Abstract
Background
Recurrent spontaneous abortion (RSA) is a challenging condition that affects the health of women both physically and mentally, but its pathogenesis and treatment have yet to be studied in detail. In recent years, Wharton’s jelly-derived mesenchymal stem cells (WJ-MSCs) have been shown to be effective in treating various diseases. Current understanding of RSA treatment using WJ-MSCs is limited, and the exact mechanisms of WJ-MSCs action in RSA remains largely unclear. In this study, we explored the decidual deficiencies in RSA and the therapeutic potential of WJ-MSCs at single-cell resolution.
Methods
Three mouse models were established: a normal pregnancy group, an RSA group, and a WJ-MSC treatment group. Decidual tissue samples were collected for single-cell RNA sequencing (scRNA-seq) and functional verification, including single-cell resolution in situ hybridization on tissues (SCRINSHOT) and immunofluorescence.
Results
We generated a single-cell atlas of decidual tissues from normal pregnant, RSA, and WJ-MSC-treated mice and identified 14 cell clusters in the decidua on day 14. Among these cell populations, stromal cells were the most abundant cell clusters in the decidua, and we further identified three novel subclusters (Str_0, Str_1, and Str_2). We also demonstrated that the IL17 and TNF signaling pathways were enriched for upregulated DEGs of stromal cells in RSA mice. Intriguingly, cell–cell communication analysis revealed that Str_1 cell-related gene expression was greatly reduced in the RSA group and rescued in the WJ-MSC treatment group. Notably, the interaction between NK cells and other cells in the RSA group was attenuated, and the expression of Spp1 (identified as an endometrial toleration-related marker) was significantly reduced in the NK cells of the RSA group but could be restored by WJ-MSC treatment.
Conclusion
Herein, we implemented scRNA-seq to systematically evaluate the cellular heterogeneity and transcriptional regulatory networks associated with RSA and its treatment with WJ-MSCs. These data revealed potential therapeutic targets of WJ-MSCs to remodel the decidual subpopulations in RSA and provided new insights into decidua-derived developmental defects at the maternal–foetal interface.
Similar content being viewed by others
Introduction
Successful pregnancy is characterized by effective embryo implantation, suitable embryonic development, appropriate decidual and placental development, and timely delivery that involves the participation of maternal decidual and placental units with many other cell types [1, 2]. Any disturbance of these processes leads to compromised pregnancy outcomes via a ripple effect. Recurrent spontaneous abortion (RSA), which occurs in 5% of fertile couples, refers to at least two consecutive spontaneous abortions before 20 weeks of gestation; however, the underlying mechanisms of RSA remain largely unknown due to its complex pathology. Maternal immune disorders, endocrine dysfunction, and genetic factors are hypothesized to be potential causes for RSA, making its clinical treatment extremely challenging [3, 4]. To date, there are no strict criteria for the diagnosis and treatment of RSA, and this greatly affects the physical and mental health of patients with RSA [5, 6]. Although a series of clinical studies regarding patients with RSA have investigated its underlying factors (including chromosomal analysis and pelvic ultrasonography), little progress has been made towards effectively preventing the development of RSA [3, 7]. Therefore, early identification of the causes of RSA will facilitate the effective early management of RSA patients and improve pregnancy outcomes in patients with RSA. It is conventionally stated that RSA is a problematic condition that affects the health of women both physically and mentally, and its pathogenesis and treatment have yet to be analyzed in depth.
There is an increasing amount of interest in elucidating maternal decidual functions as a critical signal and nutritional hub at the maternal-foetal interface to guide the successful development of the embryo and placenta by orchestrating the homeostatic balance between the mother and foetus [8,9,10,11]. After blastocystic trophoblast cells invade the endometrium, the stromal cells surrounding the embryo initially differentiate to form a primary decidual zone and then continue to proliferate and differentiate into a secondary decidual zone, which eventually transforms into decidual cells. There is also an undifferentiated stromal cell population in the decidual zone, which, together with other decidual cells, provides nutrition and protection for the embryo. Recent studies have shown the presence of in maternal decidual development in RSA, which may be related to factors such as the abnormal distribution of decidual cells and the disruption of their interactions. However, the cellular and molecular mechanisms and the details of interactions among the different cell clusters in the decidua remain enigmatic, as do the potentially therapeutic mechanisms involved [12,13,14,15,16].
Mesenchymal stem cells (MSCs) arise mainly from the fat, bone marrow, and umbilical cord and comprise a class of stem cells with self-renewal and multidirectional differentiative potential [17,18,19]. Wharton’s jelly-derived MSCs (WJ-MSCs) isolated from human umbilical cords exhibit strong tissue repair and immunomodulatory potential [20, 21]. These cells can be obtained in vitro by noninvasive methods with few ethical implications, and their usefulness has become an area of intense research interest for stem cell therapy in regenerative medicine. WJ-MSCs are considered a promising and stable source of homozygous MSCs from umbilical cord tissue and constitute an ideal cell type for therapeutic use in areas such as developmental biology and regenerative medicine [22,23,24,25]. In recent years, WJ-MSCs are shown to be an effective agent in the treatment of various diseases, such as improving cognitive impairment in patients with Alzheimer’s disease, reducing liver and kidney damage from sepsis, and accelerating wound healing in patients with diabetes [26,27,28]. Nevertheless, our current understanding of RSA treatment via WJ-MSCs is limited, and the exact underlying mechanism(s) involved remains largely unclear.
Therefore, to further investigate the pathogenesis of RSA and the therapeutic potential of WJ-MSCs, we performed 10× Genomics single-cell RNA sequencing (scRNA-seq) by collecting decidual tissues from mice in normal pregnancy, RSA model mice, and a WJ-MSC treatment group of mice. We mapped the decidual single-cell transcript profiles of the three gestational groups of mice by analyzing the differential gene expression profiles of the five stromal cell types and identified 14 major cellular subpopulations. We observed that stromal cells were the most-represented cells in the decidual tissues and that different stromal cell subpopulations possessed a variety of functions. In addition, we observed inhibited communication among decidual cells, including aberrant interactions between stromal cells and NK cells, in RSA model group samples. Notably, we demonstrated at the single-cell level that caudal vein-grafted WJ-MSCs can repair defects in decidua of mice with RSA to a certain degree. In conclusion, we elucidated the possible mechanisms by which WJ-MSCs promote improved pregnancy outcomes at the single-cell level, providing a novel direction for the future clinical application of WJ-MSCs and effective treatment of RSA.
Results
Cellular heterogeneity within the decidua on pregnancy day 14
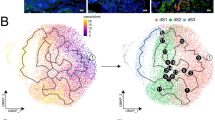
Although the cellular composition of the decidua at early and later stages has been investigated, the changes in cell types and gene expression in the RSA and WJ-MSCs-treated groups (RSA + MSCs) have not been elucidated. To analyze the cellular heterogeneity of decidual tissues in detail, cell suspensions from the mouse decidua of the normal (Nor), RSA, and RSA + MSC groups were subjected to single-cell transcriptomic sequencing on a 10× Genomics Chromium platform (Fig. 1A). There were a total of 14 distinct cell types in day 14 decidua, including five different stromal cells (Str), macrophages (Mac), endothelial cells (Endo), decidualized stromal cells (Dec), NK cells, epithelial cells (Epi), lymphatic endothelial cells (Lymph), pericytes (Peri), T-cells, and muscle cells (Mus). We visualized subpopulations of decidual cells by UMAP and annotated them according to marker genes (Fig. 1B, D and E). We noted that the proportions of stromal cell subpopulations appeared to change among the different subgroups. In the RSA group, the proportion of Str_0 cells was significantly reduced, while it was increased in the Str_1 group. After treatment with WJ-MSCs, the proportions of these subpopulations were similar to those in the normal group (Fig. 1C).
Characterization of gene expression in various stromal cells
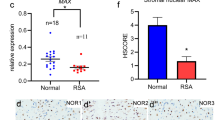
After blastocyst trophoblast cells invade the endometrium, the stromal cells surrounding the embryo differentiate into a primary decidual zone and continue to proliferate and differentiate into a secondary decidual zone that eventually transforms into decidual cells. There is also a population of undifferentiated stromal cells in the decidual zone, which, together with other decidual cells, provides nutrition and protection for the implanting embryo [29, 30]. Stromal cells were the most abundant cell type in the decidua, as demonstrated by the expression of the marker genes Rrm2 and Sfrp4 via single-cell resolution in situ hybridization on tissues (SCRINSHOT) (Fig. 1F, G). Uterine decidualization is a key event in the pregnancy process, is essential for the establishment and maintenance of pregnancy, and is characterized by massive proliferation, differentiation, and polyploidization of endometrial stromal cells at the site of implantation [31, 32]. These stromal cells are identified primarily by the expression of prolactin family 8, subfamily A, member 2 (Prl8a2), which is principally localized near the placenta (Fig. 1H).
To fully describe the molecular characteristics of these three major stromal cell types, we selected the genes that were highly expressed in each stromal cell type and performed functional enrichment analysis of the genes that were highly expressed by applying KEGG analysis (Fig. 2A–C). Str_0 cells exhibited highly expressed genes that were enriched mainly in the lysosome, proteasome, and ferroptosis pathways (Fig. 2C, D), and the genes highly expressed in Str_1 were enriched mainly in the spliceosome and endocytosis pathways (Fig. 2C, E). Commensurate with the continuous invasion of the embryo into the maternal uterus, the placental tissue requires transformation to facilitate the subsequent development of the foetus, which involves dynamic changes in trophoblasts and stromal cells. We noted that genes with high expression in the Str_2 group were associated with smooth muscle contraction and relaxin signaling pathways, indicating the relevance of this stromal subpopulation to muscle cells (Fig. 2F). This result also correlated with the expression of Oxtr in stromal cells that we observed in our previous study [33]. As muscle cells remain quiescent during pregnancy before labor with the distribution of smooth muscle cells among stromal cells, this stromal cell type might be related to these cells; the highly expressed genes in these stromal cells also appeared in the trackplot (Fig. 2G).
During the decidualization process after embryo implantation, stromal cells undergo continuous proliferation and differentiation to provide a suitable environment for subsequent embryonic development. We conducted RNA velocity and pseudotime analyses to explore the relationships among the different stromal cells involved in decidual development, and our results showed that among the three most abundant types of stromal cells, Str_0 had its own developmental trajectory, and some Str_0 cells also developed into Str_1 and finally into Str_2 (Fig. 3A–C). Additional potential fate-determining gene analysis during decidual development revealed that some specific genes were expressed during this developmental trajectory (Fig. 3D). Trackplot analysis also confirmed that the genes related to these trajectories exhibited cell type-specific expression patterns (Fig. 3E). Both the expression and developmental velocity of these genes were greater in the Str_0 cell type for Cpe and Cdo1 (Fig. 3F, H), while the expression and developmental velocity of Ptgis and Igf1 were greater in the Str_2 cell type (Fig. 3G, I).
Defective stromal development in RSA
To further characterize the differences among the three stromal cell types, we compared the cellular composition, gene expression, and functional enrichment of the stromal cells and demonstrated that the cellular composition, gene expression, and enrichment of the three groups of stromal cells were significantly altered (Fig. 4A–D). In the RSA group, the number of Str_0 cells was significantly reduced, and there was an increased proportion of Str_1 cells. After treatment with MSCs, the composition of these stromal cells was comparable to that in the normal group, indicating that the decreased number of Str_0 cells was related to RSA (Fig. 4A). Gene expression analysis of each stromal cell type suggested that there were more DEGs between the normal and RSA groups in Str_1 and Str_2 (Fig. 4B, C). Functional enrichment of these DEGs revealed that the IL17 and TNF signaling pathways were significantly enriched for RSA-related genes with upregulated expression in the Str_0, Str_1, and Str_2 groups, indicating that in RSA, defective stromal cell development was accompanied by increased inflammation (Fig. 4D). Among these DEGs, Lum and Fabp4 expression was substantially reduced in all RSA-affected stromal cells, while Parp1 and Atf4 expression (associated with apoptosis and ER stress) was significantly elevated in all RSA-affected stromal cells (Fig. 4E). The UMAP plots of the expression of Lum and Atf4 further confirmed this result (Fig. 4I, J).
Interactions among the three groups of stromal cells
We next aimed to decipher derailed the communication and homeostasis of the microenvironment in the maternal decidua. The number of interactions between different cell types was calculated with CellphongeDB depending on the expression of ligands and corresponding receptors. Our data indicated that there were fewer interactions between Str_1 cells and other cell types in the RSA group than in the normal and RSA + MSC groups (Fig. 5A-C). Str_1 cell-related gene expression was decreased in the RSA group and rescued in the RSA + MSC group (Fig. 5D–F). In addition, we noted that the interaction between NK cells and other cells decreased in the RSA group, further revealing the sub-healthy nature of the RSA decidua and reflecting an important role for NK cells in maternal decidua (Fig. 5D-F).
The aforementioned results indicated that the decreased stroma in the RSA group potentially affected the activity of NK cells. To thoroughly explore the cellular crosstalk between stromal cells and NK cells, NicheNet was utilized. We first identified the ligands released by NK cells that interact with stromal cells. Among the prioritized ligands, Spp1 was the ligand most abundantly expressed in NK cells, and the predicted target genes in the stroma include Jun, Icam1, and Nfkb1 (Fig. 6A), with potential receptors in the stroma including Cd44 and S1pr1. Interestingly, the expression of Cd44 was greater in Str_1 and Str_2 (Fig. 6B). However, when we investigated the crosstalk dominated by stroma-targeting NK cells, we observed that the ligands released by the stroma were primarily cytokines that regulate the expression of Gzmd and Gzmf, the major functional markers of NK cells in the decidua (Fig. 6C). This regulation was likely mediated by the binding of Cx3cl1 to Cx3cr1 (Fig. 6D). To investigate the global interaction of Spp1-related signaling in different cell types, we used CellChat and showed that there were strong interactions between NK cells and different stromal cells in the normal group (Fig. 6E). In contrast, except for NK cells, Spp1-related signaling became much more active in the epithelia of the RSA group and then declined to a normal level of activation in the MSC-treated group (Fig. 6F, G).
Spp1, or osteopontin (OPN), is a biomarker associated with endometrial tolerance [34]. We examined the expression of Spp1 and its receptors in different cell types, and our results revealed that Spp1 was chiefly expressed in NK cells and epithelial cells in the normal group and that its receptors Cd44, Itga3, and Itga5 were highly expressed in different stromal and other cell types (Fig. 7A-D). However, the expression of Spp1 in NK cells was significantly attenuated in the RSA group and restored after treatment with WJ-MSCs, and the distribution of expression of its receptor in the RSA group was also restored after treatment with WJ-MSCs (Fig. 7A-D). Analysis of the localization of Spp1 expression clearly revealed that Spp1 was principally expressed in NK cells through colocalization with DBA marked NK cells in the normal decidua and diminished expression of Spp1 in the RSA group (Fig. 7E). Analysis of the mRNA expression data also revealed that the receptors for Spp1 were expressed in the decidua (Fig. 7F).
Discussion
Although investigators applying single-cell sequencing have recently mapped the single-cell transcriptome of the decidua from RSA patients, the interactions between different cellular subpopulations in this tissue and the specific therapeutic mechanisms underlying WJ-MSC-related actions remain unclear [12,13,14,15,16]. Because the safety of using WJ-MSCs in pregnant women is unclear, our studies were conducted in model mice. We herein found that stromal cells were the most abundant cell type in normal maternal decidua and that the different types of stromal cells possess their own unique functional characteristics, consistent with previous studies [12]. The process of embryonic growth requires continuous and adequate decidualization of stromal cells for support [29, 33, 35]. Our study revealed that the proportion of Str_0 cells in the RSA group was lower than that in the normal group, suggesting that stromal cells do not provide adequate support for foetal development when RSA develops. There may be abnormal decidualization of stromal cells at the onset of RSA, which in turn leads to changes in the cellular composition of the decidual microenvironment.
We also demonstrated that stromal cells, particularly the Str_2 subpopulation, dominated reciprocal communication with other decidual cells and that this was principally manifested in interactions with muscle cells and endothelial cells. However, the activity of endothelial cells and Str_11 stromal cells was significantly increased in the RSA group, which was closely correlated with an imbalance in local homeostasis in the decidua during foetal absorption in dams experiencing RSA. In the present study, treatment with WJ-MSCs restored the activities of the majority of signaling pathways to emulate those of the normal pregnancy group. Since the major role of WJ-MSCs is to improve intercellular homeostasis via their secretion, the sequencing results of the present study not only supported this finding but also indicated that WJ-MSCs can effectively restore the intercellular interactions of the aforementioned signaling pathways; this finding revealed the greater therapeutic potential of WJ-MSCs for improving pregnancy outcomes in RSA at the single-cell level.
The balance of immune tolerance at the maternal–foetal interface is also critical for a successful pregnancy [36, 37]. NK cells constitute the major leukocyte population of the decidual microenvironment at the maternal–foetal interface in early pregnancy and play an important role in placental formation, trophoblast invasion, and decidual artery remodeling [38, 39]. NK cells are significant sources of cytokine secretion for the maintenance of pregnancy, and changes in their number, activity, and function are associated with pregnancy complications such as miscarriage and preeclampsia; however, the exact mechanisms underlying these effects remain unclear [40,41,42]. The activity of NK cells in mice experiencing RSA was diminished in this study, and our analysis showed that Spp1 signaling from NK cells that targeted the stroma was reduced in RSA; the interaction between epithelial cells and stromal cells then commensurately and significantly increased. Spp1 is hypothesized to be a marker of endometrial tolerance and is involved in embryonic implantation, and Spp1 expression has been detected in NK cells in the endometrium during early pregnancy [43, 44]. The present study provides strong single-cell evidence for the localization of Spp1 and its receptors Itga3, Itga5, and Cd44 in the decidua and experimentally verifies that Spp1 expression is reduced in RSA NK cells; however, the exact underlying mechanism of action of Spp1-targeted regulation of stromal cells during this process requires further investigation. Recent scRNA-seq analysis has also revealed that proinflammatory Spp1 + macrophages preferentially interact with decidual stromal cells (DSCs) in the decidua of RSA patients, that Mφ2-Spp1 is significantly enriched in inflammation-related pathways, and that there is a potential interaction between Cd44 + DSCs and Spp1 + macrophages [15]. Our study also revealed significant Spp1 expression in macrophages, but due to the small sample size, we did not further sort the macrophages. A more in-depth analysis of the single-cell sequencing data from this study will be performed in conjunction with available public databases.
Due to ethical constraints, the safety and efficacy of WJ-MSC administration in our specific group of RSA patients are still being explored. We herein demonstrated that WJ-MSCs acted on the decidua to effectively improve the decidual cell composition at the maternal–foetal interface and restore the modes of interaction between decidual cells. Overall, we have studied the use of WJ-MSCs for the treatment of RSA at the single-cell level. However, the mechanism(s) underlying the actions of WJ-MSCs still require further exploration.
Conclusions
In recent years, single-cell sequencing studies related to RSA have focused on developmental defects in stromal cells and the immune microenvironment of the decidua in RSA patients. We explored the pathogenesis of RSA to further clarify the role of WJ-MSCs in treating RSA, thereby promoting pregnancy maintenance and providing a theoretical basis for the clinical application of WJ-MSCs. In summary, through single-cell RNA sequencing, we mapped the single-cell profiles of the decidual tissues of three groups of mice: those that underwent normal pregnancy, those with RSA, and RSA mice that were treated with WJ-MSCs. We described the functional characteristics and pseudotemporal trajectories of each subpopulation of stromal cells and the major cellular population of the decidua, further resolved the characteristics of decidual heterogeneity, revealed the signaling pathways involved in the imbalance of cellular interactions in the decidual microenvironment in RSA and further illustrated the disruption of interactions between stromal cells and NK cells. Our results also revealed a potential therapeutic mechanism in which WJ-MSCs could be utilized to remodel the decidual subpopulation in RSA, providing novel insights into the developmental defects of the decidua at the maternal-foetal interface in RSA and the therapeutic potential of WJ-MSCs (Fig. 8).
Methods
Preparation of WJ-MSCs
We collected umbilical cord samples from a population of infants that had undergone uncomplicated full-term caesarean sections, and samples were obtained after the mothers signed an informed consent form. We stored the collected umbilical cord samples in DMEM/F-12 culture medium supplemented with 100 µg/ml streptomycin and 100 U/ml penicillin and isolated, cultured, and identified WJ-MSCs in the laboratory as described previously [45]. The principle of asepsis was strictly followed during the experiments.
Establishment of animal models
CBA/J (female, n = 50), BALB/c (male, n = 15), and DBA/2 (male, n = 15) laboratory mice, 6–8 weeks old, were purchased from Beijing HFK Bioscience Co. Ltd., China. The mice were housed in the Experimental Animal Center in a suitable environment (12-h light/dark cycles) and routinely fed for three days before being caged together. The CBA/J mice were mated with BALB/c and DBA/2 mice to establish our normal pregnancy model, CBA/J×BALB/c, and RSA model CBA/J×DBA/2. Vaginal plugs were checked on the morning of the second day of caging, and this day was designated the first day of pregnancy. Half of the mice with vaginal plugs observed in the RSA group constituted a model group for RSA treatment with WJ-MSCs. On the first, third, and fifth days of gestation, CBA/J mice were injected with 0.1 ml of PBS through the tail vein in the normal-gestation and RSA-model groups, and 0.1 ml of WJ-MSC suspension (concentration 1*107 cells/ml) was injected into the WJ-MSC treatment group. Mice were anaesthetised with 3% isoflurane on the 14th day of gestation, cervical dislocation was performed for tissue collection, and the decidual tissue was retained for subsequent 10× Genomics scRNA-seq and experimental validation. This study was approved by the Animal Ethics Committee of Nantong University (S20210302-913). This work has been reported in line with the ARRIVE guidelines 2.0.
10x Genomics single-cell RNA-seq of decidual tissue
The deciduae containing only maternal tissue from three individual on days 14 were surgically removed, mixed together, rinsed with ice-cold DPBS (Merck Millipore, D8537) and chopped using scalpels into small pieces and enzymatically digested in 5 mL digestion solution containing 1 mg/mL collagenase I (Worthington, LS004196) and 1 mg/mL collagenase V (Worthington, LS004188) in HBSS. The supernatant was diluted with HBSS and passed through 70-µm cell sieve and then 40-µm cell sieve (Biosharp, BS-40-CS). The flow-through was centrifuged at 500 g and resuspended in 1 mL of red blood cell lysis buffer for 3 min. The cells were collected by spinning down at 500 g for 5 min and re-suspended in DPBS and kept on ice for subsequent 10X scRNA sequencing.
Single-cell RNA-seq data processing
Reads were processed using the CellRanger (4.0.0) pipeline with the default and recommended parameters. The CellRanger output was then imported into Seurat (v4.0) for quality control and downstream analysis. All functions were run with default parameters unless otherwise specified. Low-quality cells (cells with a total UMI count per cell (library size) below 30,000, cells with < 500 genes per cell and cells with a content of mitochondrial genes > 20%) were excluded. Next, we used a cluster-level approach to remove potential doublet cells. In brief, the doublet score was calculated for each cell using the doubletCells function of the R package v.1.18.7. Cell clusters in each sample were identified by examining the top 50 principal components (PCs) across highly variable genes (HVGs), building a neighbor graph by the SNNGraph function, and then clustering using the cluster_louvain function from the igraph R package v.1.2.9. The median doublet score of each cell cluster was calculated using a median-centred MAD-variance normal distribution. Clusters with a median score above the extreme top end of this distribution were considered doublets. After filtering, the remaining 16,888 cells were retained for downstream analysis.
Cell clustering and annotation
The HVGs were selected using the highly_variable_genes function in Seurat. Nearest neighborhood graphs were built using the neighbors function, and the community algorithm was applied for clustering using the louvain function. The markers of characterized cell types in our single-cell RNA sequencing data were confirmed by FindAllMarkers. The major cell types identified in our dataset were annotated based on well-known marker genes.
Gene-clustering analysis
After averaging the expression of each gene in the different cell types using the AverageExpression function in Seurat, the genes expressed in the Str_0, Str_1, and Str_2 were clustered into distinct groups depending upon their expression patterns using the TCSeq R package. Genes highly expressed in the Str_0, Str_1, and Str_2 groups were ultimately subjected to KEGG analysis.
RNA velocity analysis
Read annotations of samples were performed by applying the velocyto.py (v0.17.17) command-line tool (velocyto run10x) using BAM, genome annotation, and duplicate annotation files. The BAM file was generated by the default parameters of Cell Ranger software (10× Genomics). Genomic annotations from the Cell Ranger prebuilt reference GRCm38 were used to count molecules while classifying them into three categories: “spliced,” “unspliced,” or “ambiguous.” Duplicate annotation files were downloaded from the UCSC Genome Browser. The velocyto.R package v0.6 was used to calculate RNA velocity values for the selected genes from each cell. Highly variably expressed genes calculated using the FindVariableFeatures function of Seurat were further filtered based on clustered expression, and the remaining highly variably expressed genes were used as the input for velocyto.R. Finally, the RNA velocity vectors were embedded in UMAP plots generated by the Seurat R package.
Cell–cell interaction analysis by CellPhoneDB
To investigate potential interactions across different cell types in the decidua, we conducted cell–cell interaction analysis using CellPhoneDB, a public database that is used to infer potential ligand–receptor interactions [1]. Enriched receptor–ligand interactions between two cell types were derived on the basis of the expression of a receptor by one cell type and the expression of the corresponding ligand by another cell type. We then identified the most relevant interactions between various cell types, and only receptors and ligands expressed in the cells in the corresponding clusters were considered. To identify potential ligands driving potential phenotypes, we used NicheNet (v.1.1.0) to investigate possible interactions and target genes between the indicated cell types.
CellChat analysis of cell–cell communication
To further analyse and compare the differences in intercellular communication among normal, RSA, and WJ-MSC-treated decidual samples, we used CellChat, an open-source R package (https://github.com/sqjin/CellChat), to analyse the scRNA-seq data. We first inferred intercellular communication among stromal cells and other cellular subsets separately for each of the three groups of datasets and then analyzed them together via joint manifold learning and classification of the inferred communication networks based on their functional similarities.
Single-cell resolution in situ hybridization
The padlock probes for the genes identified in this study were designed with PrimerQuest (https://eu.idtdna.com/PrimerQuest/Home/Index?_Display=AdvancedParams). Frozen tissue sections were removed from a − 80 °C freezer, equilibrated at 45 °C thermostat for 3 min, and fixed in 4% paraformaldehyde (PFA) for 1 h. The slides were then incubated with a blocking solution containing tRNA and oligo-dT sequences at room temperature (RT) for 30 min to block nonspecific binding of the probes to the tissue. Hybridization solution containing padlock probes (Supplementary Table 1) was added, and the mixture was incubated at 55 °C for 15 min for denaturation and at 45 °C for 2 h for hybridization. The mixture containing SplintR ligase was added, and the slides were incubated at 25 °C for 12–16 h. Phi 29 (Φ29) polymerase and RCA primer were added for rolling circle amplification (RCA) at 30 °C for 12–16 h. Finally, HRP-labelled detection probes were added and incubated at RT for 45–60 min in the dark, and the signal was displayed using TSA.
Immunofluorescence
Frozen sections of decidual tissue were fixed in 4% PFA for 1 h and washed three times for 5 min each with 0.05% PBS-Triton X-100. BSA blocking solution (5%) was then applied to the slides for 1 h at RT to block nonspecific binding within the tissue. We added primary antibodies to the sections and incubated them overnight at 4 °C, and secondary antibodies and DAPI were added and incubated for 1 h at RT (the antibodies used in this study are listed in Supplementary Table 2).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (v.9.0) (for experimental data), R (v.4.1.0), and RStudio (2021.09.1). Differences were considered significant at a p value < 0.05. Unless otherwise stated, each experiment was repeated three or more times with biologically independent samples.
Single cell transcription landscape of D14 decidua in normal, RSA and WJ-MSC mice. (A) The experimental workflow for single-cell transcriptome profiling in decidua. (B) scRNA-seq UMAP map of cell clusters in mouse decidual tissue. (C) The composition of decidual cells in normal group (Nor), RSA group (RSA), and WJ-MSCs treatment group (Tr). (D) Dot plots of specific expression molecules of different decidual cell types in the normal decidua. (E) The Pearson correlation of gene expression of different groups. (F-H) The UMAP visualization and the expression of Rrm2, Sfrp4 and Prl8a2 in situ hybridization in normal decidua
Functional characteristics of different stromal cells in normal decidua. (A) Genes highly express in Str_0, Str_1, and Str_2 by gene expression clustering analysis. (B) Heatmap of genes highly express in Str_0, Str_1, and Str_2. (C) KEGG enrichment of genes highly express in Str_0, Str_1, and Str_2. (D) Genes that are specifically highly expressed and associated with Ferroptosis in Str_0. (E) Genes that are specifically highly expressed and associated with Spliceosome in Str_1. (F) Genes that are specifically highly expressed and associated with Smooth muscle contraction in Str_2. (G) TracksPlot of genes highly express in Str_0, Str_1, and Str_2
The RNA Velocity and pseudotime sequence analysis of different stromal cells in normal decidua. (A, C) The RNA Velocity map of stromal cells in Str_0, Str_1, and Str_2. (B) Pseudotime of stromal cells in Str_0, Str_1, and Str_2. (D) Heatmap of genes highly express in the pseudotime maps of Str_0, Str_1, and Str_2. (E) TracksPlot of genes highly express in the pseudotime maps of Str_0, Str_1, and Str_2. (F-I) The RNA Velocity and expression map of Cpe, Ptgis, Cdo1, Igf1 in Str_0, Str_1, and Str_2
The RNA Velocity and pseudotime sequence analysis of different stromal cells in normal decidua. (A) Stromal cells composition in Str_0, Str_1, and Str_2. (B) Scatterplots depict differentially expressed genes in Str_0, Str_1, and Str_2 between normal and RSA group. Red and blue represent genes specifically expressed in normal and RSA group, respectively. (C) Heatmap of genes highly express in Str_0, Str_1, and Str_2. (D) KEEG enrichment of genes highly express in Str_0, Str_1, and Str_2. (E-H) Quantitative expression of Lum, Fabp4, Prap1, Atf4 in distinct stromal cells. Wilcoxon tests. (I, J) UMAP visualization of the expression of Lum and Atf4 in Str_0, Str_1, and Str_2
The interaction between NK cells and stromal cells in decidua. (A) The interaction between NK cells and stromal cells. The most left is the prioritized ligands defined by NicheNet. Pearson correlation indicates the ability of each ligand to its target genes, and better predictive ligands are thus ranked higher. The dot plots represent the expression of ligands in NK cells and their target genes in different stromal cells. Heatmap shows the predicted ligands activity by NicheNet on their target genes in different stromal cells. (B) Heatmap shows the bona fide interactions between the ligands in NK cells and their receptors in different stromal cells. Dot plots represent the average expression of ligands and their receptors in senders and receivers, respectively. (C) The interaction between NK cells and stromal cells. The most left is the prioritized ligands defined by NicheNet. The dot plots represent the expression of ligands in different stromal cells and their target genes in NK cells. (D) Heatmap shows the bona fide interactions between the ligands in different stromal cells and their receptors in NK cells. (E-G) Spp1-mediated signaling pathways in normal, RSA, and treatment group
Spp1-mediated interactions in decidua. (A-C) Expression of Spp1 and its receptors Cd44, Itga3 and Itga5 in decidua. (D) Quantitative expression of Spp1 in NK_7 cells. Wilcoxon tests. (E) In situ hybridization of Spp1 (red), DAPI (blue) and immunofluorescence staining of NG2 (green) in the normal, RSA, and WJ-MSCs groups. (F) In situ hybridization of Itga3 (red), Itga5 (red), Cd44 (red), and DAPI (blue) in the normal decidua
Data availability
Raw data generated in this study have been deposited at the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA1048538 (reviewer token: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1048538?reviewer=93v363k69m9433lo0lhij9dqov).
Abbreviations
- RSA:
-
Recurrent spontaneous abortion
- WJ-MSCs:
-
Wharton’s jelly-derived mesenchymal stem cells
- scRNA-seq:
-
Single-cell RNA-sequencing
- SCRINSHOT:
-
Single-cell resolution in-situ hybridization on tissues
- PCs:
-
Principal components
- HVGs:
-
Highly variable genes
- PFA:
-
Paraformaldehyde
- RT:
-
Room temperature
- RCA:
-
Rolling circle amplification
- Nor:
-
Normal
- Str:
-
Stromal cells
- Mac:
-
Macrophages
- Endo:
-
Endothelial cells
- Dec:
-
Decidualized stromal cells
- Epi:
-
Epithelial cells
- Lymph:
-
Lymphatic endothelial cells
- Peri:
-
Pericytes
- Mus:
-
Muscle cells
- Prl8a2:
-
Prolactin family 8, subfamily A, member 2
- OPN:
-
osteopontin
- DSCs:
-
decidual stromal cells
References
Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–53. https://doi.org/10.1038/s41586-018-0698-6.
Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018;28:819–32. https://doi.org/10.1038/s41422-018-0066-y.
Coomarasamy A, Dhillon-Smith RK, Papadopoulou A, Al-Memar M, Brewin J, Abrahams VM, et al. Recurrent miscarriage: evidence to accelerate action. Lancet. 2021;397:1675–82. https://doi.org/10.1016/s0140-6736(21)00681-4.
Dimitriadis E, Menkhorst E, Saito S, Kutteh WH, Brosens JJ. Recurrent pregnancy loss. Nat Rev Dis Primers. 2020;6:98. https://doi.org/10.1038/s41572-020-00228-z.
Quenby S, Gallos ID, Dhillon-Smith RK, Podesek M, Stephenson MD, Fisher J, et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. 2021;397:1658–67. https://doi.org/10.1016/s0140-6736(21)00682-6.
van Wely M. Series of overviews on miscarriage and recurrent miscarriage. Fertil Steril. 2023;120:932–3. https://doi.org/10.1016/j.fertnstert.2023.09.006.
Yao Y, Ye Y, Chen J, Zhang M, Cai X, Zheng C. Maternal-fetal immunity and recurrent spontaneous abortion. Am J Reprod Immunol. 2024;91:e13859. https://doi.org/10.1111/aji.13859.
Deng W, Cha J, Yuan J, Haraguchi H, Bartos A, Leishman E, et al. p53 coordinates decidual sestrin 2/AMPK/mTORC1 signaling to govern parturition timing. J Clin Invest. 2016;126:2941–54. https://doi.org/10.1172/jci87715.
Cha J, Bartos A, Egashira M, Haraguchi H, Saito-Fujita T, Leishman E, et al. Combinatory approaches prevent preterm birth profoundly exacerbated by gene-environment interactions. J Clin Invest. 2013;123:4063–75. https://doi.org/10.1172/jci70098.
Hirota Y, Cha J, Yoshie M, Daikoku T, Dey SK. Heightened uterine mammalian target of rapamycin complex 1 (mTORC1) signaling provokes preterm birth in mice. Proc Natl Acad Sci U S A. 2011;108:18073–8. https://doi.org/10.1073/pnas.1108180108.
Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest. 2010;120:803–15. https://doi.org/10.1172/jci40051.
Du L, Deng W, Zeng S, Xu P, Huang L, Liang Y, et al. Single-cell transcriptome analysis reveals defective decidua stromal niche attributes to recurrent spontaneous abortion. Cell Prolif. 2021;54:e13125. https://doi.org/10.1111/cpr.13125.
Wang F, Jia W, Fan M, Shao X, Li Z, Liu Y, et al. Single-cell Immune Landscape of Human recurrent miscarriage. Genomics Proteom Bioinf. 2021;19:208–22. https://doi.org/10.1016/j.gpb.2020.11.002.
Guo C, Cai P, Jin L, Sha Q, Yu Q, Zhang W, et al. Single-cell profiling of the human decidual immune microenvironment in patients with recurrent pregnancy loss. Cell Discov. 2021;7:1. https://doi.org/10.1038/s41421-020-00236-z.
Bao S, Chen Z, Qin D, Xu H, Deng X, Zhang R, et al. Single-cell profiling reveals mechanisms of uncontrolled inflammation and glycolysis in decidual stromal cell subtypes in recurrent miscarriage. Hum Reprod. 2023;38:57–74. https://doi.org/10.1093/humrep/deac240.
Pan D, Liu Q, Du L, Yang Y, Jiang G. Polarization disorder of decidual NK cells in unexplained recurrent spontaneous abortion revealed by single-cell transcriptome analysis. Reprod Biol Endocrinol. 2022;20:108. https://doi.org/10.1186/s12958-022-00980-9.
Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128–39. https://doi.org/10.1038/cdd.2015.168.
Lelek J, Zuba-Surma EK. Perspectives for future use of Extracellular vesicles from umbilical cord- and adipose tissue-derived mesenchymal Stem/Stromal cells in regenerative therapies-synthetic review. Int J Mol Sci. 2020;21. https://doi.org/10.3390/ijms21030799.
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. https://doi.org/10.1126/science.284.5411.143.
Shin YH, Choi SJ, Kim JK. Mechanisms of Wharton’s Jelly-derived MSCs in enhancing peripheral nerve regeneration. Sci Rep. 2023;13:21214. https://doi.org/10.1038/s41598-023-48495-6.
Chen P, Tang S, Li M, Wang D, Chen C, Qiu Y, et al. Single-cell and spatial Transcriptomics decodes Wharton’s jelly-derived mesenchymal stem cells heterogeneity and a subpopulation with Wound Repair signatures. Adv Sci (Weinh). 2023;10:e2204786. https://doi.org/10.1002/advs.202204786.
Musiał-Wysocka A, Kot M, Sułkowski M, Badyra B, Majka M. Molecular and Functional Verification of Wharton’s Jelly Mesenchymal Stem Cells (WJ-MSCs) Pluripotency. Int J Mol Sci. 2019;20. https://doi.org/10.3390/ijms20081807.
Joerger-Messerli MS, Marx C, Oppliger B, Mueller M, Surbek DV, Schoeberlein A. Mesenchymal stem cells from Wharton’s Jelly and amniotic fluid. Best Pract Res Clin Obstet Gynaecol. 2016;31:30–44. https://doi.org/10.1016/j.bpobgyn.2015.07.006.
Abbaszadeh H, Ghorbani F, Derakhshani M, Movassaghpour AA, Yousefi M, Talebi M, et al. Regenerative potential of Wharton’s jelly-derived mesenchymal stem cells: a new horizon of stem cell therapy. J Cell Physiol. 2020;235:9230–40. https://doi.org/10.1002/jcp.29810.
Pu L, Meng M, Wu J, Zhang J, Hou Z, Gao H, et al. Compared to the amniotic membrane, Wharton’s jelly may be a more suitable source of mesenchymal stem cells for cardiovascular tissue engineering and clinical regeneration. Stem Cell Res Ther. 2017;8:72. https://doi.org/10.1186/s13287-017-0501-x.
Hour FQ, Moghadam AJ, Shakeri-Zadeh A, Bakhtiyari M, Shabani R, Mehdizadeh M. Magnetic targeted delivery of the SPIONs-labeled mesenchymal stem cells derived from human Wharton’s jelly in Alzheimer’s rat models. J Control Release. 2020;321:430–41. https://doi.org/10.1016/j.jconrel.2020.02.035.
Cóndor JM, Rodrigues CE, Sousa Moreira R, Canale D, Volpini RA, Shimizu MH, et al. Treatment with Human Wharton’s jelly-derived mesenchymal stem cells attenuates Sepsis-Induced kidney Injury, Liver Injury, and endothelial dysfunction. Stem Cells Transl Med. 2016;5:1048–57. https://doi.org/10.5966/sctm.2015-0138.
Kassem DH, Kamal MM. Wharton’s Jelly MSCs: potential Weapon to sharpen for our battle against DM. Trends Endocrinol Metab. 2020;31:271–3. https://doi.org/10.1016/j.tem.2020.01.001.
Wang X, Matsumoto H, Zhao X, Das SK, Paria BC. Embryonic signals direct the formation of tight junctional permeability barrier in the decidualizing stroma during embryo implantation. J Cell Sci. 2004;117:53–62. https://doi.org/10.1242/jcs.00826.
Paria BC, Zhao X, Das SK, Dey SK, Yoshinaga K. Zonula occludens-1 and E-cadherin are coordinately expressed in the mouse uterus with the initiation of implantation and decidualization. Dev Biol. 1999;208:488–501. https://doi.org/10.1006/dbio.1999.9206.
Ramathal CY, Bagchi IC, Taylor RN, Bagchi MK. Endometrial decidualization: of mice and men. Semin Reprod Med. 2010;28:17–26. https://doi.org/10.1055/s-0029-1242989.
Lim HJ, Wang H. Uterine disorders and pregnancy complications: insights from mouse models. J Clin Invest. 2010;120:1004–15. https://doi.org/10.1172/jci41210.
Zhao H, Wang Y, Xu H, Liu M, Xu X, Zhu S, et al. Stromal cells-specific retinoic acid determines parturition timing at single-cell and spatial-temporal resolution. iScience. 2023;26:107796. https://doi.org/10.1016/j.isci.2023.107796.
Casals G, Ordi J, Creus M, Fábregues F, Carmona F, Casamitjana R, et al. Expression pattern of osteopontin and αvβ3 integrin during the implantation window in infertile patients with early stages of endometriosis. Hum Reprod. 2012;27:805–13. https://doi.org/10.1093/humrep/der432.
Akaeda S, Aikawa S, Hirota Y. Spatial and molecular anatomy of the endometrium during embryo implantation: a current overview of key regulators of blastocyst invasion. Febs j. 2024. https://doi.org/10.1111/febs.17077.
Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31:387–411. https://doi.org/10.1146/annurev-immunol-032712-100003.
Horvat Mercnik M, Schliefsteiner C, Sanchez-Duffhues G, Wadsack C. TGFβ signalling: a nexus between inflammation, placental health and preeclampsia throughout pregnancy. Hum Reprod Update. 2024;30:442–71. https://doi.org/10.1093/humupd/dmae007.
Fu B, Wei H. Decidual natural killer cells and the immune microenvironment at the maternal-fetal interface. Sci China Life Sci. 2016;59:1224–31. https://doi.org/10.1007/s11427-016-0337-1.
Moffett A, Colucci F. Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest. 2014;124:1872–9. https://doi.org/10.1172/jci68107.
Moffett A, Shreeve N. Local immune recognition of trophoblast in early human pregnancy: controversies and questions. Nat Rev Immunol. 2023;23:222–35. https://doi.org/10.1038/s41577-022-00777-2.
Hu J, Guo Q, Liu C, Yu Q, Ren Y, Wu Y, et al. Immune cell profiling of preeclamptic pregnant and postpartum women by single-cell RNA sequencing. Int Rev Immunol. 2024;43:1–12. https://doi.org/10.1080/08830185.2022.2144291.
Hou R, Huang R, Zhou Y, Lin D, Xu J, Yang L, et al. Single-cell profiling of the microenvironment in decidual tissue from women with missed abortions. Fertil Steril. 2023;119:492–503. https://doi.org/10.1016/j.fertnstert.2022.12.016.
Wang XB, Qi QR, Wu KL, Xie QZ. Role of osteopontin in decidualization and pregnancy success. Reproduction. 2018;155:423–32. https://doi.org/10.1530/rep-17-0782.
Kramer AC, Erikson DW, McLendon BA, Seo H, Hayashi K, Spencer TE, et al. SPP1 expression in the mouse uterus and placenta: implications for implantation†. Biol Reprod. 2021;105:892–904. https://doi.org/10.1093/biolre/ioab125.
Shi Q, Gao J, Jiang Y, Sun B, Lu W, Su M, et al. Differentiation of human umbilical cord Wharton’s jelly-derived mesenchymal stem cells into endometrial cells. Stem Cell Res Ther. 2017;8:246. https://doi.org/10.1186/s13287-017-0700-5.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2022YFC2704500 and 2022YFC2704600 to W.D.), the National Natural Science Foundation of China (82122026, 32171117, and 81971419 to W.D.), and the General Project of Nantong Natural Science Foundation of China (JC2023086 to X.Y.). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
W.D., B.J., X.D., J.D., C.P., C.Z., Q.W., X.C., R.Q., X.D., and H.D. performed the experiments and prepared the figures; X.Y. and W.D. designed the experiments; X.Y., W.D., B.J., and X.D. analyzed the data; and X.Y., W.D., B.J., and X.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved for implementation by the Animal Ethics Committee of Nantong University (approval number: S20210302-913, approved date: 02/03/2021). The approved project is “Effect and mechanism of human umbilical cord mesenchymal stem cells on recurrent spontaneous abortion decidua”. This study procedure was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (approval number: 2021-L106, approved date: 28/02/2021). The approved project is “A study of the role and therapeutic mechanism of uterine decidua heterogeneity in recurrent spontaneous abortion”.
Consent for publication
All authors agree to publish this manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jin, B., Ding, X., Dai, J. et al. Deciphering decidual deficiencies in recurrent spontaneous abortion and the therapeutic potential of mesenchymal stem cells at single-cell resolution. Stem Cell Res Ther 15, 228 (2024). https://doi.org/10.1186/s13287-024-03854-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-024-03854-6