Abstract
Background
Inner ear supporting cells (SCs) in the neonatal mouse cochlea are a potential source for hair cell (HC) regeneration, but several studies have shown that the regeneration ability of SCs decreases dramatically as mice age and that lost HCs cannot be regenerated in adult mice. To better understand how SCs might be better used to regenerate HCs, it is important to understand how the gene expression profile changes in SCs at different ages.
Methods
Here, we used Sox2GFP/+ mice to isolate the Sox2+ SCs at postnatal day (P)3, P7, P14, and P30 via flow cytometry. Next, we used RNA-seq to determine the transcriptome expression profiles of P3, P7, P14, and P30 SCs. To further analyze the relationships between these age-related and differentially expressed genes in Sox2+ SCs, we performed gene ontology (GO) analysis.
Results
Consistent with previous reports, we also found that the proliferation and HC regeneration ability of isolated Sox2+ SCs significantly decreased as mice aged. We identified numerous genes that are enriched and differentially expressed in Sox2+ SCs at four different postnatal ages, including cell cycle genes, signaling pathway genes, and transcription factors that might be involved in regulating the proliferation and HC differentiation ability of SCs. We thus present a set of genes that might regulate the proliferation and HC regeneration ability of SCs, and these might serve as potential new therapeutic targets for HC regeneration.
Conclusions
In our research, we found several genes that might play an important role in regulating the proliferation and HC regeneration ability of SCs. These datasets are expected to serve as a resource to provide potential new therapeutic targets for regulating the ability of SCs to regenerate HCs in postnatal mammals.
Similar content being viewed by others
Introduction
Hair cells (HCs) in the inner ear play a critical role in converting mechanical sound waves into neural signals for hearing and play a critical role in maintaining balance [1, 2]. Multiple studies have reported that HCs in non-mammalian vertebrates can be regenerated in both the auditory and vestibular systems after HC loss and thus lead to the complete recovery of hearing and balance function [3, 4]. Conversely, HCs in the mammalian cochlea can be spontaneously regenerated after damage only to a very limited extent and only in the neonatal cochlea and cannot be regenerated at all in adult animals, and thus in adult mammals, HC damage causes permanent hearing loss [1, 4,5,6,7,8,9,10]. Finding a way to regenerate HCs in mammals could possibly represent a cure for sensorineural hearing loss, which still has no treatment options other than prosthetic devices.
In the mouse organ of Corti, HCs and supporting cells (SCs) emerge from the same inner ear prosensory cells. These inner ear prosensory cells start to exit the cell cycle from the apical turn to the basal turn of the cochlea. The apical prosensory cells exit the cell cycle at around embryonic day 12.5 (E12.5), and the basal prosensory cells exit the cell cycle at around E14.5. The inner ear prosensory cells start to differentiate into HCs and SCs beginning at the mid-base of the cochlea at around E13.5 and reaching the rest of the base and up to the apex of the cochlea over the next few days [11]. SCs in the mouse inner ear have also been shown to be a reliable source for regenerating HCs after damage in vitro either through mitotic or direct differentiation [10, 12,13,14,15]. Recent studies have demonstrated that the SCs isolated from the neonatal mouse cochlea are competent to form new HCs in culture [10, 16,17,18], but the ability of SCs to form spheres in suspension cultures decreases about 100-fold during the second and third postnatal weeks [19]. In contrast, the adult mammalian cochlea has almost no HC regeneration capacity, and attempts to stimulate the dormant regenerative capacity have met with very limited success [15, 20]. Multiple factors have been reported to be involved in regulating the process by which SCs regenerate HCs, including factors in the Wnt, Notch, Hedgehog, and STAT3 signaling pathways [10, 21,22,23,24]. HC regeneration strategies have only worked at all in the neonatal mouse cochlea, and none has been able to overcome the age barrier in the adult cochlea. An obvious limitation to these previous strategies has been a lack of understanding of the age-related changes in gene expression profiles, and possible age-related genes regulating the proliferation and HC regeneration ability of SCs have not been identified.
Sox2 is a universal stem cell marker, and it is also expressed in neural progenitor cells at different stages of central nervous system development [25]. In the neonatal mouse inner ear, Sox2 labels the SCs that have been shown to be a reliable source for regenerating HCs after damage. In this study, we performed RNA-seq profiling of the Sox2+ SCs isolated from Sox2GFP/+ transgenic mice at four different postnatal time points and determined the age-related differential expression of genes that might be involved in regulating the proliferation and HC differentiation ability of Sox2+ SCs. The Sox2+ SCs we sorted included Hensen’s cells, Deiters’ cells, pillar cells, inner phalangeal cells, and the cells in the greater epithelium ridge. To further analyze the role of these age-related differentially expressed genes, we constructed a protein–protein interaction network using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins). These datasets are expected to serve as a resource to provide potential new therapeutic targets for regulating the ability of SCs to regenerate HCs in postnatal mammals.
Materials and methods
Mice and genotyping
Sox2GFP/+ mice were obtained from the Jackson Laboratory (stock no. 17592). Transgenic mice were genotyped using genomic DNA from tail tips by adding 180 μl 50 mM NaOH, incubating at 98 °C for 1 h, and then adding 20 μl 1 M Tris-HCl to neutralize the base. The genotyping primers were as follows: GFP forward: 5′-CAC ATG AAG CAG CAC GAC TT-3′; GFP reverse: 5′-TGC TCA GGT AGT GGT TGT CG-3′.
The cochleae were harvested at P3, P7, P14, and P30. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All animal procedures were performed according to protocols approved by the Animal Care and Use Committee of Southeast University and were consistent with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All efforts were made to minimize the number of animals used and to prevent their suffering.
Immunofluorescence
The dissected cochleae or the cultured cells were fixed in 4% paraformaldehyde for 1 h at room temperature, washed three times for 3 min with 1× PBST (0.1% Triton X-100 in PBS), and incubated for 1 h at room temperature in blocking medium (1% Triton X-100, 1% BSA, 10% heat-inactivated donkey serum, and 0.02% sodium azide in PBS at pH 7.2). The primary antibody was diluted in PBT-1 (10% Triton X-100, 1% BSA, 5% heat-inactivated goat serum, and 0.02% sodium azide in PBS at pH 7.2) and incubated with the samples overnight at 4 °C. The samples were washed three times for 3 min with 1× PBST, and the secondary antibody diluted in PBT-2 (0.1% Triton X-100 and 1% BSA in PBS at pH 7.2) was added for 1 h at room temperature. The samples were washed again three times with 1× PBST and then mounted on slides in a mounting medium (DAKO, S3023). Cells were imaged with an LSM700 confocal microscope. The antibodies used in this study were anti-myosin7a (Proteus Bioscience, #25-6790, 1:1000 dilution), anti-sox2 (Santa Cruz, #sc-17320, 1:500 dilution), Alexa Fluor 647 donkey anti-goat IgG (Invitrogen, A-21447, 1:500 dilution), and Alexa Fluor 555 donkey anti-rabbit IgG (Invitrogen, A-31572, 1:500 dilution).
Flow cytometry
The cochleae were dissected in cold 1× HBSS (Gibco) and transferred to 50 μl 1× PBS in 1.5-ml Eppendorf tubes. A total of 50 μl 0.25% trypsin-EDTA (Invitrogen; #25200-056) was added to the tubes, and these were incubated for 8–12 min at 37 °C. The digestion was stopped by the addition of 50 μl trypsin inhibitor (Worthington Biochem, #LS003570), and 200-μl (Eppendorf, #22491245) and 1000-μl (Eppendorf, #22491253) blunt pipette tips were used to triturate the tissues into single cell suspensions. The cells were filtered through a 40-μm strainer (BD Biosciences, 21008-949) to eliminate clumps, and the GFP+ cells were sorted on a BD FACS Aria III flow cytometer (BD Biosciences).
Sphere-forming assay and differentiation assay
For the sphere-forming assay, the flow-sorted Sox2+ SCs were cultured at a density of 2 cells/μl in Costar ultra-low attachment dishes (Costar, 3473) for 5 days in DMEM/F12 (Gibco, 11330-032), 2% B27 (Invitrogen, 17504-044), 1% N2 (Invitrogen, 17502-048), IGF (50 ng/ml, Sigma, I8779), EGF (20 ng/ml; Sigma, E9644), b-FGF (10 ng/ml, Sigma, F0291), heparan sulfate (20 ng/ml, Sigma, H4777), and 0.1% ampicillin (Sigma, A9518-5G). For the differentiation assay, we used both flow-sorted GFP+ SCs and spheres from the sphere-forming assay. In the cell-differentiation assay, the flow-sorted Sox2+ SCs were cultured at a density of 50 cells/μl on laminin-coated four-well dishes for 10 days in DMEM/F12, 1% N2, 2% B27, EGF (20 ng/ml; Sigma, E9644), IGF (50 ng/ml, Sigma, I8779), heparan sulfate (20 ng/ml, Sigma, H4777), b-FGF (10 ng/ml, Sigma, F0291), and 0.1% ampicillin. In the sphere-differentiation assay, the first-generation spheres were seeded on laminin-coated four-well dishes and cultured for 10 days in DMEM/F12 medium with 1% N2, 2% B27, and 0.1% ampicillin.
RNA extraction for RNA-seq analysis
Approximately 5000 GFP+ SCs were isolated by FACS and split into three fractions for separate replicates. RNA-seq libraries of FACS-purified cells were generated using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing and the Illumina mRNA-Seq Sample Prep Kit. FACS-purified cells were suspended in 10× lysis buffer. First-strand and second-strand cDNA synthesis, adaptor ligation, and PCR amplification were performed using the Illumina mRNA-Seq Sample Prep Kit. SPRI beads (Ampure XP, Beckman) were used in each purification step after RNA fragmentation for size selection. All libraries were analyzed for quality and concentration using an Agilent Bioanalyzer. Sequencing was performed using the Illumina HiSeq2500 150-bp Paired-End Platform, and FASTQ files of paired-end read files were generated.
Quantitative real-time PCR
We used the RNeasy Micro Kit (QIAGEN, 74004) to extract the total RNA from ~ 20,000 FACS-sorted GFP+ SCs, and the RevertAid First Strand cDNA Synthesis Kit (Thermo, K1622) was used to synthesize cDNA. Real-time PCR was carried out by using the FastStart Universal SYBR Green Master (Rox) (Roche, 04913914001) on a Bio-Rad C1000 Touch thermal cycler. The expression levels of the target genes were normalized to Gapdh and the q-PCR primers are listed in Additional file 1.
Data analysis
The trimmomatic software was used to trim the RNA-seq reads in the FASTQ files. Clean reads were mapped to the mouse reference genome (mm9) using TopHat followed by transcript assembly and differential gene expression analysis using Cufflinks [26]. Genes and transcripts were annotated using the RefGene database (NCBI). Genes with a p value of less than or equal to 0.05 were considered significant. Gene ontology (GO) analysis with the functional annotation tool DAVID 6.7 was performed to assess the extent of functional enrichment [27], which determines whether biological processes are enriched within a list of genes. Protein functional association analysis was performed using STRING on genes in top enriched GO categories.
Statistical analysis
All of the data presented in the text are means ± standard deviations, and we used GraphPad Prism 6 for statistical analysis. For all experiments, n represents the number of replicates, and at least three individual experiments were conducted. Two-tailed, unpaired Student’s t tests were used to determine statistical significance when comparing two groups, and one-way ANOVA followed by a Dunnett’s multiple comparisons test was used when comparing more than two groups. A p value < 0.05 was considered to be statistically significant.
Results
Neonatal SCs have higher sphere-forming ability compared with older SCs in vitro
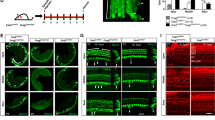
First, we performed an immunofluorescence assay to observe the GFP expression pattern in Sox2GFP/+ mice, and we found that GFP was mainly expressed in Hensen’s cells, Deiters’ cells, pillar cells, inner phalangeal cells, and the greater epithelium ridge in the P3 mouse cochlea (Fig. 1a, b). We then used flow cytometry to sort the Sox2+ SCs from cochleae dissected from mice at P3, P7, P14, and P30, and these made up 6.19% of the viable cells in the P3 mice, 4.59% of the viable cells in P7 mice, 2.07% of the viable cells in the P14 mice, and 1.11% of the viable cells in the P30 mice (Fig. 1c). We observed that the proportion of Sox2+ cells gradually decreased with age, and this might be because the increasing ossification with age made the dissection and dissociation of the organ of Corti more difficult at older ages. We then performed immunofluorescence to double confirm the sorted cells and found that at P3 94.9 ± 2.3% and 94.5% ± 2.31% of the sorted cells were Sox2+ and GFP+, respectively, while none of the sorted cells was Myo7a+ (Fig. 1d, e), suggesting that the flow-sorted cells were almost all Sox2+ SCs and that the sorted cells were highly pure.
The purity of sorted GFP+ cells. a Immunostaining in the HC layer showed no GFP+ cells co-labeled with HCs. b In the SC layer, GFP+ cells co-labeled with Sox2+ SCs. c Different ages of Sox2GFP/+ mouse cochleae were dissected and dissociated into single cells, and the Sox2+ SCs were sorted via flow cytometry. The proportions of Sox2+ cells were 6.19% at P3, 4.59% at P7, 2.07% at P14, and 1.11% at P30. d, e Immunostaining of flow-sorted Sox2+ SCs of different ages showed a high percentage of Sox2+ and GFP+ cells, and no Myo7a+ cells were found. Scale bars are 20 μm in a and b and 50 μm in d and e
We next performed a sphere-forming assay using P3, P7, P14, and P30 SCs. A total of 200 isolated cells were plated onto a 96-well ultra-low attachment plate at a density of 2 cells/μl for 5 days (Fig. 2a). We evaluated the proliferation capacity of the SCs by quantifying the numbers and diameters of the spheres. Consistent with previous reports [19], we found that 200 P3 Sox2+ SCs could form around 7 spheres/well and that the diameter of each sphere was more than 70 μm (Fig. 2b). However, the spheres were fewer and smaller from P7 Sox2+ SCs and were even fewer and smaller from P14 Sox2+ SCs (Fig. 2b, c). No spheres were observed from the P30 Sox2+ SCs (Fig. 2b, c). The greater sphere-forming ability of P3 SCs suggests that the neonatal (P3) SCs possess greater proliferation ability than aged (P7, P14, P30) SCs.
The neonatal SCs have greater sphere-forming ability than the older SCs. a The Sox2GFP/+ mice were harvested at P3, P7, P14, and P30. Flow cytometry was used to isolate the different ages of Sox2+ SCs, and these cells were cultured for 5 days. b, c P3 Sox2+ SCs generated significantly more and larger spheres than P7 and P14 Sox2+ SCs, while the P30 Sox2+ SCs could not form spheres. d The cultured cells in the first generation were used for the differentiation assay. e The spheres formed by P3 Sox2+ SCs stained with the HC marker Myo7a. f The spheres formed by P7 Sox2+ SCs stained with the HC marker Myo7a. g The spheres formed by P30 Sox2+ SCs stained with the HC marker Myo7a. h The average number of HCs generated by spheres of each age Sox2+ SCs. i The total number of HCs generated by P3, P7, P14, and P30 Sox2+ SCs. ***p < 0.001. Scale bars are 50 μm in b and 10 μm in e–g
In order to further evaluate the HC regeneration ability of these spheres, we isolated the spheres derived from P3, P7, and P14 SCs and differentiated those spheres for 10 days and then immunostained them with the HC marker Myo7a (Fig. 2d). We counted the Myo7a+ HCs in each differentiated sphere and calculated the total Myo7a+ HCs that were generated from the original 200 flow cytometry-isolated Sox2+ SCs. We found that the P3 Sox2+ SC spheres generated significantly more Myo7a+ HCs than the P7 and P14 Sox2+ SC spheres (Fig. 2e–i). In summary, these results support prior findings that neonatal (P3) SCs have a greater capacity to form spheres than aged (P7, P14, P30) SCs and that the spheres formed from neonatal SCs can generate more HCs than spheres formed from aged SCs.
Neonatal SCs have a greater capacity to regenerate HCs compared with aged SCs in vitro
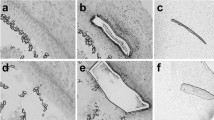
Most inner ear cell differentiation occurs during embryonic development, but the neonatal mouse retains a limited ability to regenerate HCs through the differentiation of SCs. This ability is quickly lost, however, and by the first week after birth, there is a notable decline in this regenerative activity. We cultured 5000 isolated Sox2+ P3, P7, P14, and P30 SCs in laminin-coated four-well dishes at a density of 50 cells/μl for 10 days and then immunostained them with the HC marker Myo7a (Fig. 3a). We found that the P3 SCs generated significantly more Myo7a+ colonies than the P7 SCs, while no colonies were seen to develop from P14 and P30 SCs (5000 P3 SCs and P7 SCs generated 146.75 ± 12.71 and 76.5 ± 5.22 HCs inside of the colonies, respectively, p < 0.001, n = 3) (Fig. 3b–e). At P14 and P30, we only found the HCs outside of the colonies, suggesting that they were directly trans-differentiated from SCs. The total number of Myo7a+ HCs inside and outside of the colonies decreased with age, suggesting that the ability of SCs to regenerate HCs was significantly decreased with age (Fig. 3f).
P3 Sox2+ SCs generated more HCs compared with the other three ages of SCs in vitro. a We used the FITC channel to sort P3, P7, P14, and P30 Sox2+ SCs, and we cultured the sorted GFP+ cells at 50 cells/μl for 10 days. b P3 Sox2+ SCs generated a large number of Myo7a+ cells. c P7 Sox2+ SCs also could form colonies and generate Myo7a+ cells. d, e Both P14 and P30 Sox2+ SCs could not form colonies, but the single cells could generate Myo7a+ cells. f P3 Sox2+ SCs formed more Myo7a+ cells compared with P7, P14, and P30 Sox2+ SCs. g Both inside and outside of the colony, P3 SCs formed more Myo7a+ cells compared with P7, P14, and P30 Sox2+ SCs. ***p < 0.001. Scale bars are 10 μm in b–h
RNA-seq analysis of SCs isolated at different ages
To determine the gene expression profiles of SCs at different ages, RNA-seq analysis was performed on flow cytometry-isolated Sox2+ SCs from P3, P7, P14, and P30 basilar membranes. Three biological replicates were prepared for each time point. After alignment to the reference genome (Mouse mm10, UCSC), the gene expression abundance was normalized to FPKM (fragments per kilobase of transcript per million fragments mapped). We next explored the data set with principal component analysis and sample clustering analysis. Replicates from the same group were well clustered and no outliers were found (Fig. 4). We next carried out pairwise comparison among all time points, and the genes that were differentially expressed within any two groups were marked. In total, we found 1296 differentially expressed genes.
Cell cycle analysis
The neonatal Sox2+ SCs had significantly greater proliferation and mitotic HC regeneration ability than the aged SCs; however, the detailed mechanism behind this difference remains unknown. To identify the possible genes regulating the age-dependent cell cycling of SCs, we used RNA-seq analysis to compare the expression of genes regulating the cell cycle and cell proliferation in P3, P7, P14, and P30 SCs. A prior study suggested that over 1000 cell cycle genes might exist in the average mammalian cell [28], some of which had significant expression differences between SCs at different ages. We found that Rad17, Ppm1d, Skp2, Abl1, Cdk4, E2f3, Terf1, Cks1b, Cdk5rap1, Atr, Cdk1, Birc5, Ccna2, Cdkn3, Nek2, Ccnc, Ccnb2, and Tfdp1 were highly expressed in the neonatal SCs compared with adult SCs and that Ccnf, Rad9a, Ddit3, Pmp22, Cdc6, Itgb1, Stmn1, Ccnd2, Smc1a, Brca2, and Tsg101 were highly expressed in the adult SCs compared with neonatal SCs (Fig. 5a). Among them, Skp2 [29,30,31], E2f3 [32, 33], Cdk1 [34, 35], Birc5 [36], Ddit3 [37, 38], and Itgb1 [39] have already been reported in the inner ear. The results of the qPCR were consistent with RNA-seq results, thus confirming the expression difference in the cell cycle genes (Fig. 5d). However, most of the differentially expressed cell cycle genes that we identified at different ages of SCs have not been characterized before in the inner ear and need to be further studied in the future.
Cell cycle genes, Wnt pathway genes, and TGFβ pathway genes in P3, P7, P14, and P30 Sox2+ SCs. a The expression of 72 genes involved in the cell cycle in P3, P7, P14, and P30 Sox2+ SCs. b The differentially expressed genes in P3, P7, P14, and P30 Sox2+ SCs that are involved in Wnt signaling pathways. c The differentially expressed genes in P3, P7, P14, and P30 Sox2+ SCs that are involved in TGFβ signaling pathways. d Quantitative RT-PCR analysis of some cell cycle and Wnt pathway genes that are differentially highly expressed in P3, P7, P14, and P30 Sox2+ SCs as identified by RNA-seq analysis. Student’s paired t test; * = P7, P14, and P30 Sox2+ SCs vs. P3 Sox2+ SCs; & = P14 and P30 Sox2+ SCs vs. P7 Sox2+ SCs; ^ = P30 Sox2+ SCs vs. P14 Sox2+ SCs. *p < 0.05, **p < 0.01, ***p < 0.001, &p < 0.05, &&&p < 0.001, ^p < 0.05, ^^^p < 0.001
Wnt signaling analysis
The Wnt signaling pathway is a highly conserved pathway and has been reported to be involved in multiple processes including proliferation, cell fate determination, differentiation, and cell protection [40, 41]. In the inner ear, activation of the Wnt signaling pathway is important for HC regeneration and survival [8, 10, 12, 23, 42,43,44,45,46,47]. To determine which Wnt pathway factors are involved in regulating the age-dependent proliferation and HC regeneration ability of SCs, we measured the expression of over 147 genes, some of which had significant expression differences between SCs at different ages. We found that Daam1, Fzd6, Frat1, Wnt4, Kremen1, Fzd3, Ctbp1, Jun, Aes, Wisp1, Csnk2a1, Wnt2b, Ctnnbip1, Strp4, Ruvbl1, Rhoa, and Fgf4 were significantly upregulated in adult mice compared with neonatal mice, while Prickle1, Ctnnb1, Fzd1, Tle1, Fzd9, and Dixdc1 were highly expressed in neonatal mice compared with adult mice (Fig. 5b). Among them, Jun [48], Wnt2b [49, 50], Strp4 [51], Fgf4 [52, 53], Fzd1 [54], and Fzd3 [55, 56] have already been reported in the inner ear. We performed qPCR to confirm the RNA-seq data, and the results were consistent with the RNA-seq analysis (Fig. 5d).
TGFβ signaling analysis
TGFβ signaling plays an important role in inner ear development and HC regeneration [57, 58], but studies of TGFβ signaling in HC regeneration are still limited. To determine which TGFβ pathway factors might be involved in regulating HC regeneration, we examined the expression of TGFβ pathway genes in the mouse genome in P3, P7, P14, and P30 SCs. We found that Srebf2, Crebbp, Ptk2, Gtf2i, Rad21, Id2, Txnip, Nfib, Nfkbia, Ptk2b, Brd2, Id3, Smad1, S100a8, Atf4, Dnaja1, Cryab, Bcl2l1, and Smad6 were significantly upregulated in adult mice compared with neonatal mice, while Fn1, Ephb2, and Bach1 were highly expressed in neonatal mice compared with adult mice (Fig. 5c). Among them, Ephb2 [59], Bdnf [60], and Pdgfa [61] have already been reported in the inner ear.
Notch signaling analysis
Notch signaling plays an important role during the development and patterning of sensory HCs. The activation of Notch signaling promotes the development of progenitor cells but prevents the differentiation of SCs into HCs. Inhibition of Notch signaling or Notch ligands such as Dll1 and Jagged2 results in the generation of supernumerary HCs in the mouse inner ear [62,63,64]. To determine which Notch pathway genes are involved in regulating the age-dependent proliferation and HC regeneration ability of SCs, we measured over 1000 genes, some of which had significant expression differences among SCs at different ages. We found that the expression of Maml2, Numb, Smo, Notch1, Tle1, and Lor decreased with increasing age and that Hey2, Ncstn, Hes1, Runx1, Wisp1, Nfkb1, Snw1, Figf, Lfng, Id1, Psenes, Adam10, and Notch2 were highly expressed in adult SCs (Fig. 6a). Among these, Numb [65], Smo [21], Notch1 [43, 66, 67], Hey2 [68, 69], Hes1 [70, 71], Gsk3b [72], Lfng [73, 74], Id1 [75, 76], and Adam10 [77,78,79] have already been reported in the inner ear. We also performed qPCR to confirm the RNA-seq data, and the results were consistent with the RNA-seq analysis data (Fig. 6c).
Transcription factor and Notch signaling pathway genes in P3, P7, P14, and P30 Sox2+ SCs. a The expression of 96 transcription factor genes in P3, P7, P14, and P30 Sox2+ SCs. b The differentially expressed genes in P3, P7, P14, and P30 Sox2+ SCs that are involved in Notch signaling pathways. c Quantitative RT-PCR analysis of some transcription factor genes and Notch signaling pathway genes that are differentially highly expressed genes in P3, P7, P14, and P30 Sox2+ SCs as identified by RNA-seq analysis. Student’s paired t test; * = P7, P14, and P30 Sox2+ SCs vs. P3 Sox2+ SCs; & = P14 and P30 Sox2+ SCs vs. P7 Sox2+ SCs; ^ = P30 Sox2+ SCs vs. P14 Sox2+ SCs. *p < 0.05, **p < 0.01, ***p < 0.001, &p < 0.05, &&p < 0.01, &&&p < 0.001, ^p < 0.05, ^^p < 0.01, ^^^p < 0.001
Transcription factor analysis
Transcription factors (TFs) are regulatory proteins that control the expression of targeted genes by binding either to enhancer or promoter regions. TFs are involved in various processes, including inner ear development and HC regeneration. To determine which TFs might be involved in regulating HC regeneration, we examined the expression of 1324 TFs in the mouse genome in P3, P7, P14, and P30 SCs. We found that 9 TF genes (Zfp286, E2f6, Dlx1, Cebpb, Zfp275, Nfic, Irf3, Junb, and Zfp651) were highly expressed in adult mice compared to neonatal mice, while there were 28 TF genes (Zfp454, Zfp41, Zfp641, Zfp846, Zeb2, Zfp72, Foxf2, Elf5, Klf1, Tbx18, Zscan20, Zfp354b, Otx2, Irx2, Zfp52, Dlx2, Zfp865, Mycn, Zxdb, Crebl2, Zfp566, AU041133, Zfp429, Egr3, Rfx1, Zfp707, Zfp667, and Six5) that were highly expressed in neonatal mice compared with adult mice (Fig. 6b). Some of the TF genes that are highly expressed in neonatal SCs have been reported to play roles in promoting HC fate and patterning regulation during inner ear development, including Rfx1 [80], Tbx18 [81], Otx2 [82, 83], Dlx2 [84], and Mycn [85]. We also performed qPCR to confirm the RNA-seq data, and the results were consistent with the RNA-seq analysis data (Fig. 6c). We have identified many TFs that have not been characterized before, and their involvement in the differential regeneration capacity in mouse cochlear SCs at different ages should be investigated in the future.
Gene ontology analysis of the genes that are differentially expressed in SCs of different ages
After clustering the expression of all 1296 differentially expressed genes in P3, P7, P14, and P30 Sox2+ SCs in a heatmap (fold change > 2.0, q < 0.05), we applied GO analysis to the gene clusters. GO terms with the greatest enrichment fold are shown on the right of Fig. 7a, which also shows the protein interaction network of these GO enriched genes (Fig. 7b). GO analysis was applied to the genes that were upregulated in SCs at different ages (fold change > 2.0, p < 0.01). The genes with altered expression in P3 Sox2+ SCs were highly enriched in functional categories such as auditory receptor cell fate determination, neuron fate determination, signaling, and extracellular matrix formation and maintenance. Genes upregulated in P30 SCs were highly enriched in functional categories such as biosyntheic processes and positive regulation of programmed cell death.
Global comparisons of differentially expressed genes among four time points by hierarchical clustering and gene ontology analysis. a Hierarchical clustering of FPKM of all differentially expressed genes. Red denotes above-average expression levels, and blue denotes below-average levels. Each row represents one gene, and each column represents one time point. Gene ontology analysis was performed on the highly expressed gene clusters in the P3, P7, and P30 groups. b STRING network analysis of genes present in GO categories
Discussion
Several previous studies have shown that the ability of SCs to regenerate lost or damaged HCs decreases dramatically with age; however, the detailed transcriptome profiles of SCs at different ages have not been studied. Here, we isolated SCs from P3, P7, P14, and P30 mice and compared their transcriptome expression profiles. We identified a set of differentially expressed genes, including cell cycle genes, signaling pathway genes, and TFs, that might be involved in regulating the proliferation and HC differentiation ability of SCs. Most of the differentially expressed genes identified in this study have not been investigated in the inner ear before and need to be further studied in the future.
In order to find the key genes regulating inner ear HC regeneration, our previous studies have reported the transcriptome profiles of SCs or Lgr5+ inner ear progenitors, which are a sub-population of SCs, at different locations and under different treatment conditions [13, 14, 86, 87]. We characterized the transcriptomes of Lgr5+ progenitor cells in the apical and basal turns of the mouse cochlea [14]. Compared with our current results, we found that the cell cycle genes Ccnc, Cdk4, Nek2, and Skp2 were highly expressed both in the Lgr5+ progenitor cells in the apical turn of the cochlea and in neonatal mouse inner ear SCs. Also, the TF genes Irx2 and Zfp667 were highly expressed both in the Lgr5+ progenitor cells in the apical turn of the cochlea and in neonatal mouse inner ear SCs, while Junb was highly expressed both in the Lgr5+ progenitor cells in the basal turn of cochlea and in adult mouse inner ear SCs.
We also characterized the transcriptomes of Lgr5+ progenitor cells and other Lgr5− SCs [13]. Compared with our current results, we found that the cell cycle genes Skp2 and Terf1 were highly expressed both in the Lgr5+ progenitor cell and in neonatal mouse inner ear SCs, while Ccnf, Notch2, Ppm22, Ccnd2, and Tsg101 were highly expressed both in the Lgr5− SCs and in adult mouse inner ear SCs. The TF gene Zfp667 was highly expressed both in the Lgr5+ progenitor cells and neonatal mouse inner ear SCs, while Junb was highly expressed both in the Lgr5− SCs and adult mouse inner ear SCs. Among Wnt signaling pathway genes, Wisp1 and Rhoa were highly expressed both in the Lgr5− SCs and the adult mouse inner ear SCs.
Next, we characterized the transcriptomes of Lgr5+ progenitor cells with or without neomycin injury to show the damage-induced transcriptome changes in the Lgr5+ progenitors [87]. Compared with our current results, we found that the cell cycle gene Tfdp1 was highly expressed both in the neomycin-treated Lgr5+ progenitors and neonatal mouse inner ear SCs, while Stmn1 was highly expressed both in the untreated Lgr5+ progenitors and in adult mouse inner ear SCs. The TF gene Zfp52 was highly expressed both in the neomycin-treated Lgr5+ progenitors and in neonatal mouse inner ear SCs, while Junb was highly expressed both in the untreated Lgr5+ progenitors and in adult mouse inner ear SCs. Among Notch, Wnt, TGFβ signaling pathway genes, Hes1, Ctnnbip1, Id2, and Id3 were highly expressed both in the untreated Lgr5+ progenitors and in adult mouse inner ear SCs.
Lastly, we characterized the transcriptomes of Lgr5+ progenitor cells and Lgr6+ progenitor cells [86]. Compared with our current results, we found that the TF genes Ilx2 and AU041133 were highly expressed both in the Lgr6+ progenitors and in neonatal mouse inner ear SCs; while the cell cycling genes Rad17 and Skp2 were highly expressed both in the Lgr5+ progenitors and in neonatal mouse inner ear SCs. Among Notch signaling pathway genes, Maml2 was highly expressed both in the Lgr6+ progenitors and in neonatal mouse inner ear SCs, while Hey2, Hes1, and Id1 were highly expressed both in the Lgr5+ progenitors and in adult mouse inner ear SCs. These candidate genes might play important roles in regulating HC regeneration in the inner ear.
Cell cycle analysis
Among the differentially expressed cell cycle-related genes, Skp2, E2f3, Cdk1, Birc5, Ddit3, and Itgb1 have been reported in the inner ear before. Skp2 is an F-box protein that regulates the G1 to S transition by controlling the stability of several G1 regulators, including p27, and it is expressed in the auditory epithelia and neurons at early stages of development. In the mature auditory epithelium, overexpression of Skp2 alone can induce SC proliferation but cannot induce new HC formation, while overexpression of Skp2 combined with overexpression of Atoh1 generates new HCs [29,30,31]. This suggests that regulation of HC regeneration requires multi-gene coordination. Skp2 is also highly expressed in tumor cells and promotes cell proliferation [88,89,90]. E2f3 is a member of the E2F transcription factor family and is involved in regulating cell proliferation. In isolated human islets, it can induce proliferation of β cells [91]. E2f3 is barely expressed in the inner ear, but its expression increases in outer HC nuclei upon excessive noise exposure [32, 33]. Cdk1 is ubiquitously expressed throughout the organ of Corti and spiral ganglion cells, and inhibition of Cdk1 and other cyclin-dependent kinases can induce differentiation of supernumerary HCs and Deiters’ cells in the developing organ of Corti in cultured rat cochleae [34, 35]. Birc5 is expressed during embryonic development and cannot be detected in most terminally differentiated tissues, and it is also highly expressed in many tumors such as pancreatic ductal adenocarcinoma [92]. Birc5 is widely expressed in the organ of Corti, and it provides protection against ototoxin-induced cytotoxicity [36]. Ddit3 is an endoplasmic reticulum stress marker gene. In the animal model of acute hearing loss, the expression of Ddit3 is upregulated in the lateral wall of the cochlea, and this high expression of Ddit3 might lead to hearing loss because of endoplasmic reticulum stress [37, 38]. Itgb1 is involved in the regulation of cell migration and invasion of hepatoma carcinoma, breast cancer, and gallbladder cancer [93,94,95]. It is expressed throughout the otic area, including the fusion plate epithelium and the periotic mesenchyme [39]. Rad17, Ppm1d, Abl1, Cdk4, Terf1, Cks1b, Cdk5rap1, Atr, Ccna2, Cdkn3, Nek2, Ccnc, Ccnb2, Tfdp1, Ccnf, Rad9a, Pmp22, Cdc6, Stmn1, Ccnd2, Smc1a, Brca2, and Tsg101 have not been reported previously in the inner ear and need to be further studied in the future.
Wnt signaling analysis
Among the differentially expressed Wnt signaling-related genes, Jun, Wnt2b, Strp4, Fgf4, Fzd1, Fzd3, and Fzd6 have been previously reported in the inner ear. Jun has been implicated in the regulation of cell proliferation, differentiation, and apoptosis. It plays a critical role during inner ear development by mediating apoptosis through the JNK pathway [48]. Wnt2b is expressed in the endolymphatic duct; however, the role of Wnt2b in inner ear development has not been reported [49, 50]. Sfrp4 is a Wnt pathway inhibitor that is involved in many diseases including obesity, type 2 diabetes, cancer, and psoriasis [96]. In the inner ear, Sfrp4 can be directly targeted by miR-124 to regulate HC differentiation and polarization in the organ of Corti [97]. Fgf4 is present in many cancerous and noncancerous tissues, indicating that Fgf4 plays an important role in cell differentiation and proliferation [98]. In zebrafish, Fgf4 can be mediated by miR-194 to regulate the development and differentiation of sensory patches [52, 53]. Frizzled signaling is involved in diverse tissue closure processes, and defects in frizzled signaling result in some of the most common congenital anomalies in humans. In the organ of Corti at E18, Fzd1 is weakly expressed in the three outer rows of sensory HCs and is strongly expressed in the flanking non-sensory epithelial cells and the underlying phalangeal and pillar cells, and Fzd1 mutations cause mis-orientation of inner ear sensory HCs [54]. Fzd3 and Fzd6 are key regulators of planar cell polarity in the mammalian cochlea. In the inner ear, both Fzd3 and Fzd6 are localized on the lateral faces of sensory and SCs in all sensory epithelia, and this localization overlaps with Vangl2 and suggests that Fzd3 and Fzd6 might play an important role in the planar polarity of HCs because Vangl2 plays an important role in regulating hair bundle orientation [55, 56, 99]. This suggests that different Frizzled genes in the inner ear have different functions. Although Jun, Wnt2b, Strp4, Fgf4, Fzd1, Fzd3, and Fzd6 have been previously reported in the inner ear, the function of these genes in HC regeneration still need to be studied further. Daam1, Frat1, Wnt4, Kremen1, Ctbp1, Wisp1, Csnk2a1, Ctnnbip1, Ruvbl1, Rhoa, Prickle1, Ctnnb1, Tle1, Fzd9, and Dixdc1 have not been reported previously in the inner ear and need to be further studied in the future.
TGFβ signaling analysis
Among the differentially expressed TGFβ signaling-related genes, Ephb2, Bdnf, and Pdgfa have previously been reported in the inner ear. Ephb2 is a member of the largest group of transmembrane receptor tyrosine kinases, and deletion of Ephb2 leads to vestibular dysfunction because of the reduced production of endolymph [59]. Bdnf acts as a nerve growth factor, and it promotes the growth and survival of neurons in the central and peripheral nervous systems [100]. In the inner ear, it supports spiral ganglion neuron survival [60]. Pdgfa is a growth factor with restricted otic expression, and it overlaps with Fgf16 in the anterior and posterior cristae in the chick inner ear [61]. Srebf2, Crebbp, Ptk2, Gtf2i, Rad21, Id2, Txnip, Nfib, Nfkbia, Ptk2b, Brd2, Id3, Smad1, S100a8, Atf4, Dnaja1, Cryab, Bcl2l1, Smad6, Fn1, and Bach1 have not been reported previously in the inner ear and need to be further studied in the future.
Notch signaling analysis
Among the differentially expressed Notch signaling-related genes, Numb, Smo, Notch1, Hey2, Hes1, Gsk3b, Lfng, Id1, and Adam10 have previously been reported in the inner ear. Numb is a cell fate determinant gene that regulates cardiac progenitor cell differentiation and cardiac morphogenesis [101]. In the auditory epithelium, Numb expression has different patterns, which suggests that Numb plays an important role in cochlear development [65]. Smo encodes a membrane protein that is essential for the transduction of Hedgehog signals into the cytoplasm. Activation of Smo inhibits prosensory cell differentiation into HCs or SCs and maintains their properties as prosensory cells, and conditional knockout of the Smo gene in the cochlea delays HC and SC differentiation in the apical region [21]. Notch1 is the primary Notch receptor expressed in the mouse inner ear, and activation of Notch1 in developing auditory HCs causes profound deafness, while deletion of Notch1 leads to limited mitotic HC generation [43, 66]. Hey2 is a putative Notch target gene and functions in cell fate specification. Hey2 is expressed in the cochlear epithelium prior to terminal differentiation, and its overexpression overlaps with that of Hes1 in the developing cochlea. The genetic inactivation of Hey2 leads to increased numbers of mis-patterned inner HCs and outer HCs [70, 71], and activation of Hey2 by FGF signaling blocks HC differentiation [68, 69]. Gsk3 plays an important role in the regulation of apoptosis and proliferation in the inner ear, and activation of Gsk3 causes the release of inflammatory factors that can eventually lead to hearing loss, while inactivation of Gsk3 increases the total number of HCs [72, 102]. The Lfng gene is expressed in non-sensory SCs in the mouse cochlea, but there is no noticeable effect on HC differentiation in Lfng mutant mice. However, mutation of Lfng suppresses the effects of the Jag2 mutations on inner HCs [73, 74]. Id1 is able to prevent differentiation of pluripotent cells, and in bone marrow transplantation assays, reducing Id1 enhanced hematopoietic stem cells’ self-renewal potential [103]. Id1 is expressed within the cochlear duct in a pattern that is consistent with a role in the regulation of HC development. However, there is no hearing deficit in the absence of the Id1 gene, and the reason for this might be compensatory effects by other Ids like Id3, which has a similar expression pattern as Id1 in the cochlea [75, 76]. Adam10 is abundantly expressed in the brain and is linked to epilepsy, Alzheimer’s disease, Hunting’s disease, and developmental disorder Fragile X syndrome because of its role in regulating the activity of excitatory synapses [104, 105]. Adam10 is also expressed in the cochlea and vestibule, and inhibition of Adam10 after HC loss increases the proliferation of SCs in the vestibular system [77,78,79]. Although Numb, Smo, Notch1, Hey2, Hes1, Gsk3b, Lfng, Id1, and Adam10 have been reported in the inner ear, the functions of some of these genes in HC regeneration still need to be further studied. Maml2, Tle1, Lor, Ncstn, Runx1, Wisp1, Nfkb1, Snw1, Figf, Psenes, and Notch2 have not been reported previously in the inner ear and need to be further studied in the future.
Transcription factor analysis
Among the differentially expressed TFs, Rfx1, Tbx18, Otx2, Dlx2, and Mycn have previously been reported in the inner ear. Rfx1 has an important function in brain tumors and sensorineural hearing loss. Together with Atho1, Rfx1/3 can induce HC-like cell differentiation, and the conditional knockout of Rfx1/3 leads to severe hearing loss and OHC damage [80, 106, 107]. Tbx18 is a critical TF for cell proliferation and cell fate determination, and it is also a target gene of the Hippo pathway [108]. The expression of Tbx18 during inner ear development is restricted to the sub-region of the otic mesenchyme that is fated to differentiate into fibrocytes, and Tbx18-deficient mice show profound deafness and a complete disruption of the endocochlear potential that is essential for the transduction of sound by sensory HCs [81]. Otx2 is a regulator of embryonic development and embryonic neurogenesis. And it plays a role in brain, craniofacial, and sensory organ development [109, 110]. In the inner ear, Otx2 can induce Hes5 and lead to the differentiation of both cochlear and macular neuroepithelium [82, 83]. In the chick inner ear, the expression of Dlx1 and Dlx2 during the later stages of inner ear morphogenesis is limited to cochlear and vestibular nerves, and expression levels are lower than in the early stages of morphogenesis [84]. Mycn is a critical factor in the development of the central and peripheral nervous systems [111]. In humans, the mutation of Mycn can cause structural and functional abnormalities of the inner ear [85]. Zfp286, E2f6, Dlx1, Cebpb, Zfp275, Nfic, Irf3, Junb, Zfp651, Zfp454, Zfp41, Zfp641, Zfp846, Zeb2, Zfp72, Foxf2, Elf5, Klf1, Zscan20, Zfp354b, Irx2, Zfp52, Zfp865, Zxdb, Crebl2, Zfp566, AU041133, Zfp429, Egr3, Zfp707, Zfp667, and Six5 have not been reported previously in the inner ear and need to be further studied in the future.
Conclusion
Consistent with previous reports, in this study we also found that neonatal SCs have significantly greater proliferation and HC regeneration ability than adult SCs. We systematically investigated the transcriptome differences between P3, P7, P14, and P30 SCs and identified several significantly differentially expressed genes that might regulate the proliferation and HC regeneration capacity of SCs of different ages. The transcriptomes of different ages of SCs reported here establish a framework for future characterization of the genes that regulate the proliferation and HC regeneration ability of SCs, and these genes might represent new therapeutic targets for HC regeneration.
Availability of data and materials
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- E:
-
Embryonic day
- GO:
-
Gene ontology
- HC:
-
Hair cell
- P:
-
Postnatal day
- SC:
-
Supporting cell
- STRING:
-
Search Tool for the Retrieval of Interacting Genes/Proteins
- TFs:
-
Transcription factors
References
Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240(4860):1774–6.
Lombarte A, Yan HY, Popper AN, Chang JS, Platt C. Damage and regeneration of hair cell ciliary bundles in a fish ear following treatment with gentamicin. Hear Res. 1993;64(2):166–74.
Cruz RM, Lambert PR, Rubel EW. Light microscopic evidence of hair cell regeneration after gentamicin toxicity in chick cochlea. Arch Otolaryngol Head Neck Surg. 1987;113(10):1058–62.
Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240(4860):1772–4.
Balak KJ, Corwin JT, Jones JE. Regenerated hair-cells can originate from supporting cell progeny - evidence from phototoxicity and laser ablation experiments in the lateral line system. J Neurosci. 1990;10(8):2502–12.
Brigande JV, Heller S. Quo vadis, hair cell regeneration? Nat Neurosci. 2009;12(6):679–85.
Bermingham-McDonogh O, Rubel EW. Hair cell regeneration: winging our way towards a sound future. Curr Opin Neurobiol. 2003;13(1):119–26.
Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141(4):816–29.
Warchol ME. Sensory regeneration in the vertebrate inner ear: differences at the levels of cells and species. Hearing Res. 2011;273(1–2):72–9.
Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, et al. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci U S A. 2012;109(21):8167–72.
Basch ML, Brown RM 2nd, Jen HI, Groves AK. Where hearing starts: the development of the mammalian cochlea. J Anat. 2016;228(2):233–54.
Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32(28):9639–48.
Cheng C, Guo L, Lu L, Xu X, Zhang S, Gao J, et al. Characterization of the transcriptomes of Lgr5+ hair cell progenitors and Lgr5- supporting cells in the mouse cochlea. Front Mol Neurosci. 2017;10:122.
Waqas M, Guo L, Zhang S, Chen Y, Zhang X, Wang L, et al. Characterization of Lgr5+ progenitor cell transcriptomes in the apical and basal turns of the mouse cochlea. Oncotarget. 2016;7(27):41123–41.
Feghali JG, Lefebvre PP, Staecker H, Kopke R, Frenz DA, Malgrange B, et al. Mammalian auditory hair cell regeneration/repair and protection: a review and future directions. Ear Nose Throat J. 1998;77(4):276 80, 82–5.
Doetzlhofer A, White P, Lee YS, Groves A, Segil N. Prospective identification and purification of hair cell and supporting cell progenitors from the embryonic cochlea. Brain Res. 2006;1091(1):282–8.
Golub JS, Tong L, Ngyuen TB, Hume CR, Palmiter RD, Rubel EW, et al. Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J Neurosci. 2012;32(43):15093–105.
Sinkkonen ST, Chai RJ, Jan TA, Hartman BH, Laske RD, Gahlen F, et al. Intrinsic regenerative potential of murine cochlear supporting cells. Sci Rep. 2011;1:26.
Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, Geleoc GS, et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8(1):18–31.
Corwin JT, Oberholtzer JC. Fish n' chicks: model recipes for hair-cell regeneration? Neuron. 1997;19(5):951–4.
Chen Y, Lu X, Guo L, Ni W, Zhang Y, Zhao L, et al. Hedgehog signaling promotes the proliferation and subsequent hair cell formation of progenitor cells in the neonatal mouse cochlea. Front Mol Neurosci. 2017;10:426.
Chen Q, Quan Y, Wang N, Xie C, Ji Z, He H, et al. Inactivation of STAT3 signaling impairs hair cell differentiation in the developing mouse cochlea. Stem Cell Rep. 2017;9(1):231–46.
Wu J, Li W, Lin C, Chen Y, Cheng C, Sun S, et al. Co-regulation of the Notch and Wnt signaling pathways promotes supporting cell proliferation and hair cell regeneration in mouse utricles. Sci Rep. 2016;6:29418.
Li W, Wu J, Yang J, Sun S, Chai R, Chen ZY, et al. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci U S A. 2015;112(1):166–71.
Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105(47):18396–401.
Trapnell C, Schatz MC. Optimizing data intensive GPGPU computations for DNA sequence alignment. Parallel Comput. 2009;35(8):429–40.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
Forrest ARR, Taylor D, Grimmond S, Grp RG, Members G. Exploration of the cell-cycle genes found within the RIKEN FANTOM2 data set. Genome Res. 2003;13(6B):1366–75.
Minoda R, Izumikawa M, Kawamoto K, Zhang H, Raphael Y. Manipulating cell cycle regulation in the mature cochlea. Hear Res. 2007;232(1–2):44–51.
Dong Y, Nakagawa T, Endo T, Kim TS, Iguchi F, Yamamoto N, et al. Role of the F-box protein Skp2 in cell proliferation in the developing auditory system in mice. Neuroreport. 2003;14(5):759–61.
Nakayama KI, Hatakeyama S, Nakayama K. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem Biophys Res Commun. 2001;282(4):853–60.
Jamesdaniel S, Hu B, Kermany MH, Jiang H, Ding D, Coling D, et al. Noise induced changes in the expression of p38/MAPK signaling proteins in the sensory epithelium of the inner ear. J Proteome. 2011;75(2):410–24.
Pang J, Xiong H, Yang H, Ou Y, Xu Y, Huang Q, et al. Circulating miR-34a levels correlate with age-related hearing loss in mice and humans. Exp Gerontol. 2016;76:58–67.
Liu YY, Wang GP, Peng Z, Guo JY, Wu Q, Xie J, et al. E2F1-CDK1 pathway activation in kanamycin-induced spiral ganglion cell apoptosis and the protective effect of CR8. Neurosci Lett. 2016;617:247–53.
Malgrange B, Knockaert M, Belachew S, Nguyen L, Moonen G, Meijer L, et al. The inhibition of cyclin-dependent kinases induces differentiation of supernumerary hair cells and Deiters' cells in the developing organ of Corti. FASEB J. 2003;17(14):2136–8.
Habtemichael N, Heinrich UR, Knauer SK, Schmidtmann I, Bier C, Docter D, et al. Expression analysis suggests a potential cytoprotective role of Birc5 in the inner ear. Mol Cell Neurosci. 2010;45(3):297–305.
Fujinami Y, Mutai H, Mizutari K, Nakagawa S, Matsunaga T. A novel animal model of hearing loss caused by acute endoplasmic reticulum stress in the cochlea. J Pharmacol Sci. 2012;118(3):363–72.
Fujinami Y, Mutai H, Kamiya K, Mizutari K, Fujii M, Matsunaga T. Enhanced expression of C/EBP homologous protein (CHOP) precedes degeneration of fibrocytes in the lateral wall after acute cochlear mitochondrial dysfunction induced by 3-nitropropionic acid. Neurochem Int. 2010;56(3):487–94.
Matilainen T, Haugas M, Kreidberg JA, Salminen M. Analysis of Netrin 1 receptors during inner ear development. Int J Dev Biol. 2007;51(5):409–13.
Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15(1):28–32.
Pinto D, Clevers H. Wnt, stem cells and cancer in the intestine. Biol Cell. 2005;97(3):185–96.
Lu X, Sun S, Qi J, Li W, Liu L, Zhang Y, et al. Bmi1 regulates the proliferation of cochlear supporting cells via the canonical Wnt signaling pathway. Mol Neurobiol. 2017;54(2):1326–39.
Ni W, Lin C, Guo L, Wu J, Chen Y, Chai R, et al. Extensive supporting cell proliferation and mitotic hair cell generation by in vivo genetic reprogramming in the neonatal mouse cochlea. J Neurosci. 2016;36(33):8734–45.
Yu X, Liu W, Fan Z, Qian F, Zhang D, Han Y, et al. c-Myb knockdown increases the neomycin-induced damage to hair-cell-like HEI-OC1 cells in vitro. Sci Rep. 2017;7:41094.
Liu L, Chen Y, Qi J, Zhang Y, He Y, Ni W, et al. Wnt activation protects against neomycin-induced hair cell damage in the mouse cochlea. Cell Death Dis. 2016;7:e2136.
Chai R, Xia A, Wang T, Jan TA, Hayashi T, Bermingham-McDonogh O, et al. Dynamic expression of Lgr5, a Wnt target gene, in the developing and mature mouse cochlea. J Association Res Otolaryngol. 2011;12(4):455–69.
Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci U S A. 2013;110(34):13851–6.
Sanz C, Leon Y, Canon S, Alvarez L, Giraldez F, Varela-Nieto I. Pattern of expression of the Jun family of transcription factors during the early development of the inner ear: implications in apoptosis. J Cell Sci. 1999;112(Pt 22):3967–74.
Lin ZS, Cantos R, Patente M, Wu DK. Gbx2 is required for the morphogenesis of the mouse inner ear: a downstream candidate of hindbrain signaling. Development. 2005;132(10):2309–18.
Choo D, Ward J, Reece A, Dou H, Lin Z, Greinwald J. Molecular mechanisms underlying inner ear patterning defects in kreisler mutants. Dev Biol. 2006;289(2):308–17.
Sai XR, Yonemura S, Ladher RK. Junctionally restricted RhoA activity is necessary for apical constriction during phase 2 inner ear placode invagination. Dev Biol. 2014;394(2):206–16.
Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228(2):267–72.
Cao H, Shi J, Du J, Chen K, Dong C, Jiang D, et al. MicroRNA-194 regulates the development and differentiation of sensory patches and statoacoustic ganglion of inner ear by Fgf4. Med Sci Monit. 2018;24:1712–23.
Yu H, Smallwood PM, Wang Y, Vidaltamayo R, Reed R, Nathans J. Frizzled 1 and frizzled 2 genes function in palate, ventricular septum and neural tube closure: general implications for tissue fusion processes. Development. 2010;137(21):3707–17.
Wang Y, Guo N, Nathans J. The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J Neurosci. 2006;26(8):2147–56.
Montcouquiol M, Sans N, Huss D, Kach J, Dickman JD, Forge A, et al. Asymmetric localization of Vangl2 and Fz3 indicate novel mechanisms for planar cell polarity in mammals. J Neurosci. 2006;26(19):5265–75.
Du XP, Li W, Gao XS, West MB, Saltzman WM, Cheng CJ, et al. Regeneration of mammalian cochlear and vestibular hair cells through Hes1/Hes5 modulation with siRNA. Hearing Res. 2013;304:91–110.
Kawamoto K, Yagi M, Stover T, Kanzaki S, Raphael Y. Hearing and hair cells are protected by adenoviral gene therapy with TGF-beta1 and GDNF. Mol Ther. 2003;7(4):484–92.
Raft S, Andrade LR, Shao D, Akiyama H, Henkemeyer M, Wu DK. Ephrin-B2 governs morphogenesis of endolymphatic sac and duct epithelia in the mouse inner ear. Dev Biol. 2014;390(1):51–67.
McGuinness SL, Shepherd TK. Exogenous BDNF rescues rat spiral ganglion neurons in vivo. Otol Neurotol. 2005;26(5):1064–72.
Chapman SC, Cai Q, Bleyl SB, Schoenwolf GC. Restricted expression of Fgf16 within the developing chick inner ear. Dev Dyn. 2006;235(8):2276–81.
Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132(19):4353–62.
Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–89.
Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77(1):58–69.
Gao Z, Chi FL, Huang YB, Yang JM, Cong N, Li W. Expression of Numb and Numb-like in the development of mammalian auditory sensory epithelium. Neuroreport. 2011;22(2):49–54.
Liu Z, Owen T, Fang J, Zuo J. Overactivation of Notch1 signaling induces ectopic hair cells in the mouse inner ear in an age-dependent manner. PLoS One. 2012;7(3):e34123.
Savoy-Burke G, Gilels FA, Pan W, Pratt D, Que J, Gan L, et al. Activated Notch causes deafness by promoting a supporting cell phenotype in developing auditory hair cells. PLoS One. 2014;9(9):e108160.
Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16(1):58–69.
Benito-Gonzalez A, Doetzlhofer A. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J Neurosci. 2014;34(38):12865–76.
Li S, Mark S, Radde-Gallwitz K, Schlisner R, Chin MT, Chen P. Hey2 functions in parallel with Hes1 and Hes5 for mammalian auditory sensory organ development. BMC Dev Biol. 2008;8:20.
Korrapati S, Roux I, Glowatzki E, Doetzlhofer A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One. 2013;8(8):e73276.
Park HJ, Kim HJ, Bae GS, Seo SW, Kim DY, Jung WS, et al. Selective GSK-3beta inhibitors attenuate the cisplatin-induced cytotoxicity of auditory cells. Hear Res. 2009;257(1–2):53–62.
Zhang N, Martin GV, Kelley MW, Gridley T. A mutation in the Lunatic fringe gene suppresses the effects of a Jagged2 mutation on inner hair cell development in the cochlea. Curr Biol. 2000;10(11):659–62.
Basch ML, Brown RM, Jen HI, Semerci F, Depreux F, Edlund RK, et al. Fine-tuning of Notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates. eLife. 2016;5.
Jones JM, Montcouquiol M, Dabdoub A, Woods C, Kelley MW. Inhibitors of differentiation and DNA binding (Ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J Neurosci. 2006;26(2):550–8.
Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17(12):7317–27.
Yan X, Lin JT, Wang H, Markus A, Wree A, Rolfs A, et al. Regional expression of the ADAMs in developing chicken cochlea. Dev Dyn. 2010;239(8):2256–65.
Lin J, Yan X, Wang C, Talabattula VA, Guo Z, Rolfs A, et al. Expression patterns of the ADAMs in early developing chicken cochlea. Develop Growth Differ. 2013;55(3):368–76.
Warchol ME, Stone J, Barton M, Ku J, Veile R, Daudet N, et al. ADAM10 and gamma-secretase regulate sensory regeneration in the avian vestibular organs. Dev Biol. 2017;428(1):39–51.
Elkon R, Milon B, Morrison L, Shah M, Vijayakumar S, Racherla M, et al. RFX transcription factors are essential for hearing in mice. Nat Commun. 2015;6:8549.
Trowe MO, Maier H, Schweizer M, Kispert A. Deafness in mice lacking the T-box transcription factor Tbx18 in otic fibrocytes. Development. 2008;135(9):1725–34.
Palombo R, Porta G, Bruno E, Provero P, Serra V, Neduri K, et al. OTX2 regulates the expression of TAp63 leading to macular and cochlear neuroepithelium development. Aging. 2015;7(11):928–36.
Vendrell V, Lopez-Hernandez I, Alonso MBD, Feijoo-Redondo A, Abello G, Galvez H, et al. Otx2 is a target of N-myc and acts as a suppressor of sensory development in the mammalian cochlea. Development. 2015;142(16):2792.
Brown ST, Wang JM, Groves AK. Dlx gene expression during chick inner ear development. J Comp Neurol. 2005;483(1):48–65.
Chen CP, Lin SP, Chern SR, Wu PS, Chang SD, Ng SH, et al. A de novo 4.4-Mb microdeletion in 2p24.3 --> p24.2 in a girl with bilateral hearing impairment, microcephaly, digit abnormalities and Feingold syndrome. Eur J Med Genet. 2012;55(11):666–9.
Zhang YP, Guo L, Lu XL, Cheng C, Sun S, Li W, et al. Characterization of Lgr6+ cells as an enriched population of hair cell progenitors compared to Lgr5+ cells for hair cell generation in the neonatal mouse cochlea. Front Mol Neurosci. 2018;11:147.
Zhang S, Zhang Y, Yu P, Hu Y, Zhou H, Guo L, et al. Characterization of Lgr5+ progenitor cell transcriptomes after neomycin injury in the neonatal mouse cochlea. Front Mol Neurosci. 2017;10:213.
Du S, Wang S, Zhang F, Lv Y. SKP2, positively regulated by circ_ODC1/miR-422a axis, promotes the proliferation of retinoblastoma. J Cell Biochem. 2019;121:322–31.
He Z, Chen L, Wang Q, Yin C, Hu J, Hu X, et al. MicroRNA-186 targets SKP2 to induce p27(Kip1)-mediated pituitary tumor cell cycle deregulation and modulate cell proliferation. Korean J Physiol Pharmacol. 2019;23(3):171–9.
Zhao H, Pan H, Wang H, Chai P, Ge S, Jia R, et al. SKP2 targeted inhibition suppresses human uveal melanoma progression by blocking ubiquitylation of p27. Onco Targets Ther. 2019;12:4297–308.
Rady B, Chen Y, Vaca P, Wang Q, Wang Y, Salmon P, et al. Overexpression of E2F3 promotes proliferation of functional human beta cells without induction of apoptosis. Cell Cycle. 2013;12(16):2691–702.
Liu SH, Hong Y, Markowiak S, Sanchez R, Creeden J, Nemunaitis J, et al. BIRC5 is a target for molecular imaging and detection of human pancreatic cancer. Cancer Lett. 2019;457:10–9.
Huang L, Li X, Gao W. Long non-coding RNA linc-ITGB1 promotes cell proliferation, migration, and invasion in human hepatoma carcinoma by up-regulating ROCK1. Biosci Rep. 2018;38(5).
Wang L, Zhang YJ, Lv WJ, Lu JH, Mu JS, Liu YB, et al. Long non-coding RNA Linc-ITGB1 knockdown inhibits cell migration and invasion in GBC-SD/M and GBC-SD gallbladder cancer cell lines (retracted article. See vol. 92, pg. 1815, 2018). Chem Biol Drug Des. 2015;86(5):1064–71.
Yan MD, Zhang LN, Li GQ, Xiao SW, Dai J, Cen XY. Long noncoding RNA linc-ITGB1 promotes cell migration and invasion in human breast cancer. Biotechnol Appl Bioc. 2017;64(1):5–13.
Guo HY, Xing YZ, Deng F, Yang K, Li YH. Secreted Frizzled-related protein 4 inhibits the regeneration of hair follicles. Peerj. 2019;6.
Huyghe A, Van den Ackerveken P, Sacheli R, Prevot PP, Thelen N, Renauld J, et al. MicroRNA-124 regulates cell specification in the cochlea through modulation of Sfrp4/5. Cell Rep. 2015;13(1):31–42.
Terada M, Yoshida T, Sakamoto H, Miyagawa K, Katoh O, Hattori Y, et al. Biological significance of the hst-1 gene. Princess Takamatsu Symp. 1989;20:71–80.
Fukuda T, Kominami K, Wang S, Togashi H, Hirata K, Mizoguchi A, et al. Aberrant cochlear hair cell attachments caused by Nectin-3 deficiency result in hair bundle abnormalities. Development. 2014;141(2):399–409.
Acheson A, Conover JC, Fandl JP, DeChiara TM, Russell M, Thadani A, et al. A BDNF autocrine loop in adult sensory neurons prevents cell death. Nature. 1995;374(6521):450–3.
Wu M, Li J. Numb family proteins: novel players in cardiac morphogenesis and cardiac progenitor cell differentiation. Biomol Concepts. 2015;6(2):137–48.
Ellis K, Driver EC, Okano T, Lemons A, Kelley MW. GSK3 regulates hair cell fate in the developing mammalian cochlea. Dev Biol. 2019;453:191–205.
Singh SK, Singh S, Gadomski S, Sun L, Pfannenstein A, Magidson V, et al. Id1 ablation protects hematopoietic stem cells from stress-induced exhaustion and aging. Cell Stem Cell. 2018;23(2):252–65 e8.
Vezzoli E, Caron I, Talpo F, Besusso D, Conforti P, Battaglia E, et al. Inhibiting pathologically active ADAM10 rescues synaptic and cognitive decline in Huntington's disease. J Clin Investig. 2019;129(6):2390–403.
Prox J, Bernreuther C, Altmeppen H, Grendel J, Glatzel M, D'Hooge R, et al. Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J Neurosci. 2013;33(32):12915–28 28a.
Chen YC, Tsai CL, Wei YH, Wu YT, Hsu WT, Lin HC, et al. ATOH1/RFX1/RFX3 transcription factors facilitate the differentiation and characterisation of inner ear hair cell-like cells from patient-specific induced pluripotent stem cells harbouring A8344G mutation of mitochondrial DNA. Cell Death Dis. 2018;9(4):437.
Feng CZ, Zhang Y, Yin JB, Li J, Abounader R, Zuo ZY. Regulatory factor X1 is a new tumor suppressive transcription factor that acts via direct downregulation of CD44 in glioblastoma. Neuro-Oncology. 2014;16(8):1078–85.
Singh A, Ramesh S, Cibi DM, Yun LS, Li J, Li L, et al. Hippo signaling mediators Yap and Taz are required in the epicardium for coronary vasculature development. Cell Rep. 2016;15(7):1384–93.
Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14(10):5725–40.
Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, Czerny T, et al. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004;131(9):2037–48.
Pession A, Tonelli R. The MYCN oncogene as a specific and selective drug target for peripheral and central nervous system tumors. Curr Cancer Drug Tar. 2005;5(4):273–83.
Acknowledgements
Not applicable
Funding
This work was supported by grants from the National Key R&D Program of China (2017YFA0103903), the Strategic Priority Research Program of the Chinese Academy of Science (XDA16010303), the National Natural Science Foundation of China (81622013, 81970882, 81900941, 81570919, 81870721, 81771019, 81700913, 81670928, 81570921), Jiangsu Province Natural Science Foundation (BK20190121), Boehringer Ingelheim Pharma GmbH, the Fundamental Research Funds for the Central Universities (2242018k1G011), the Open Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University (SKLGE1809), and the Project of Invigorating Health Care through Science, Technology and Education.
Author information
Authors and Affiliations
Contributions
CC, YW, LG, RC, XG, and HL designed the study. CC, XL, WZ, WM, LZ, and LL performed the laboratory experiments. CC, YW, LG, XL, JG, ML, LL, FC, and MT contributed to critical discussion and data analysis. CC, XG, MT, HL, and RC wrote the paper. All authors read and approved the final manuscript.
Authors’ information
Not applicable
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal procedures were performed according to protocols approved by the Animal Care Committee of Southeast University and were consistent with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All efforts were made to minimize the number of animals used and to prevent their suffering.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
q-PCR primers
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cheng, C., Wang, Y., Guo, L. et al. Age-related transcriptome changes in Sox2+ supporting cells in the mouse cochlea. Stem Cell Res Ther 10, 365 (2019). https://doi.org/10.1186/s13287-019-1437-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-019-1437-0