiopia: a systematic review and meta-analysis. J Global Antimicrob Resist. 2022;29(4):83–498.
Siegel J. Healthcare Infection Control Practices Advisory Committee 2007 Guideline for isolation precautions: preventing transmission of infectious agents in healthcare settings. 2007.
CDC. Centers for Disease Control Prevention reminds clinical laboratories and healthcare infection preventionists of their role in the search and containment of vancomycin-resistant Staphylococcus aureus (VRSA). 2012.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. 2020.
EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 10th edition. European Society of Clinical Microbiology and Infectious Diseases Basel; 2020.
IntHouta J, Ioannidis JPA, Borma GF, Goeman JJ. Small studies are more heterogeneous than large ones: a meta-meta-analysis. J Clin Epidemiol. 2015;68:860e9.
Jackson DWI. When should meta-analysis avoid making hidden normality assumptions? Biom J. 2018;60:1040–58.
Acknowledgements
Not applicable.
Funding
This study did not receive any specific grant from funding agencies.
Author information
Authors and Affiliations
Contributions
M.A.B. conceived and designed the study. M.A.B., A.G., E.A., and M.T. participated in article search, and data extraction. M.A.B., A.G., H.D., and D.G. conduct a quality assessment of the included studies and perform the statistical analysis and interpretation of the data. M.A.B. drafts manuscript. M.A.B., A.G., O.M. and E.A. check the validity and monitor the overall process. M.T., D.G., H.E., O.M., and H.D. critically reviewed the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Belete, M.A., Gedefie, A., Alemayehu, E. et al. The prevalence of vancomycin-resistant Staphylococcus aureus in Ethiopia: a systematic review and meta-analysis. Antimicrob Resist Infect Control 12, 86 (2023). https://doi.org/10.1186/s13756-023-01291-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-023-01291-3