Abstract
Background
Epidemiological studies suggested that determinants for antibiotic resistance have originated in aquaculture. Recently, the integrated agriculture-aquaculture system has been implemented, where fish are raised in ponds that receive agriculture drainage water. The present study aims to investigate the occurrence of β-lactamase and carbapenemase-producing Enterobacteriaceae in the integrated agriculture-aquaculture and the consequent public health implication.
Methods
Samples were collected from fish, fishpond water inlets, tap water, outlet water, and workers at sites of integrated agriculture-aquacultures. Samples were also taken from inhabitants of the aquaculture surrounding areas. All samples were cultured on MacConkey agar, the Enterobacteriaceae isolates were tested for susceptibility to cephalosporins and carbapenems, and screened for blaCTX-M-15, blaSHV, blaOXA-1, blaTEM, blaPER-1, blaKPC, blaOXA-48, and blaNDM. Strains having similar resistance phenotype and genotype were examined for the presence of Incompatible (Inc) plasmids.
Results
A major proportion of the Enterobacteriaceae isolates were resistant to cephalosporins and carbapenems. Among the 66 isolates from fish, 34 were resistant to both cephalosporin and carbapenem groups, 26 to carbapenems alone, and 4 to cephalosporins alone. Of the 15 isolates from fishpond water inlets, 8 showed resistance to both groups, 1 to carbapenems alone, and 5 to cephalosporins alone. Out of the 33 isolates from tap water, 17 were resistant to both groups, and 16 to cephalosporins alone. Similarly, of the 16 outlet water isolates, 10 were resistant to both groups, and 6 to cephalosporins alone. Furthermore, of the 30 examined workers, 15 carried Enterobacteriaceae resistant strains, 10 to both groups, and 5 to cephalosporins alone. Similar strains were isolated from the inhabitants of the aquaculture surrounding areas. Irrespective of source of samples, strains resistant to all examined antibiotics, carried predominantly the carbapenemase gene blaKPC either alone or with the β-lactamase genes (blaCTX-M-15, blaSHV, blaTEM, and blaPER-1). The isolates from fish, water, and workers harboured a wide-range of multi-drug-resistance Inc. plasmids, which were similar among all isolates.
Conclusion
The present findings suggest transmission of the resistance genes among Enterobacteriaceae strains from different sources. This reiterates the need for control strategies that focus on humans, animals, water, and sewage systems to solve the antibiotic resistance problem.
Similar content being viewed by others
Introduction
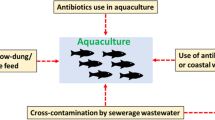
A link between aquaculture and the development of antibiotic resistance has been demonstrated by an increasing body of research [1, 2]. Worldwide, there is a massive increase in fish farming, which is associated with intensive use of antibiotics to combat bacterial infections [3]. Other potential explanations are contamination of aquaculture with antibiotic residues and antibiotic resistant bacteria coming from animals through using the so-called livestock integrated aquaculture system [4,5,6]. Recently, an integrated agriculture-aquaculture system has been implemented to save water resources [7]. In such system, fish are reared in ponds receiving water from crop farms through irrigation canals.
The major concern with the use of antibiotics in aquaculture is that fish do not effectively metabolize antibiotics and pass them largely unused in faeces [8]. This encourages the development of antibiotic resistance in bacteria present in fish and the surrounding environment [1, 2, 9]. Fishponds are not subject to frequent water exchange, resulting in accumulation of antibiotic-residues and -resistant bacteria, which are then disseminated at harvest time.
Fish bacteria particularly Enterobacteriaceae can exchange antibiotic resistance genes with human and animal bacteria [10, 11]. Among these, β-lactam resistant bacteria are of great concern, as they are becoming non-susceptible to nearly all available antibiotics [12, 13]. They include extended-spectrum β-lactam (ESBL) and carbapenem-resistant (CRE) Enterobacteriaceae. ESBL carry a broad spectrum of β-lactamase enzymes that hydrolyse a wide range of penicillin and cephalosporin antibiotics, but not carbapenems [14]. However, CRE carry carbapenemase enzymes that confer resistance to nearly all β-lactam antibiotics, including carbapenems [15]. The β-lactamase enzymes encompass members of the TEM and SHV families and other groups, such as CTX-M, OXA, and PER β-lactamases [16]. The carbapenemase enzymes are a diverse group of β-lactamases, the most remarkable of which are the big five enzymes KPC, NDM, OXA-48, IMP, and VIM [17]. There is strong evidence that the β-lactamase and carbapenemase encoding genes are found on plasmids that facilitate their transfer among bacteria of different genera and kingdoms [15, 16]. Plasmids are extra-chromosomal, self-replicating genetic elements, carried by the bacteria. Plasmids having the same origin of replication cannot stably co-reside within the same bacterial cells, are known as incompatible (Inc) [18]. The Inc. plasmids can positively select for the presence of resistance genes [19]. Identification of the Inc. plasmids in the bacterial isolates helps to trace the transfer of the resistance genes. Currently, a PCR method is accepted to classify the Inc. resistance plasmids based on their type of replicon [20]. Thirty replicons are now recognised, which represent the major replicase genes of the Inc. plasmids circulating among Enterobacteriaceae, including HI1, HI2, I1, I2, X1, X2, X3, X4, L, M, N, FIA, FIB, FIC, FII, FIIS, FIIK, FIB-KN, FIB-KQ, W, Y, P1, A/C, T, K, U, R, B/O, HIB-M, and FIB-M [21].
Recently in Egypt, small scale use of the integrated agriculture-aquaculture system began to be used. The aims of the present study are, therefore, to investigate the extent of antibiotic resistance in aquacultures integrated with agriculture in Egypt and its public health significance. Accordingly, this was performed through (i) assessing the prevalence of β-lactamase (βLPE) and carbapenemase-producing Enterobacteriaceae (CPE) in the water inlets of aquaculture originating from agriculture and supply fishponds, in fish, tap water, and the outlet water of the aquaculture; (ii) determination of the occurrence of βLPE and CPE in aquaculture workers and inhabitants of the aquaculture surrounding areas, those having a history of eating fish from these aquaculture and also use the same tap water source as the aquaculture (iii) identifying the presence and type of Inc. plasmids carried by the resistant strains isolated from different sources.
Materials and methods
Sample collection
Four integrated agriculture-aquaculture farms were randomly selected from the Giza governorate, the third-largest city in Egypt. The selected farms are located near each other (approx. 5 km away from each other), they combine agriculture with fish farming As illustrated in Fig. 1, the fish are raised in ponds, each fishpond receives water from agriculture drainage water through a separate water pipe (Water inlet). Tap water is another source of water for the aquaculture, which is used to dissolve medicines given to the fish and sometimes as a supply for the fishponds in case of shortage of agriculture water. Tap water is also used by the workers for drinking and hand washing. Outlet water represents the waste drained from fishponds through pipes.
Samples were aseptically collected from internal organs (liver, spleen, and kidney) from 105 apparently healthy fresh Tilapia fish (Oreochromis niloticus). Additionally, samples were collected from fishpond water inlets (n = 30), tap water (n = 44), and outlet water (n = 26). The water samples were placed in sterile glass bottles containing sodium thiosulphate and were filtered through 0.4 μm pore size nitrocellulose filters (Sartorius, Aubagne, France) placed on tubes containing peptone broth [22].
Faecal and hand swab samples were collected from humans working in the aquaculture farms (n = 30), having contact with the fish, and who agreed to participate in the study.
Additionally, faecal samples were collected from inhabitants of the aquaculture surrounding areas (n = 45), who use the same source of tap water as the aquacultures and were admitted to the local medical centre with diarrhoea after eating fish from the tested aquacultures.
Bacterial isolation and identification
All samples were inoculated into Trypticase soy broth tubes and were incubated at 37 °C for 24 h. A loop-ful from the previously incubated tubes were streaked onto MacConkey agar plates (Oxoid Ltd., Cairo, Egypt) and Eosin methylene blue agar (EMB) (Oxoid) and were incubated aerobically at 37 °C for 24 h. Suspected colonies were purified through subculture on MacConkey agar plates and were examined for morphology and phenotype traits according to Collee et al. [23].
Biochemical identification
Pure colonies were subjected to API 20E kits (BioMerieux, Marcy-l’Étoile, France) according to the manufacturer’s instructions. The accuracy of API 20E results was 100% as determined by API web software (BioMérieux).
Antibiotic susceptibility test
Confirmed strains of Enterobacteriaceae were examined for susceptibility to different antibiotics (Oxoid) using the disk diffusion test on Mueller-Hinton agar (Oxoid). The antibiotics included were cephalosporins [Cefoxitin (FOX, 30 μg), Ceftazidime (CAZ, 30 μg) Cefotaxime (CTX, 30 μg), Ceftriaxone (CRO, 30 μg)] and the carbapenems [Imipenem (IPM, 10 μg), Meropenem (MEM, 10 μg), Ertapenem (ETP, 10 μg)] groups. The results were interpreted according to the clinical breakpoints recommended by CLSI (Clinical laboratory Standard Institute) [24].
Based on CDC definitions [25], Enterobacteriaceae isolates resistant to at least one of the carbapenem drugs (Imipenem, Meropenem, Ertapenem, or Doripenem) or produce a carbapenemase gene are called carbapenem-resistant Enterobacteriaceae (CRE). Accordingly, in the present study, isolates resistant to at least one of the tested carbapenem drugs were considered CRE and were tested for the presence of carbapenemase genes.
PCR for detection of β-lactamase- and carbapenemase- encoding genes
Genomic DNA from resistant isolates was extracted using a QIAamp® mini kit (Qiagen, Hombrechtikon, Switzerland). PCR was performed to identify β-lactamase-encoding genes (blaCTX-M-15, blaSHV, blaTEM, blaPER-1) using specific oligonucleotide primers as previously described [26]. The PCR for blaOXA-1 was done using specific oligonucleotide primers according to [27].
Multiplex PCR [28] was performed to identify carbapenemase-encoding genes (blaKPC,blaOXA-48, and blaNDM) using specific oligonucleotide primers [29]. The three genes were chosen to include one representative candidate from each of the three carbapenemase groups (Serine, OXA, and MLBs) and the selection was based on previous findings in Egypt [28]. The sequences of the primers used in the present study are shown in S1.
Characterisation of Inc. plasmids present in the resistant isolates using PCR-based-replicon-typing
Genomic DNA from the isolates that were resistant to all tested antibiotics and showed similar resistance genotypes (n = 30) were subjected to plasmid typing using the PBRT 2.0 kit (PCR based replicon typing Ver.01/06/2017, Diatheva, Fano, Italy). The kit contains eight multiplex PCR, each of which amplifies three to four targets, allowing detection of a total of 30 replicons. It also includes positive control plasmids for the 8 multiplex PCR. The kit was used following the manufacturer’s instructions. Interpretation of the results was performed according to the manufacture instruction with IncF plasmids designated as multi-replicons when they showed a combination of FII replicons with FIA or FIB [30, 31].
Statistical analysis
PASW Version 18 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Descriptive statistics were run and showed that the data are normally distributed. A Chi square (X2) test was used to determine whether there is a difference in the prevalence of the isolated Enterobacteriaceae strains and type of samples (fish, fishpond water inlets, tap water, and outlet water) (Table 1). Chi-square (X2) test was also performed to examine whether the prevalence of antibiotic resistance is influenced by the type of samples or species of bacteria, or both combined. On each occasion, the significance of differences was set at p value < 0.05.
Results
Prevalence of Enterobacteriaceae in apparently healthy Nile tilapia fish and water from integrated agriculture-aquacultures
As demonstrated in Table 1, of the 105 tested apparently healthy Nile tilapia fish, 66 (63%) showed the presence of Enterobacteriaceae. Additionally, the occurrence of Enterobacteriaceae was also high among the collected samples from fishpond water inlets (15/30; 50%), tap water (33/44; 75%), and outlet water (16/26; 62%) from the aquacultures. E. coli represented the major species of Enterobacteriaceae isolated from fish and water. Enterobacter cloacae complex (ECC) and Klebsiella (K.) pneumoniae were also detected, but with a significantly lower prevalence than E. coli.
Occurrence of cephalosporin- and carbapenem-resistant Enterobacteriaceae in the integrated agriculture-aquacultures
The Enterobacteriaceae isolates from fish and water samples of the aquacultures were screened for resistance against carbapenems (CRE) and cephalosporins (CEPH). The resistant strains were examined for carbapenemase and β-lactamase genes (Figs. 2,3, 4).
Number of carbapenem- and cephalosporin- resistant Enterobacteriaceae isolates among fish. The Enterobacteriaceae isolates from fish (n = 66), including E. coli (n = 45), ECC (n = 12), and K. pneumoniae (n = 9) were screened for resistance against carbapenems (CRE) and cephalosporins (CEPH: FOX, CAZ, CTX, and CRO). The strains are grouped according to their resistance phenotypes as follow: CRE and all CEPH (isolates resistant to at least one of the tested carbapenems and all tested cephalosporins); CRE and 2 CEPH (isolates resistant to at least one of the tested carbapenems and two of the tested cephalosporins); CEPH (isolates resistant to 2 or more of the tested cephalosporins but not to carbapenems); CRE (isolates resistant to at least one of the tested carbapenems but not to any of the tested cephalosporins). Each phenotype is marked with colour and its resistance genotypes are also provided; carbapenemase (blaKPC, blaOXA-48, and blaNDM) and β-lactamase (blaCTX-M-15, blaSHV, blaOXA-1, blaTEM, and blaPER-1) genes. (n) represents numbers of resistant strains
Number of carbapenem- and cephalosporin- resistant Enterobacteriaceae isolates among fishpond water inlets and tap water. The Enterobacteriaceae isolates from a fishpond water inlets (n = 14), including E. coli (n = 9), ECC (n = 3), and K. pneumoniae (n = 2), as well as from b tap water (n = 33), including E. coli (n = 22), ECC (n = 6), and K. pneumoniae (n = 5) were screened for resistance against carbapenems (CRE) and cephalosporins (CEPH: FOX, CAZ, CTX, and CRO). The strains are grouped according to their resistance phenotypes as follow: CRE and all CEPH (isolates resistant to at least one of the tested carbapenems and all tested cephalosporins); CRE and 2 CEPH (isolates resistant to at least one of the tested carbapenems and two of the tested cephalosporins); CEPH (isolates resistant to 2 or more of the tested cephalosporins but not to carbapenems); CRE (isolates resistant to at least one of the tested carbapenems but not to any of the tested cephalosporins). Each phenotype is marked with colour and its resistance genotypes are also provided; carbapenemase (blaKPC, blaOXA-48, and blaNDM) and β-lactamase (blaCTX-M-15, blaSHV, blaOXA-1, blaTEM, and blaPER-1) genes. (n) represents numbers of resistant strains
Number of carbapenem- and cephalosporin- resistant Enterobacteriaceae isolates among outlet water. The Enterobacteriaceae isolates from outlet water (n = 16), including E. coli (n = 12), ECC (n = 2), and K. pneumoniae (n = 2) were screened for resistance against carbapenems (CRE) and cephalosporins (CEPH: FOX, CAZ, CTX, and CRO). The strains are grouped according to their resistance phenotypes as follow: CRE and all CEPH (isolates resistant to at least one of the tested carbapenems and all tested cephalosporins); CRE and 2 CEPH (isolates resistant to at least one of the tested carbapenems and two of the tested cephalosporins); CEPH (isolates resistant to 2 or more of the tested cephalosporins but not to carbapenems). Each phenotype is marked with colour and its resistance genotypes are also provided; carbapenemase (blaKPC, blaOXA-48, and blaNDM) and β-lactamase (blaCTX-M-15, blaSHV, blaOXA-1, blaTEM, and blaPER-1) genes. (n) represents numbers of resistant strains
The Chi-square test showed that resistance to carbapenems and CEPH was significantly influenced by the source of samples (fish or water), but not by species of bacteria (E. coli, ECC, and K. pneumoniae). Indeed, Figs. 2, 3, 4 demonstrated that the resistance phenotype was similar between the three species of bacteria.
As displayed in Figs. 2, 3, 4, the isolates were grouped according to the resistance phenotype into; CRE and all CEPH (isolates resistant to at least one of the tested carbapenems and all tested cephalosporins); CRE and 2 CEPH (isolates resistant to at least one of the tested carbapenems and two of the tested cephalosporins); CRE (isolates resistant to at least one of the tested carbapenems, but not to any of the tested cephalosporins); CEPH (isolates resistant to 2 or more of the tested cephalosporins, but not to carbapenems).
Of the 66 fish isolates, 64 (45 E. coli, 12 ECC, 7 K. pneumoniae) showed resistance as follow; 29 to carbapenems and all tested cephalosporins, 5 to carbapenems and two of the tested cephalosporins, 26 to carbapenems alone, and 4 to cephalosporins alone (Fig. 2). Additionally, of the 15 isolates from fishpond water inlets, 7 were resistant to carbapenems and all cephalosporins, 1 to carbapenems and two cephalosporins, 1 to carbapenems alone, and 5 to cephalosporins alone (Fig. 3a). All the 33 isolates from tap water were resistant. Among them, 17 were resistant to carbapenems and all cephalosporins, and 16 were resistant to cephalosporins alone (Fig. 3b). Similarly, the 16 outlet water isolates were resistant, 9 to carbapenems and all cephalosporins, 1 to carbapenems and two cephalosporins drugs, and 6 to cephalosporins alone (Fig. 4).
Irrespective of the source of samples or species of bacteria, we found that strains resistant to all examined antibiotics, carry predominantly blaKPC either alone or with the β-lactamase genes (blaCTX-M-15, blaSHV, blaTEM, blaOXA-1, and blaPER-1). However, strains resistant to carbapenems and two cephalosporins harbour only carbapenemase genes (blaKPC, blaOXA-48, blaNDM). Of the 26 fish isolates resistant only to carbapenems, 17 were carbapenemase producing, while 9 were negative to the three screened carbapenemase genes (Figs. 2, 3, 4).
High numbers of aquaculture workers carried cephalosporin- and carbapenem-resistant Enterobacteriaceae
Hand swab and faecal samples were collected from humans working in the integrated agriculture-aquacultures. A relatively high proportion of the workers showed the presence of Enterobacteriaceae in hand swabs (20 / 30) and faecal samples (15 / 30) (Fig. 5, Panel I). Like in fish and water isolates, E. coli was the predominant species isolated from the human samples (Panel I A and B). Among the 20 isolates from hand swabs, 5 were resistant to carbapenems and all cephalosporins, 2 to carbapenems and two of the tested cephalosporins, and 3 to cephalosporins alone (Panel I C). Of the 15 faecal isolates, 10 were resistant to carbapenems and all cephalosporins, and 5 to cephalosporins alone (Panel I D). Additionally, the resistance phenotype and genotype were similar among E. coli, ECC, and K. pneumoniae. Strains resistant to all examined antibiotics harboured predominantly blaKPC either with the other carbapenemase genes (blaNDM and blaOXA-48) or with the β-lactamase genes (blaCTX-M-15, blaSHV, blaTEM, and blaPER-1) (Panel I C and D).
Number of cephalosporins- and carbapenems-resistant Enterobacteriaceae isolates from aquaculture workers and inhabitants of aquaculture surrounding areas. Panel I: Hand swab and faecal samples from aquaculture workers (n = 30) were examined. a and b illustrate the number (n) and species of Enterobacteriaceae isolated from hand swabs and faecal samples, respectively. c and d illustrate the number (n) of resistant Enterobacteriaceae isolates from hand swabs and faecal samples, respectively. Panel II: Faecal samples from inhabitants (n = 45) were examined. e illustrates the number (n) and species of Enterobacteriaceae isolated. f shows the number (n) of resistant Enterobacteriaceae. The strains are grouped based on resistance against carbapenems (CRE) and cephalosporins (CEPH: FOX, CAZ, CTX, and CRO) as follow: CRE and all CEPH (isolates resistant to at least one of the tested carbapenems and all tested cephalosporins); CRE and 2 CEPH (isolates resistant to at least one of the tested carbapenems and two of the tested cephalosporins); CEPH (isolates resistant to 2 or more of the tested cephalosporins but not to carbapenems). Each phenotype is marked with colour and its resistance genotypes are also provided; carbapenemase (blaKPC, blaOXA-48, and blaNDM) and β-lactamase (blaCTX-M-15, blaSHV, blaOXA-1, blaTEM, and blaPER-1) genes. (n) represents numbers of resistant strains
Faecal samples were also collected from inhabitants of the aquaculture surrounding areas. Enterobacteriaceae was isolated from 15 of the examined 45 samples with a predominance of E. coli (Fig. 5, Panel II A). Of the 15 isolates, 6 were resistant to carbapenems and all cephalosporins, and 7 were resistant to cephalosporins alone (Fig. 5, Panel II B). Consistent with the results of the aquaculture workers, the strains from the inhabitants were resistant to carbapenems and all cephalosporins carried predominantly blaKPC with other carbapenemase genes (blaNDM,blaOXA-48) or with the β-lactamase genes (blaCTX-M-15, blaSHV, blaTEM, and blaPER-1) (Fig. 5, Panel II B).
Similar types of incompatible (Inc) plasmids were carried by the Enterobacteriaceae resistant strains isolated from different sources as determined by the PBRT-kits
CRE isolates that were resistant to all tested antibiotics and showed similar resistance genotypes were examined for the presence and type of Inc. plasmids using PCR-Based-Replicon-Typing (S2, Fig. 6).
Types of Incompatible (Inc) plasmids carried by the Enterobacteriaceae-resistant isolates. a illustrates the types of Inc. plasmids found in the examined CRE isolates (n = 30) that were resistant to all tested CEPH and showed similar genotypes. The red colour in the histogram of the IncF plasmid indicates number of isolates that harbour multi-replicon. b shows the differences between the CRE isolates producing-carbapenemase genes alone (CPE, n = 11) and those producing both carbapenamese and β-lactamase (CPE + βLPE, n = 19) genes, in the proportion and types of Inc. plasmids
There were no differences in the type of Inc. plasmids, based on the source of the isolates (fish, fishpond water inlets, tap water, outlet water, and workers) (S2). As demonstrated in Fig. 6a, of the 30 examined resistant strains, 29 showed the presence of the IncF and the IncHI groups of plasmids, 28 carried the IncX, 21 harboured the IncI complex group, 17 had the epidemic plasmid IncA/C, and 14 carried the wide-host range IncP and the IncW groups. Other Inc. plasmids found in minority of the isolates were as follows; IncN (n = 9), IncN2 (n = 7), IncR (n = 5), IncU (n = 4), and IncT (n = 4).
It is important to note that, the majority of the IncF plasmids present (22 / 29) were multi-replicon (Fig. 6a), carrying the FII with FIA and or FIB replicons (S2) [30, 31]. A divergence in FIB (FIB-KN and FIB-KQ) and FII (FIIS and FIIK) replicons was observed. The FIB-KN was mainly found in the K. pneumoniae isolates, while the FIB-KQ was present in K. pneumoniae and E. coli isolates. Moreover, the FIIS and FIIK divergences were found in E. coli, K. pneumoniae, and ECC isolates (S2).
Some differences were noticed in the type of Inc. plasmids between strains producing carbapenemase genes alone (CPE) and those producing both carbapenemase and β-lactamase genes (CPE + βLPE) (Fig. 6b). Among them, the IncI complex plasmid was found in the higher proportion of the CPE + βLPE strains (15/19) than the CPE strains (6/11). The IncI complex contains five groups IncI, IncL, IncM, IncK, and IncB/O combined, or at least two of them. Another difference was the presence of the IncW plasmid only in CPE + βLPE strains (14 /19).
Discussion
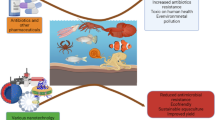
For the past 60 years, β-lactam antibiotics have been amongst the most successful drugs used for the treatment of bacterial infections in humans, animals, and fish [32,33,34]. In the current study, we investigated the occurrence of β-lactamase (βLPE)- and carbapenemase (CPE)- producing Enterobacteriaceae in fresh fish, fishpond water inlets, tap water, outlet water, and workers at four integrated agriculture-aquacultures. Additionally, faecal samples were collected from inhabitants of the aquaculture surrounding areas. The prevalence of Enterobacteriaceae isolated from apparently healthy fresh Nile Tilapia fish was 63%, which is higher than that reported by previous studies performed on fish raised in earthen pond in Egypt (44–53%) [35,36,37]. Like in fish, the aquaculture water showed a high prevalence of Enterobacteriaceae, particularly the tap water (75%) as compared to fishpond water inlets (50%) and outlet water (62%). Furthermore, of the 30 examined workers, 15 showed the presence of Enterobacteriaceae in the hand and faecal samples, whereas, 5 had Enterobacteriaceae only in the hand swabs. The species of Enterobacteriaceae isolated from fish, water samples and the workers included E. coli, ECC, and K. pneumoniae, with E. coli being the predominant species. Although the species of Enterobacteriaceae found could be fish pathogens [35,36,37,38,39,40], it is not the most common found bacteria in fish [2]. This together with the high prevalence of Enterobacteriaceae found in water, suggests faecal contamination in the aquaculture environment.
A high proportion of the Enterobacteriaceae isolates from the aquaculture showed resistance to the tested antibiotics, as found in fish (64 / 66), water inlets (14 / 15), tap water (33 / 33), outlet water (16 /16), and the workers (15 / 20). Regardless of the source of samples, a major number of these isolates were CRE, which encompass predominantly strains resistant to carbapenems and all tested cephalosporins, followed by strains resistant to carbapenems and some cephalosporins, while a few strains were resistant to carbapenems alone. The three phenotypes of CRE isolates had different resistance genotypes. However, CRE isolates resistant to all cephalosporins carried predominantly both carbapenemase and β-lactamase genes, CRE isolates resistant to some cephalosporins or to carbapenems alone harboured only carbapenemase genes. According to CLSI 2020, CRE isolates that produce one or more carbapenemase genes usually test resistant to one or more of cephalosporins, however, some CPE isolates can still test susceptible to cephalosporins [41]. This can be a possible explanation for our finding of CRE isolates which carry one or more of the carbapenemase genes (blaKPC, blaOXA-48, blaNDM), but susceptible to the tested cephalosporins (Figs. 2,3). In this regard, a study from South Africa also found some CPE-CRE isolates which were susceptible to cephalosporins [42].
It is important to note that among the 26 CRE fish isolates that showed resistance only to carbapenems, 9 isolates did not carry any of the tested carbapenemase genes, suggesting that they might harbour other carbapenemases which were not screened in the present study.
Enterobacteriaceae isolates resistant to cephalosporins alone were also found but in a lower proportion than the CRE. Slight differences were noticed between the four examined aquacultures, in the numbers of CRE and cephalosporins-resistant Enterobacteriaceae, which were not statistically different. Cephalosporins-resistant Enterobacteriaceae isolates have previously been found in fish and aquatic environments from different countries, including Egypt [40, 43,44,45,46,47,48]. However, a few recent studies showed the presence of CRE strains among fish [49, 50], indicating an increase in the prevalence of CRE in aquaculture. Irrespective of the species of bacteria or source of samples, the carbapenemase-producing isolates showed a predominance of blaKPC over the other two genes blaOXA-48 and blaNDM, which agrees with other study that detected the presence of blaKPC in seafood isolates [51]. Furthermore, all the isolates that were positive for β-lactamase genes harboured blaCTX-M-15 in combination with one or more of the other β-lactamase genes (blaOXA-1, blaSHV, blaTEM, and blaPER-1). This is consistent with findings that showed a predominance of blaCTX-M-15 among β-lactam-resistant Enterobacteriaceae isolated from wild fish in Algeria [44], from frozen Mackerel in Saudi Arabia [45], and water collected from fish farms in Egypt [46]. Moreover, the blaCTX-M was shown to be commonly associated with blaTEM on the same plasmid [52], and possibly with blaSHV and the broad spectrum blaOXA-1 [15, 51]. Our findings then raised the question of how such CRE strains have reached the aquacultures. The most likely explanation is a co-resistance to carbapenems by selective pressure from overuse of other antibiotics, for which resistance determinants are co-localised with carbapenemase on the same plasmid [53]. Another possible explanation might be unrestricted use of carbapenems to treat fish, which is consistent with our results that strains resistant only to carbapenems were found mainly among fish isolates. Unfortunately, this cannot be clarified, as no data is available on quantities and types of antibiotics used in aquaculture in Egypt. Furthermore, the resistant strains might access the aquaculture from agricultural sources, where animal manure is known to be used as fertilizers [6, 54, 55]. CRE has been isolated from dairy farms in Egypt [56]. Another possible source of the resistant strains could be tap water, which showed the presence of high amounts of CRE and cephalosporins-resistant Enterobacteriaceae, indicating faecal contamination. The main source of tap water in Egypt [57] is from surface water like rivers, lakes or canals. In many Egyptian governorates, sewage is usually discharged untreated into the surface water [58, 59]. Since sewage act as a pool of antibiotic residues as well as resistant bacteria from different sources [58, 59], this can result in contamination of tap water. This is supported by our results that showed the occurrence of Enterobacteriaceae in faecal samples from inhabitants of the aquaculture surrounding areas, which have similar resistance phenotype and genotype as the isolates from the aquaculture workers with a predominance of blaKPC and blaCTX-M-15. In this regard, previous studies reported preponderance of Enterobacteriaceae carrying blaKPC and blaCTX-M-15 among humans in Middle East [60] and North Africa [61, 62].
Characterisation of the Inc. plasmid types harboured by the CRE isolates that were resistant to all tested cephalosporins, revealed a similar profile among the isolates from fish, water, and workers. The major plasmid groups found were as follow; IncF, IncHI, IncX, IncI-complex, IncA/C, IncP, and IncW, which agrees with findings that IncF, IncHI, IncX, IncI-complex, and IncA/C are commonly associated with CRE isolated from humans and animals [19,20,21]. Crucially, the IncA/C is considered as an epidemic plasmid, as it has been detected in different countries and has a wide-host range of different sources [63]. IncP and IncW have been categorised among rarely reported plasmids [21, 64], however, we found them in a relatively high number of the isolates from different sources. Since these two plasmids have a broad-host range and can carry multiple resistance genes [64, 65], their presence in our isolates indicates a role in the spread of resistance genes among bacteria of different genera and sources. Other plasmids like IncN, IncN2, IncL, IncU, IncT, and IncR were also present, but in minority of the current isolates. This is consistent with [19], who stated that these plasmids are not often reported.
One of the interesting results is a predominance of the multi-replicon IncF among the tested CRE isolates, with divergence in FII (FIIS, FIIK) and FIB (FIB-KQ, FIB-KN), but not in FA (S2), suggesting that such plasmids are rapidly emerging with the subsequent increase in dissemination of resistance genes. IncF plasmid is unique, as it can carry many replicons, one replicon is strongly conserved for plasmid replication, while the others are free to diverge as a mechanism of evolution [19, 30]. Additionally, the IncF plasmid can co-integrate with other plasmids found in the current study, like IncI1, IncA/C, and IncN forming a mega-plasmid with multi-resistance regions [19, 31, 66, 67]. Taken together, the current CRE isolates carry a wide range of Inc. plasmids which are known to be associated with multi-drug resistance determinants. The profile of these plasmids was similar between isolates from fish, water, and humans working in contact with fish, suggesting a possible transfer among bacteria from different sources. To our knowledge, no data is available regarding the prevalent type of Inc. plasmids in Egypt.
Conclusion
The present study reveals the occurrence of CRE which are resistant to all tested cephalosporins among fish, water, and workers at aquacultures integrated with agriculture systems in Egypt. The fish included in the study were apparently healthy, indicating they could act as reservoirs for potentially pathogenic bacteria in humans. Such resistant strains might have occurred in the aquaculture due to unregulated use of antibiotics to treat fish, or through agricultural sources, faecal contaminated tap water, or the workers. The resistant strains carry a variety of resistance Inc. plasmids which are similar between the strains from different sources. Most of these plasmids have a broad-host range and pose a great risk for the possible spread of resistance. Therefore, we recommend that studies on antimicrobial resistance should consider thorough, systematic studies of animals, fish, humans, tap water, and sewage, as all these factors are linked together. The current study has some limitations, including examination of integrated aquaculture from only one governorate in Egypt and the limited number of the tested carbapenemase genes (blaKPC, blaNDM-1, and blaOXA-48). Therefore, further work is required to elucidate the extent of antibiotic resistance in more integrated aquaculture from other governorates. Particularly, to screen for carbapenems and extended-spectrum-cephalosporins, with including a wide panel of carbapenemase genes.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- βLPE:
-
β-lactamase-producing Enterobacteriaceae
- CAZ:
-
Ceftazidime
- CDC:
-
Centre for Disease Control and Prevention
- CEPH:
-
Cephalosporins
- CLSI:
-
Clinical and Laboratory Standards Institute
- CPE:
-
Carbapenemase-producing Enterobacteriaceae
- CRE:
-
Carbapenem-resistant Enterobacteriaceae
- CRO:
-
Ceftriaxone
- CTX:
-
Cefotaxime
- ECC :
-
Enterobacter cloacae complex
- EMB:
-
Eosin methylene blue agar
- ESBL:
-
Extended-spectrum β-lactam
- ETP:
-
Ertapenem
- FOX:
-
Cefoxitin
- Inc. plasmids:
-
Incompatible plasmids
- IPM:
-
Imipenem
- K. pneumoniae :
-
Klebsiella pneumoniae
- MEM:
-
Meropenem
References
Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ. Aquaculture as yet another environmental gateway to the development and globalization of antimicrobial resistance. Lancet Infect Dis. 2016;16:127–33.
Schmidt AS, Bruun MS, Dalsgaard I, Larsen JL. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl Environ Microbiol. 2001;67:5675–82.
Defoirdt T, Sorgeloos P, Bossier P. Alternative to antibiotics for the control of bacterial disease in the aquaculture. Curr Opin Microbiol. 2011;14:251–8.
Hoa PT, Managaki S, Nakada N, Takada H, Shimiz A, Anh DH. Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci Total Environ. 2011;409:2894–901.
Little DC, Edwards P. Integrated livestock-fish farming systems. Rome: Food and Agriculture Organization of the United Nations; 2003. http://www.fao.org/3/a-y5098e.
Neela FA, Banu NA, Rahman A, Rahman MH, Alam MF. Occurrence of antibiotic resistant bacteria in pond water associated with integrated poultry-fish farming in Bangladesh. Sains Malaysiana. 2015;44:371–7.
Seiler C, Berendonk TU. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:399.
Burridge L, Weis JS, Cabello F, Pizarro J, Bostick K. Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture. 2010;306:7–23.
Buschmann AH, Tomova A, Lopez A, Maldonado MA, Henriquez LA, Ivanova L, et al. Salmon aquaculture and antimicrobial resistance in the marine environment. PLoS One. 2012;7:42724.
Heuer OE, Kruse H, Grave K, Collignon P, Karunasagar I, Angulo FJ. 2009. Human health consequences of use of antimicrobial agents in aquagculture. Clin Infect Dis. 2009;49:1248–53.
Rhodes G, Huys G, Swings J, Mcgann P, Hiney M, Smith P, et al. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant Tet A. Appl Environ Microbiol. 2000;66:3883–90.
Sheu CC, Chang YT, Lin SY, ChengYH HPR. Infections caused by carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front Microbiol. 2019;10:80.
Wilson H, Török ME. Extended spectrum β-lactamase producing and carbapenamese producing Enterobacteriaceae. Microb Genom. 2018;4:7.
Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–76.
Dandachi I, Chaddad A, Hanna J, Matta J, Daoud Z. Understanding the epidemiology of multi-drug resistant gram-negative bacilli in the Middle East using a one health approach. Front Microbiol. 2019;10:1941.
Exner M, Bhattacharya S, Christiansen B, Gebel J, Goroncy-Bermes P. Hartemann p. Antibiotic resistance: What is so special about multidrug-resistant gram-negative bacteria?. GMS Hyg Infect Control. 2017;12:1–24.
Canton R, Ruiz-Garbajosa P. Co-resistance: an opportunity for the bacteria and 2 resistance genes. Curr Opin Pharmacol. 2011;11:477–85.
Austin S, Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–4.
Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerrat B. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73:1121–37.
Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38.
Carloni E, Andreoni F, Omiccioli E, Villa L, Magnani M, Carratoli A. Comparative analysis of the standard PCR-based replicon typing (PBRT) with the commercial PBRT-Kit. Plasmid. 2017;90:10–4.
Bezuidenhout CC, Mthembu N, Puckree T, Lin J. Microbiological evaluation of the Mhlathuze River, Kwazulu-Natal (RSA). Water SA. 2002;28:281–6.
Collee JG, Fraser AG, Marmion BP, Simmons A. Practical medical microbiology. New York: Churchill Livingstone; 1996.
CLSI, Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing (M100-S24);2016.
CDC, Centre for Disease Control and Prevention. Patient safety atlas. 2018. https://www.cdc.gov/hai/surveillance/ar-patient-safety-atlas.html. Accessed 13 July 2018.
Elhariri M, Hamza D, Elhelw R, Dorgham SM. Extended-spectrum beta-lactamase -producing Pseudomonas aeruginosa in camel in Egypt: potential human hazard. Ann Clin Microbiol Antimicrob. 2017;16:1–6.
Sugumar M, Kumar KM, Manoharan A, Anbarasu A, Ramaiah S. Detection of OXA-1 β-lactamase gene of Klebsiella pneumoniae from blood stream infections by conventional PCR and in-Silico analysis to understand the mechanism of OXA mediated resistance. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0091800.
Hamza E, Dorgham SM, Hamza DA. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. J Global Antimicrob Resist. 2016;7:8–10.
Li B, Yi Y, Wang Q, Woo PCY, Jing H, Gao GF. Analysis of drug resistance determinants in Klebsiella pneumoniae isolates from a tertiary-care hospital in Beijing, China. PLoS One. 2012. https://doi.org/10.1371/journal.pone.0042280.
Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. Replicon typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–29.
Osborn AM, da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiol. 2000;146:2267–75.
Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49–61.
Prestinaci F, Pezzoti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109:309–18.
Sarmah AK, Meyer MT, Boxall AB. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65:725–59.
Abdel-Latif HMR, Sedeek EK. Diversity of Enterobacteriaceae retrieved from diseased cultured Oreochromis niloticus. Int J Fish Aqua Stud. 2017;5:29–34.
Aly SM, Nouh WG, Salem-Bekhit MM. Bacteriological and histopathological studies on Enterobacteriaceae in Nile Tilapia, Oreochromis niloticus. J Pharm Biomed Sci. 2012;2:94–104.
Elsherief MF, Mousa MM, Abd El-Galil H, El-Bahy EF. Enterobacteriaceae associated with farm fish and retailed ones. Alex J Vet Sci. 2014;42:99–104.
Hassan AHM, Noor-El Deen AE, Galal HM, Dorgham SM, Bakry MA, Hakim AS. Further characterization of Enterobacteriaceae isolated from cultured freshwater fish in Kafr El Shiek governorate: clinical, biochemical and histopathological study with emphasis on treatment trials. Glob Veter. 2012;9:617–29.
Saswathi R, Sumithra P, Sivakami R. Identification of Enterobacteriaceae studies in carps during rearing a freshwater pond. Int J Curr Microbiol Appl Sci. 2015;4:661–9.
Abgottspon H, Nüesch-Inderbinen MT, Zurfluh K, Althaus D, Hächler H, Stephan R. Enterobacteriaceae with extended-Spectrum- and pAmpC-type -lactamase-encoding genes isolated from freshwater fish from two lakes in Switzerland. Antimicrob Agents Chemother. 2014;58:2482–4.
CLSI, Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing (M100, 30th Edition); 2020.
Singh-Moodley A, Perovic O. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis. 2016;16:536.
Akinbowale OL, Peng H, Barton MD. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J Appl Microbiol. 2006;100:1103–13.
Brahmi S, Tuoati A, Dunyach-Remy C, Sotto A, Pantel A, Lavigne JP. High prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in wild fish from the Mediterranean Sea in Algeria. Microb Drug Resist. 2018;24:290–8.
Elhadi N, Alsamman K. Incidence and antimicrobial susceptibility pattern of extended-spectrum-β-lactamase-producing Escherichia coli isolated from retail imported mackerel fish. Afr J Biotech. 2015;14:1954–60.
Ishida M, Ahmed AM, Mahfouz NB, Kimura T, El-Khodery SA, Moawad AA. Molecular analysis of antimicrobial resistance in gram-negative bacteria isolated from fish farms in Egypt. J Vet Med Sci. 2010;72:727–34.
Sarter S, Nguyen K, Hung LT, Lazart J, Montet D. Antibiotic resistance in gram-negative bacteria isolated from farmed catfish. Food Control. 2007;18:1391–6.
Singh AS, Lekshmi M, Prakasan S, Nayak BB, Kumar S. Multiple antibiotic-resistant, Extended Spectrum-Lactamase (ESBL)-producing Enterobacteriaceae in fresh seafood. Microorganisms. 2017;5(53):1–10.
Janecko N, Martz SL, Avery BP, Daignault D, Desruisseau A, Boyd D. Carbapenem-resistant Enterobacter spp. in retail seafood imported from Southeast Asia to Canada. Emerg Infect Dis. 2016;22:1675–7.
Mangat CS, Boyd D, Janecko N, Martz SL, Desruisseau A, Carpenter M. Characterization of VCC-1, a novel ambler class a carbapenemase from Vibrio cholerae isolated from imported retail shrimp sold in Canada. Antimicrob Agents Chemother. 2016;60:1819–25.
Sabate M, Tarrago R, Navarro F, Miro E, Verges C. Barbe, et al. cloning and sequence of the gene encoding a novel cefotaxime-hydrolysing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother. 2000;44:1970–3.
Karim A, Poirel L, Nagarajan S, Nordmann P. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Banner. 2006;201:237–41.
Barron SA, Diene SM, Rolain JM. Human microbiomes and antibiotic resistance. Hum Microb J. 2018;10:43–52.
Dang STT, Petersen A, Truong DV, Chu HTT, Dalsgaard A. Resistance in bacteria at integrated pig-fish farms in Vietnam. Appl Environ Microbiol. 2011;77:4494–8.
Petersen A, Andersen JS, Kaewmak T, Somsiri T, Dalsgaard A. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl Environ Microbiol. 2002;68:6036–42.
Braun S, Ahmed MFE, El-Adway H, Hotzel H, Engelmann I, Weiss D, et al. Surveillance of extended Spectrum β-lactamase-producing Escherichia coli in dairy cattle farms in the Nile Delta, Egypt. Front Microbiol. 2016;7(1020):1–14.
El-Kowrany SI, El-Zamarany EA, El-Nouby KA, El-Mehy DA, Abo Ali EA, Othman AA. Water pollution in the middle Nile Delta, Egypt: an environmental study. J Adv Res. 2016;7:781–9.
Ezzat SM, Mahdy HM, Abo-State MA, Abd-Elshakour EH, El-Bahnasawy MA. Water quality assessment of River Nile at Rosetta branch Middle East. J Sci Res. 2012;12:413–23.
Amine AEK. Extended spectrum Beta-lactamase producing bacteria in sewage water, Alexandria, Egypt. Int J Biosc Bioch Bioinf. 2013;3:605–8.
Sonnevend A, Al-Dhaheri K, Mag T, Herpay M, Kolodziejek J, Nowotny N. CTX-M-15-producing multidrug-resistant enteroaggregative Escherichia coli in the United Arab Emirates. Clin Microbiol Infect. 2006;12:582–285.
Iabadene H, Messai Y, Ammari H, Alouache S, Verdet C, Bakour R, Arlet G. Prevalence of plasmid-mediated AmpC β-lactamases among Enterobacteriaceae in Algiers hospitals. Int J Antimicrob Agents. 2009;34:340–2.
Mamlouk K, Boubaker IB, Gautier V, Vimont S, Picard B, Ben RS. Emergence and outbreaks of CTX-M β-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol. 2006;44:4049–56.
Hancok S, Phan MD, Peters KM, Forde BM, Chong TM, Yin WF, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multi-locus sequence typing scheme. Antimicrob Agents Chemother. 2017;61(2):1–17.
Almeida AC. de Sa Cavalcanti FL, Vilela MA, gales AC, de Morais MA, Jr Camargo de Morais MM. Escherichia coli ST502 and Klebsiella pneumoniae ST11 sharing an IncW plasmid harbouring the blaKPC-2 gene in an intensive care unit patient. Int J Antimicrob Agents. 2012;40:374–6.
Yano H, Rogers LM, Knox MG, Heuer H, Smalla K, Brown CJ, et al. Host range diversification within the IncP1 plasmid group. Microbiol. 2013;159:2303–15.
Froehlich B, Parkhill J, Sanders M, Quail MA, Scott JR. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic conitegrate. J Bacteriol. 2005;187:6509–16.
Desmet S, Nepal S, van Dijl JM, van Ranst M, Chlebowicz MA, Rossen JW, et al. Antibiotic resistance plasmids cointegrated into a megaplasmid harbouring the blaOXA-427 carbapenemase gene. Antimicrob Agents Chemother. 2018;62(3):1–6.
Acknowledgements
We are grateful to the aquaculture workers and people living in the areas surrounding the fish farms for participating in the current study. The cooperation of the aquaculture owners is highly appreciated.
Funding
This research did not receive any grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Dalia Hamza participated in the design and coordination of the study, performed the experiments, and assisted in the editing of the manuscript. Sohad Dorgham participated in the design of the study, collection of samples and performed the experiments. Elshaima Ismael performed the statistical analysis and preparation of tables. Sherein Ismail Abd El-Moez participated in the design of the study and collection of samples. Rehab Elhelw and Mahmoud Elhariri performed the experiments and preparation of the manuscript. Eman Hamza participated in the design of the study, assisted in the statistical analysis and preparation of the figures, and edited the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Protocols for collection of samples as well as the experiment plan and all methods were performed in accordance with the guidelines and regulations of Cairo University Council and were approved by the Scientific Research Committee and Bioethics Board of Cairo University, Faculty of Veterinary Medicine, Cairo, Egypt. Ethical clearance to use human subjects was obtained from the designated health facility (National Research Centre, Giza, Egypt) and written informed consent was obtained from each person upon information of the use of samples.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hamza, D., Dorgham, S., Ismael, E. et al. Emergence of β-lactamase- and carbapenemase- producing Enterobacteriaceae at integrated fish farms. Antimicrob Resist Infect Control 9, 67 (2020). https://doi.org/10.1186/s13756-020-00736-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-020-00736-3