Abstract
Hyperspectral image (HSI) classification has been long envisioned in the remote sensing community. Many methods have been proposed for HSI classification. Among them, the method of fusing spatial features has been widely used and achieved good performance. Aiming at the problem of spatial feature extraction in spectral-spatial HSI classification, we proposed a guided filter-based method. We attempted two fusion methods for spectral and spatial features. In order to optimize the classification results, we also adopted a guided filter to obtain better results. We apply the support vector machine (SVM) to classify the HSI. Experiments show that our proposed methods can obtain very competitive results than compared methods on all the three popular datasets. More importantly, our methods are fast and easy to implement.
Similar content being viewed by others
1 Introduction
Hyperspectral imaging sensors have been widely used in remote sensing, biology, chemometrics, and so on [1]. Hyperspectral imaging sensors can obtain spatial and spectral information of materials, which is called the hyperspectral image (HSI), for the same time. Due to abundant spectral information, HSI is widely applied to material recognition and classification, such as land cover [2], environmental protection [3], and agriculture [4]. Hence, HSI classification has attracted increasing attention and became a hot topic in the remote sensing community.
The task of classification is to assign a unique label to each pixel vector of HSI. For this problem, many pixel-wise (spectral-based) methods were employed, including k-nearest neighbors (KNN) [5], support vector machine (SVM) [6], and sparse representation [7] in the last two decades. SVM has shown good performance for classifying high-dimensional data when a limited number of training samples are available [8]. It can effectively overcome the Hughes phenomenon [9] and the problem of limited training samples in HSI classification. Therefore, SVM and its improved algorithms get better performance than other methods. However, they still have a wide gap for expectations. After all, it is a universal phenomenon that different materials have the same spectrum and the same material has different spectrum.
To overcome the above problem and improve the performance of classification, recent studies have suggested incorporating spatial information into a spectral-based classifier [10], which is called the spectral-spatial HSI classification. Because of the continuous improvement in spatial resolution, the spatial features of materials become more representative. Many papers show that spectral method is a very effective way for HSI classification. Various types of classification approaches have been proposed, including morphology feature extraction [11], kernel combination [3, 12], and joint representation [13]. By using geodesic opening and closing operations with fixed shape structuring elements of different sizes, morphological profiles significantly improve the classification accuracy. The main idea of a joint representation model is to exploit both spectral and spatial features by treating the test sample as a collection of its neighboring pixels (including the test pixel itself).
For SVM methods, the mainstream approaches of fusing spectral and spatial features are used by kernel combination [14]. The paper [15] proposed a series of composite kernels to fuse spectral and spatial features directly in the SVM kernels. And Li et al. [16] presented a generalized composite kernel framework to classify HSI. The aforementioned methods fused spectral and spatial features before classification. There is also a small number of ways to fuse feature after the SVM classification. Tarabalka [17] integrated into the SVM classification probabilities into a Markov random field for classifying the HSI. Moreover, Kang [18] proposed a spectral-spatial HSI classification method with edge-preserving filtering, which extracted the spatial features after the SVM classification and got competitive results.
Motivated by the paper [18], we adopt the guided filter to extract spatial features. Compared to the reference [18], our contribution can be concluded as extracting spatial features before classification. In more detail, the main contributions are listed as follows.
-
1)
We adopt the guided filter to smooth HSI, which is similar to de-noising in image processing. By this method, a fusion which consists of a pixel and its neighboring pixel information is generated. It is proved to be simple and effective.
-
2)
We attempt different spectral and spatial fusion methods, which makes sense for future work.
-
3)
The proposed methods are applied to three widely used hyperspectral datasets. We compare with two methods by three evaluation metrics.
2 Related methodology and work
2.1 SVM and HSI classification
SVM is a supervised machine learning method, proposed by Vapnik [19], which is based on the statistical learning theory. Essentially, SVM attempts to find a hyperplane in the multidimensional feature space to separate the two classes. And this hyperplane is the best decision surface which maximizes the distance between the hyperplane and two classes, called the margin. Generally, the larger the margin, the better the classifier is. From a given set of the training set, obtaining an SVM model is equivalent to an optimization problem for finding a hyperplane. For this optimization, SVM introduces a structural minimum principle that prevents over-fitting problems.
SVM is suitable for high-dimensional data with the limited training set. And a lot of researches have addressed that SVM classifier presents superior performance on HSI classification [6, 20], compared with other popular classifiers such as decision tree classifier, k-nearest neighbor classifier, and neural networks. The power of SVM is mainly due to its kernel function, especially radial basis function. However, the single kernel is not enough for all cases. Some researchers proposed a composite kernel [21], which integrates both spectral and spatial features, to improve classification performance.
2.2 Guided filter and HSI classification
The guided filter,Footnote 1 proposed by He [22] for the first time, has been widely used in the fields of noise reduction, image dehazing, and so on. We can get a new image that obtains the feature of the guided filter. Given image p as an input, and a guided filter image g, we can obtain an output image q. Generally, q is a linear transform of g in a window ωk centered at the pixel k. If the radius of k is r, the size of local window ωk is (2r + 1) × (2r + 1).
where ak and bk are a linear coefficient and bias, respectively. From the model, we can see that ∇q = a ∇ g, which means that the output q will have a similar gradient with guidance image g. The coefficient and bias, which need to be known, are solved by a minimum cost function as follows:
Here, ϵ is a regularization parameter. According to the paper [22], a solution can be derived from Eq. (2) as follows.
where μk and \( {\sigma}_k^2 \) are the mean and variance of g in ωk, ∣ω∣ is the number of pixels in ωk, and \( {\overline{p}}_k \) is the mean of p in ωk. After obtaining the coefficient ak and bk, we can compute the filtering output qi. Through the above process, we can get a linear transform image q.
The guided filter was first used for HSI classification by Kang [18]. They considered the HSI classification as a probability optimization process. They firstly obtained the initial probability by SVM. Then, they applied a guided filter to optimize the initial probability maps. They got a state-of-the-art result. Subsequently, Wang [23] adopted a guided filter to extract the spatial features from HSI. Then, a stacked autoencoder was used to classify each pixel. Guo [24] proposed a method, which combines a guided filter, joint representation, and k-nearest neighbor to improve HSI classification. Inspired by the methods mentioned above, we propose a novel method for fusing spectral and spatial information. The experimental results clearly show that our approach can be executed more rapidly than conventional methods.
3 HSI classification with SVM and guided filter
3.1 The proposed Guided Filter SVM Edge Preserving Filter (GF-SVM-EPF)
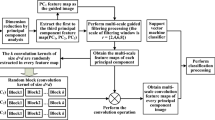
We propose a novel method for HSI classification with SVM and guided filter. First, we extract the spatial features of HSI by the guided filter, which is obtained from the original HSI by a principal component analysis (PCA) method. Then, we classify the spatial features by SVM. Finally, we employ a guided filter again to optimize the classification. The process is shown in Fig. 1.
Framework of HSI classification by the proposed method. Original HSI was used to get the guidance image (filter) and decomposed HSI by PCA firstly. Then, decomposed HSI was filtered by a guidance filter. Subsequently, a SVM classifier was adopted to get the classification map. Finally, edge preserving filter was applied to optimize the classification map
3.1.1 Extracting spatial features by guided filter
First, we obtain a guidance image by PCA. We take the first three principal components as a color guidance image. Given a dataset D = {d1, d2, ⋯, dS}, we adopt PCA to obtain the following result. Here di is the information of the ith band, and S denotes the number of bands.
So, the guidance image is G = [g1, g2, g3]. Then, based on formula (1), using input image d1 and guidance image G, we can get the output u1 by filtering. In the same way, we can yield all the ui which constructs a new hyperspectral image U = {u1, u2, ⋯, uS}.
3.1.2 Classifying HSI by SVM
After obtaining the image U = {u1, u2, ⋯, uS} by the guided filter, we can rewrite it as V = {v1, v2, ⋯, vN}, where vn = {vn, 1, vn, 2, ⋯, vn, S} is the spectral feature vector.
Then we adopt an SVM classifier to classify all the feature vector vn. We can get a classification map C as the original result.
3.1.3 Optimizing the classification map
First, we convert the classification map C into a probability map P = {p1, p2, ⋯, pn}, where pi, n is the initial probability with a value of 0 or 1. And n denotes the number of categories to classify. If a pixel i belongs to the nth class, pi, n is set to 1. Other else, pi, n is set to 0.
Then, we employ the guided filter in section 3.1.1 to filter each pi. So we get a new P as a final probability map. For each pixel, we get n probability values and choose the class label with the biggest probability value as the final label.
3.2 Other methods
In order to verify the effectiveness of our method, we also proposed another three methods with SVM and guided filter. We want to research the efficacy of spatial feature extraction and propose a GF-SVM method, which firstly extracts spatial features and then classifies them by SVM. The implementation is the same as the steps 1 and 2 in Section 3.1.
If spatial features and spectral features are fused together, can the increasement of information improve classification accuracy of the HSI? We proposed another method called Co-SVM (Connected SVM). We choose the top half of [g1, g2, ⋯, gS] as the original feature. And we choose the top half of U as the spatial features. Then we join the original and the spatial features together as a fusion feature to classify. After being classified by SVM, we get the final classification. By optimizing the result of Co-SVM as step 3 in Section 3.1, we got another method called Co-SVM-EPF.
4 Results and discussion
4.1 Experimental setup
4.1.1 Datasets
Three hyperspectral data,Footnote 2 including Indian Pines, University of Pavia, and Salinas, are employed to draw a convincing conclusion. The Indian Pines dataset was gathered by an AVIRIS sensor. The image scene, with a spatial coverage of 145 × 145 pixels, is covering the woods, grass-pasture, and so on. We choose 200 spectral channels from 220 bands in the 0.4- to 2.45-μm region of the visible and infrared spectrum.
The University of Pavia dataset was captured by the ROSIS (Reflective Optics System Imaging Spectrometer) sensor. The dataset contains 610 × 340 pixels with 115 spectral bands. After removing water absorption and low SNR bands, 103 bands were used for the analysis. And there are nine categories to be classified.
The third dataset was collected by the AVIRIS sensor, capturing an area over Salinas Valley, California, with a high spatial resolution of 3.7 m. Salinas comprises 512 × 217 pixels in all and contains vegetables, bare soils, and vineyard fields. We also selected 200 bands for experiments by discarding 20 water absorption.
4.1.2 Evaluation metrics
Three widely used indexes for HSI classification were adopted to evaluate the performance of experimental methods, including the overall accuracy (OA), the average accuracy (AA), and the kappa coefficient (KA).
4.1.3 Parameter settings
In this experiment, we use libSVM designed by Lin [25]. The libSVM has two main parameters C and g to be set. The C and g are determined by cross validation. And C changes from 10−2 to 104, and g ranges from 2−1 to 24.
For guided filter, there are two key parameters to be set. One is the radius r of the filter which represents the size of spatial feature scale. The other parameter of guided filter is the regularization parameter ɛ which controls the degree of smoothness. In our experiments, we set r = 3 and ɛ = 0.001.
4.2 Experimental results and discussion
In this section, the proposed methods are compared with two widely used classification methods, SVM [18] and EPF [18], which are typical examples of pixel-wise methods and spectral-spatial methods respectively.
4.2.1 Experimental results and discussion on Indian Pines
The Indian Pines dataset is the most commonly used and the most difficult to classify. In this experiment, the classification accuracy for each class, OA, AA, and KA is adopted to evaluate the classification performance. Figure 2 shows the classification maps obtained by different methods associated with the corresponding OA scores. From this figure, we can see that the classification accuracy obtained by SVM is the worst since lots of noisy estimations are visible. The best one is the classification accuracy obtained by GF-SVM-EPF, which is almost the same as the ground truth.
Classification performance of each class is shown in Table 1. All our proposed methods outperform the SVM and EPF significantly on all the indexes. They are higher than EPF by 2%, 2.5%, 3.5%, and 4.6%, respectively. Especially, the proposed method GF-SVM-EPF achieves 99.22%, which is the best result we have seen so far. In 16 categories, there are 11 classes to reach the highest results. Unlike our expectations, the result obtained by Co-SVM which employs the fusion of guided information and spectral information is worse than that obtained by GF-SVM which only employs the guided information. This shows that the superposition of information cannot bring better classification results. Because each pixel in the filtered image is a linear transformation of its own and neighbor pixels, the filtered feature is enough for HSI classification. We can draw a conclusion that the spectral-spatial fusion method (Co-SVM) can improve classification accuracy (compared with SVM and EPF). Using the guided filter twice can improve classification accuracy, such as GF-SVM-EPF.
4.2.2 Experimental results and discussion on the University of Pavia dataset
The University of Pavia only has nine categories. It is easier to classify. Classification maps of different methods are illustrated in Fig. 3. It can be seen from this figure that the proposed methods (Co-SVM, GF-SVM, and GF-SVM-EPF) achieve better classification performance than other compared approaches. Especially, in the map of GF-SVM-EPF, we can hardly see the difference between it and the ground truth.
And the results of different methods are shown in Table 2. It can be seen from Table 2 that our proposed methods (Co-SVM, Co-SVM-EPF, GF-SVM) are similar, and they are slightly higher than SVM-EPF (98.51%). GF-SVM-EPF obtains the result of 99.7%, which outperforms state-of-the-art methods. There are six categories to reach the highest results in nine categories. In this experiment, there is a strange phenomenon that Co-SVM is better than Co-SVM-EPF. That is because some pixels of the thin edges are divided into the background, as seen from Fig. 3. As in the previous experiment, the method of extracting spatial features by the guided filter twice (GF-SVM-EPF) is the most effective classification method.
4.2.3 Experimental results and discussion of the Salinas dataset
The last experiment is performed on the Salinas dataset, which is the biggest one we have chosen. The qualitative results are shown in Fig. 4. It is apparent from this figure that the map of GF-SVM-EPF has the fewest noise points and obtains the best results.
The detailed results are illustrated in Table 3. All the methods perform well on this dataset. The worst is about 92.21% by SVM. The proposed methods (Co-SVM-EPF, GF-SVM, and GF-SVM-EPF) are all over 99%, especially GF-SVM-EPF which reaches 99.8%, outperforming other methods greatly. There are 12 categories to achieve the best result in 16 categories. The conclusion of this experiment is consistent with that of Indian Pines. Spectral features with spatial features can improve classification accuracy. The result of extracting spatial features twice is better than that of extracting spatial features once.
From the above three experiments, we can draw a conclusion that SVM with guided filter can be well used for HSI classification. The guided filter is an effective way to fuse spatial information and spectral information. Especially for datasets with a regular shape, a guided filter is more effective to extract spatial features. Because the filtered pixels contain not only neighbor information but also their own information, they can be directly used for classification without adding other information.
5 Conclusion
In this paper, we propose several spectral-spatial HSI classification methods which combined SVM with guided filter. Two spectral and spatial fusion methods are adopted for the SVM. Moreover, the guided filter was used for extracting spatial information and optimizing the classification results, respectively. Our proposed methods can improve the classification accuracy significantly in a short time. Consequently, the proposed methods can be effective to real applications.
From this work, we can draw the following conclusions: (a) The guided filter is an effective way to extract spatial information in HSI. (b) The extracted feature by the guided filter is good enough for HSI classification, without original information. (c) The method of SVM with twice filtrations is a simple and effective way to classify HSI.
Abbreviations
- AA:
-
Average Accuracy
- Co-SVM:
-
Connected SVM
- GF-SVM-EPF:
-
Guided Filter SVM Edge Preserving Filter
- HSI:
-
Hyperspectral Image
- KA:
-
Kappa Coefficient
- KNN:
-
k-nearest neighbors
- OA:
-
Overall Accuracy
- PCA:
-
Principal Component Analysis
- ROSIS:
-
Reflective Optics System Imaging Spectrometer
- SVM:
-
Support Vector Machine
References
A.F.H. Goetz, Three decades of hyperspectral remote sensing of the earth: A personal view. Remote Sens. Environ. 113, S5–S16 (2009)
G.P. Petropoulos, C. Kalaitzidis, K. Prasad Vadrevu, Support vector machines and object-based classification for obtaining land-use/cover cartography from Hyperion hyperspectral imagery. Comput. Geosci. 41(2), 99–107 (2012)
R.L. Lawrence, S.D. Wood, R.L. Sheley, Mapping invasive plants using hyperspectral imagery and Breiman cutler classifications (RandomForest). Remote Sens. Environ. 100(3), 356–362 (2006)
L.M. Dale, A. Thewis, C. Boudry, et al., Hyperspectral imaging applications in agriculture and agro-food product quality and safety control: A review. Appl. Spectrosc. Rev. 48(2), 142–159 (2013)
L. Bruzzone, R. Cossu, A multiple-cascade-classifier system for a robust and partially unsupervised updating of land-cover maps. IEEE Trans. Geosci. Remote Sens. 40(9), 1984–1996 (2002)
F. Melgani, L. Bruzzone, Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 42(8), 1778–1790 (2004)
Y. Chen, N.M. Nasrabadi, T.D. Tran, Hyperspectral image classification via kernel sparse representation. IEEE Trans. Geosci. Remote Sens. 51(1), 217–231 (2013)
G. Camps-Valls, L. Bruzzone, Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 43(6), 1351–1362 (2005)
G. Hughes, On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 14(1), 55–63 (1968)
A. Plaza, J. Plaza, G. Martin, in Machine Learning for Signal Processing (MLSP), 2009. 2009 IEEE international workshop on. Incorporation of spatial constraints into spectral mixture analysis of remotely sensed hyperspectral data, vol 1 (IEEE, 2009), pp. 1–6
M. Dalla Mura, J.A. Benediktsson, B. Waske, et al., Morphological attribute profiles for the analysis of very high resolution images. IEEE Trans. Geosci. Remote Sens. 48(10), 3747–3762 (2010)
M. Fauvel, J. Chanussot, J.A. Benediktsson, A spatial–spectral kernel-based approach for the classification of remote-sensing images. Pattern Recogn. 45(1), 381–392 (2012)
Y. Chen, N.M. Nasrabadi, T.D. Tran, Hyperspectral image classification using dictionary-based sparse representation. IEEE Trans. Geosci. Remote Sens. 49(10), 3973–3985 (2011)
G. Camps-Valls, N. Shervashidze, K.M. Borgwardt, Spatio-spectral remote sensing image classification with graph kernels. IEEE Geosci. Remote Sens. Lett. 7(4), 741–745 (2010)
G. Camps-Valls, L. Gomez-Chova, J. Muñoz-Marí, et al., Composite kernels for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 3(1), 93–97 (2006)
J. Li, P.R. Marpu, A. Plaza, et al., Generalized composite kernel framework for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 51(9), 4816–4829 (2013)
Y. Tarabalka, M. Fauvel, J. Chanussot, et al., SVM-and MRF-based method for accurate classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 7(4), 736–740 (2010)
X.D. Kang, S. Li, J.A. Benediktsson, Spectral–spatial hyperspectral image classification with edge-preserving filtering. IEEE Trans. Geosci. Remote Sens. 52(5), 2666–2677 (2014)
C. Cortes, V. Vapnik, Support-vector networks. Mach. Learn. 20(3), 273–297 (1995)
G. Camps-Valls, L. Gomez-Chova, J. Calpe-Maravilla, J.D. Martin-Guerrero, E. Soria-Olivas, L. Alonso-Chorda, J. Moreno, Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Trans. Geosci. Remote Sens. 42(7), 1530–1542 (2004)
D. Tuia, F. Ratle, A. Pozdnoukhov, G. Camps-Valls, Multisource composite kernels for urban-image classification. IEEE Geosci. Remote Sens. Lett. 7(1), 88–92 (2010)
K.M. He, J. Sun, X.O. Tang, Guided image filtering. IEEE Trans. Pattern Anal. Mach. Intell. 35(6), 1397–1409 (2013)
L. Wang, J. Zhang, P. Liu, Spectral---spatial multi-feature-based deep learning for hyperspectral remote sensing image classification. Soft. Comput. 21(1), 213–221 (2017)
Y. Guo, H. Cao, S. Han, Spectral-spatial hyperspectral image classification with K-nearest neighbor and guided filter. IEEE Access 6, 18582–18591 (2018)
C.C. Chang, C.J. Lin, LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2(3), 27–27 (2011)
Funding
The project was supported by Open Research Fund of Shandong Provincial Key Laboratory Of Infectious Disease Control and Prevention, Shandong Center for Disease Control and Prevention (No. 2017KEYLAB01), the Science and Technology Project for the Universities of Shandong Province (No. J18KB171), Laboratory of Data Analysis and Prediction in Shandong Women’s University.
Availability of data and materials
The simulations were performed using matlab2014a and libsvm3.0 in Intel Core I7 (64bit). All data generated or analyzed during this study are included in this published article. The hyperspectral image datasets can be downloaded at http://www.ehu.eus/ccwintco/index.php?title=Hyperspectral_Remote_Sensing_Scenes
Author information
Authors and Affiliations
Contributions
YHG is the main writer of this paper. He proposed the main idea and designed the experiment. XJY and XCZ completed the analysis of the results. DXY assisted in the collection and pre-processing of the data. YB refined the idea and designed the structure of the whole manuscript. Both authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Guo, Y., Yin, X., Zhao, X. et al. Hyperspectral image classification with SVM and guided filter. J Wireless Com Network 2019, 56 (2019). https://doi.org/10.1186/s13638-019-1346-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13638-019-1346-z