Abstract
Purpose
Streptococcus alactolyticus strain FGM is used to ferment Astragalus membranaceus to develop a novel feed additive for animals in China. This study aimed at characterizing the safety and potential probiotic features of the strain FGM in vitro.
Methods
The genome of S. alactolyticus strain FGM was sequenced and used for genomic in silico studies. It was evaluated for morphology, antibiotic susceptibility, hemolytic activity, acid tolerance, bile salt tolerance, adherence ability to Caco-2, and inhibitory pathogens activity.
Result
The GC content of the strain FGM was 40.38% and composed of 29 contigs. The annotation of coding genes revealed important characteristics of the germs, especially 151 genes annotated to biological adhesion. The strain FGM forecasted 43 amino acid sequences to be VF, but did not have a hemolytic gene, and neither did it show hemolytic activity in phenotypic analysis. Although 30 amino acid sequences were predicted to aid in resisting some antibiotics, the strain FGM just showed the resistance to trimoxazole and oxytetracycline, and intermediate resistance to kanamycin. FGM cells were showed the tolerance to pH 2 broth within 4 h, and 0.15~0.30% bile salt medium with the latter being attributed to the presence of bile-salt hydrolase. The strain FGM was shown to have the ability to adhere to Caco-2 cells and the adherence rate of 1.0 × 109 CFU/mL bacterial suspensions was 37.51%. Compared with Lactobacillus acidophilus, Lactobacillus reuteri, and Lactobacillus casei, the strain FGM showed a high capability to inhibit the diffusion of Escherichia coli O78 and reduce its adhesion on Caco-2 cells.
Conclusion
The results demonstrated the presence of probiotic potential and absence of adverse effects for the Streptococcus alactolyticus strain FGM in vitro, thus contributing to develop a safety and effective fermentation feed for animals.
Similar content being viewed by others
Background
Lactic acid bacteria (LAB) are gram-positive, anaerobic, and catalase-negative rods or cocci bacteria which can ferment carbohydrate and produce plenty of lactic acids (Rinkinen et al. 2004). It was reported that many LAB increase the stability of intestinal environment and contribute to the health of the host by enhancing the gut microbiota balance through secretion of antibacterial substance (organic acids, bacteriocins) and competitive adhesion to the epithelium or by the stimulation of the GI immune system (Tallon et al. 2007). LAB of the genera Enterococcus, Lactobacillus, Lactococcus, Bifidobacteria, Pediococcus, Saccharomyces, and Streptococcus, are being widely used as veterinary drugs and feed additives (Vitetta et al. 2017; Aponte et al. 2020; Shin et al. 2019; Maria et al. 2014). These beneficial bacteria possess anti-inflammatory activity and antimicrobial properties, protect against oxidative stress and diabetes, as well as reducing the accumulation of reactive oxygen metabolites (Todorov 2009; Mazzoli et al. 2014; Lossa and Pollice 2019).
S. alactolyticus, a member of the genera Streptococcus, was originally described among isolates obtained from the intestinal tract of pigs and chickens and has been reported no hemolytic or α-hemolytic activity (Farrow et al. 1984). It has been proved as a dominant culturable LAB species of the commensal gut microbiota in animals such as chicken, pigs, dogs, ducks, pigeons, and fishes (Czerwinski et al. 2010; Baele et al. 2002). However, its characteristics related to survive and bioactivity in the animal guts were rarely reported.
In the broiler chicken gastrointestinal tract, the cecum is the main site of fermentation (Jozefiak et al. 2006). S. alactolyticus strain FGM was domesticated, screened, and isolated from bacterial load in the chicken cecum to ferment Astragalus membranaceus and produce fermented Astragalus polysaccharides (FAPS) in China (Hao et al. 2013). Previous studies showed that polysaccharide content of A. membranaceus fermented by S. alactolyticus strain FGM is four-fold higher than that before fermentation, and the biological effect of A. membranaceus after fermentation is the similar as that before fermentation, such as body weight gain and immunopotentiation of broiler chicken, obvious antagonism against hepatic fibrosis induced by CCl4 exposure on rats (Qin et al. 2012; Zhang et al. 2011). In view of the good fermentation performance and application prospect of S. alactolyticus strain FGM, its own characteristics and safety were further studied. The comparison against GenBank of the 16S rRNA gene showed that the strain belongs to S. alactolyticus, and a similarity index higher than 96% was presented between the strain FGM and S. alactolyticus EU728776.1 (Wang 2012). The strain was proved without toxic reaction and causing death on mice in acute oral toxicity test (Wang et al. 2020), and could improve the health of broilers infected by E. coli O78 through protecting the intestinal villi structure, balancing the intestinal immunity, reducing the number of E. coli, improving the anti-inflammatory and anti-oxidation (Wu et al. 2020). From these pieces of evidence, S. alactolyticus appears to be a promising strain. So we characterized its safety and potential probiotic properties in vitro using genomic analyses and phenotypic tests in this study.
Materials and methods
Strains used and growth conditions
S. alactolyticus strain FGM (GenBank accession No. JX435470; China Patent No. 20120141827.5) was isolated from chicken cecum in Lanzhou City of China and preserved in the Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences. The strain FGM, Lactobacillus reuteri (BeNa Culture Collection, China, No. 192190), Lactobacillus acidophilus (BeNa Culture Collection, China, No. 185342), and Lactobacillus casei (BeNa Culture Collection, China, No.134415) were cultured anaerobically in Macconkey broth (Hopebio, China) at 2% (v/v) volume at 37 °C for 24 h. E. coli O78 (China Veterinary Culture Catalogue, No. 1418) was cultured in Nutrient broth (Hopebio, China) at 1% (v/v) volume at 37 °C for 24 h.
Genomic analysis and bioinformatics
The genomic DNA of S. alactolyticus strain FGM was extracted by Bacterial genomic DNA Extraction Kit (OMEGA, USA) following the manufacturer’s instructions. The DNA was sent to Beijing Novogene Science and Technology Co., Ltd. for full gene sequencing by Illumina Hiseq and Miseq platform. After assembling and optimizing the reads by SOAPdenovo software, the genome components of strain FGM were predicted using five softwares: GeneMarkS (verison 4.17), RepeatMasker (verison 4.0.5), rRNAmmer(verison 1.2), tRNAscan-SE (verison 1.3.1), and Rfam (verison 11.0). The database of Gene Ontology (GO) was used to predict and assign the function of genes from strain FGM. To further investigate biological properties and safety of relevance, the antibiotic resistance genes were detected using the Comprehensive Antibiotic Resistance Database (CARD) (Jia et al. 2017). The presence of genes associated with bacterial pathogenesis was then checked against the virulence factors database (VFDB) (Liu et al. 2018).
Colony and strain morpologhy
S. alactolyticus strain FGM was inoculated on a blood agar plate (Hopebio, China) at 37 °C for 20~24 h anaerobically, before being analyzed for colony morphology and gram staining. The strain FGM cells were collected and washed 3 times by sterile PBS (Solarbio, China). They were fixed with 2.5% glutaraldehyde solution (Ameko, USA) at 4 °C for 1 h. After washing 3 times by sterile PBS (pH 7.2–7.4), the cells were dehydrated in sequence by 30%, 50%, 60%, 70%, 80%, 90%, 95%, and 100% ethyl alcohol. The specimens were critical-point dried using liquid CO2 and sputter-coated with gold before the examination on a scanning electron microscope (SEM, JSM-5600, JEOL, Japan).
Antibiotic susceptibility test
Antibiotic susceptibility test was evaluated by the disk diffusion method using drug-sensitive slips (Hangwei, China). Kanamycin (30 μg/table), ciprofloxacin (5 μg/table), penicillin (10 μg/table), trimoxazole (23.75/1.25 μg/table), erythromycin (15 μg/table), norfloxacin (10 μg/table), cefalotin (30 μg/table), lincomycin (2 μg/table), oxytetracycline (30 μg/table), and amoxicillin (20 μg/table) antibiotics were used in this test. A total of 100 μL 1.0 × 108 CFU/mL FGM suspensions were evenly spread on blood agar plates and the drug-sensitive papers put on them. After incubation for 24 h at 37 °C, the inhibitory zone diameters (IZD) of drugs were determined by vernier calipers.
Hemolytic activity test
Fresh culture of S. alactolyticus strain FGM was streaked on blood agar plates, incubated at 37 °C for 48 h, and then evaluated for the presence of hemolytic haloes. S. gallolyticus (ATCC9809) was included as a positive control. The experiment was set three replicates.
Acid tolerance
Acid tolerance of S. alactolyticus strain FGM was tested as previously described (Li et al. 2016). A 2% (v/v) volume of the overnight cultures of strain FGM were added to MRS broth with the pH adjusted to 2.0 by 2 mol/L HCl. In the control group, the same counts of cells were added to MRS broth, and the pH value of the initial suspension was 6.0. All the groups were incubated for 4 h in an incubator at 37 °C and 100 r/min. Samples were taken at 0, 1, 2, 3, and 4 h, respectively. Three repeats were set up at each observation point. After serial twofold dilutions, the suspensions were counted to calculate the survival rate of live cells.
Bile salt tolerance
The method used to assess bile salt tolerance was referenced from literature (Kaushik et al. 2009). A total of 2% (v/v) volume of the overnight cultures of S. alactolyticus strain FGM were inoculated in MRS broth supplemented with 0.15% and 0.3% (w/v) bile salts (Sigma-Aldrich, USA) followed by incubation at 37 °C and 100 r/min for 4 h. Samples were taken at 0, 1, 2, 3, and 4 h, respectively. Three repeats were set up at each observation point. After serial twofold dilutions, the cells were counted to calculate the survival rate of S. alactolyticus strain FGM.
Adherence assays to Caco-2 cells
Microscopic inspection of strain FGM’s adherence ability was referenced from literature (Zhang 2014). Caco-2 cell lines were routinely cultured in MEM/EBSS full medium (Hyclone, USA) supplemented with 10% (v/v) fetal bovine serum (AusgeneX, Australia) and 1% antibiotic penicillin/streptomycin (Sigma-Aldrich, USA). Aliquots of 6 ml containing 5 × 105 Caco-2 cells/mL were seeded on a 6-well Corning tissue culture plate and incubated at 37 °C in a 5% CO2 humid atmosphere until a complete monolayer of cells was obtained. After washing 4 times using sterile PBS, the wells were filled with 2 mL 1.0 × 108 CFU/mL FGM suspensions in MEM/EBSS medium (without antibiotics) and incubated at 37 °C in 5% CO2 humid atmosphere for 2 h. The medium of the wells was discarded, and the wells washed with sterile PBS for 4 times to clean out the unbound bacteria. The cells in wells were then fixed with 4% neutral formalin (Solarbio, China) for 15 min. After removing the fixative, the gram staining procedure was done as described in the protocol of the Gram staining kit (Solarobio, China). Finally, the wells were evaluated under a microscope (LZXS-YQ-D03-003, Olympus, Japan) with a × 100 objective lens.
The adherence ability of strain FGM was assessed more exactly following the method previously described (Bianchi et al. 2010; Li et al. 2016). FGM cells were harvested after overnight cultivation and adjusted to 1.0 × 109 CFU/mL, 6.0 × 108 CFU/mL, and 3.0×108 CFU/mL concentrations with sterile PBS. The bacterial solutions were labeled by FITC (Solarbio, China) working solution at 37 °C, 120 r/min, 1 h. After washing with sterile PBS for 4 times, the samples were resuspended with MEM/EBSS full medium without antibiotics. The bacterial solution at each concentration without FITC tagging was prepared as background values.
Caco-2 cells were seeded in each well of a 6-well tissue culture plate as described in section 2.8.1 above. A total of 1 mL of FGM-FITC suspensions of different concentrations and 1 mL MEM/EBSS full medium (without antibiotics) were added to the wells. The mixtures were incubated at 37 °C for 2 h in a 5% CO2 humid atmosphere. The cells in the wells were washed 4 times with sterile PBS and then digested by 1 mL 0.25% EDTA (Solarbio, China) for 5 min. One milliliter MEM/EBSS full medium (without antibiotics) was then added to each well to stop digestion. The fluorescence intensity of the collected cells was tested by Microplate Reader (SpectraMax M2, USA) at the absorption and emission wavelength of 485 nm and 530 nm, respectively. The unlabeled FGM at each concentration incubated with Caco-2 cells were prepared as background values. Four replications were set up at each observational point.
Bacterial adhesion rate
The bacterial adhesion rate was calculated using the formula:
Where: CFGM-FITC + Caco-2 means the fluorescence intensity of Caco-2 cells after incubated with FGM tagged by FITC
CFGM + Caco-2 means the fluorescence intensity of Caco-2 cells after incubated with FGM without tagged by FITC
CFGM-FITC means the fluorescence intensity of FGM tagged by FITC
CFGM means the fluorescence intensity of FGM without tagged by FITC.
Inhibitory pathogens activity
Inhibitory diffusion of E. coli O78
E. coli O78 solution was cultured in Macconkey broth (Hopebio, China) at 37 °C for 24 h at 2% (v/v) volume. The cells were harvested after overnight cultivation and adjusted to 1.0 × 108 CFU/mL with sterile PBS. The sterile swabs were used to spread E. coli O78 on Macconkey agar plates (Hopebio, China) evenly. After drying at room temperature for 5 min, the Oxford cups were placed at the marked location on the plates and were filled with 200 μL 1.0 × 109 CFU/mL probiotic suspension (L. reuteri, L. acidophilus or L. casei) or FGM cells suspension, respectively. The Oxford cups in the control group were filled with 200 μL MRS broth. The plates were incubated at 37 °C for 24 h and then IZD of germs were determined by Vernier calipers. The experiment was done in four replicates.
Inhibitory adherence of E. coli O78
E. coli O78 cells were harvested after overnight cultivation and adjusted to 1.0 × 109 CFU/mL with sterile PBS. After labeling with FITC, the bacterial suspension was washed with sterile PBS and resuspended in MEM/EBSS full medium. The bacterial suspension without FITC tagging was prepared as background values. Caco-2 cells were seeded in each well of the 6-well tissues culture plate as described in section 2.8.1 above. A total of 1 mL E. coli-FITC suspension was put in each well, and 1 mL 6.0 × 108 CFU/mL probiotic samples (L. reuteri, L. acidophilus or L. casei) or FGM cells suspension added respectively. The mixtures were incubated at 37 °C for 2 h in a 5% CO2 humid atmosphere. The cells were washed 4 times with sterile PBS before being digested by 1 mL 0.25% EDTA for 5 min. Digestion was stopped by the addition of 1 mL MEM/EBSS full medium (without antibiotics) to the wells. The fluorescence intensity of the collected cells was tested by Microplate Reader at the absorption wavelength is 485 nm and the emission wavelength of 530 nm. The unlabeled E. coli O78 at each concentration incubated with Caco-2 cells were used as background values. Four repeats were set up at each observational point.
The loss rate of adhering E. coli (%)
This was calculated using the formula:
Where: CEC-FITC + Caco-2 means the fluorescence intensity of Caco-2 cells after incubated with E. coli tagged by FITC.
CEC + Caco-2 means the fluorescence intensity of Caco-2 cells after incubated with E. coli without tagged by FITC.
CEC-FITC + CB + Caco-2 means the fluorescence intensity of Caco-2 cells after incubated with E. coli tagged by FITC and the competitive bacteria.
CEC + CB + Caco-2 means the fluorescence intensity of Caco-2 cells after incubated with E. coli without tagged by FITC and the competitive bacteria.
Statistical analysis
Data were analyzed using one-way analysis of variance (ANOVA) and Tukey’s test was used to perform post hoc analysis using the GraphPad Prism software (version 7, GraphPad Software, Inc., San Diego, CA, USA). Statistical significance was set at P < 0.05.
Results
Genomic analysis and bioinformatics
The genome sequence of S. alactolyticus strain FGM was uploaded to DDBJ/EMBL/GenBank (Accession No. JAAXMT000000000). An overview of the genome assemblies and annotations of the strain FGM are summarized in Table 1. The germ was assembled into 17 scaffolds (> 500 bp) and 29 contigs which were predicted from the scaffolds (> 500 bp). The guanine-cytosine (GC) content and core-genome size of strain FGM was 40.38% and 1.7 Mbp, respectively. The length of the longest contig and scaffold were 600 Kb and 972 Kb, respectively, which indicated fine continuity of assembly. The whole-genome component of strain FGM was predicted to consist of 1768 coding genes (which make up 87.7% of the whole genome), 127 scattered repeats, 38 tandem repeats, 21 minisatellite DNA, 36 tRNA, 4 rRNA, and 1 sRNA.
The database of the Gene Ontology system (GO) was used to annotate the predicted genes of strain FGM (Fig. 1). A total of 2655 genes were describing molecular function, 1359 genes describing cellular component, and 1902 genes describing biological processes in the strain.
Putative resistance-related genes of strain FGM are identified and listed in Table 2. Thirty amino acid sequences were predicted to aid in resisting some antibiotics in the strain FGM including β-lactam, macrolide, lincomycin, polypeptide, aminoglycoside, quinolone, streptogramin, tetracycline, and sulfonamide. Besides, the strain was potentially tolerant to phenicol which promotes the bile secretion and thus, might be related to the bile salt tolerance of the strain.
Finally, we used the VFanalyzer software to predict potential virulence factors (VF) and filtered the comparison results according to identity and E-value. As shown in Table 3, 43 amino acid sequences were forecasted to be VF in the strain FGM. These amino acids could be contributing to the overall adaptability of bacteria to harsh environments and various stresses. The most related genes were attributed to the capsule which has been proved to inhibit the binding of the activated complement factor C3b to the surface of germ, prevent the activation of the alternative complement pathway, and inhibit complement-mediated opsonophagocytosis. According to the result, strain FGM had 10 genes that were properly involved in the biological process of adhering host cells. The strain also had several other genes that proved to participate in multi-biological functions. However, the hemolytic gene was missing in the strain.
Morphological features
As shown in Fig. 2a, the strain FGM formed opaque, white, circular, and flat glistening colonies with neat edges on the MRS agar plate. The microscopic and SEM observation of strain FGM in Fig. 2b, c showed that the cells were gram-positive, oval-shaped, always in pairs or chains, and without spores or flagella. The Feret’s diameter of strain FGM was 1.40 ± 0.11 μm.
Antibiotic susceptibility test
The antibiotic susceptibility of S. alactolyticus strain FGM was tested using ten antibiotics which were classified into β-lactam, aminoglycosides, quinolone, sulfonamides, macrolides, lincosamides, and tetracyclines (Table 4). It was found that the strain FGM is susceptible to Ciprofloxacin, Penicillin, Trimoxazole, Cephalothin Lincomycin, Erythromycin, Erythromycin, and Amoxicillin, and resistant to Trimoxazole (sulfonamides) and Oxytetracycline (tetracyclines). Furthermore, Kanamycin (aminoglycosides) showed intermediate inhibition on the strain FGM.
Hemolysin detection
Based on hemolytic activity, streptococcus is divided into α-hemolytic streptococcus, β-hemolytic streptococcus, and γ-streptococcus (without hemolytic and pathogenic). In this study, no hemolysis ring was observed on the blood plates of S. alactolyticus strain FGM after 48 h of culturing (Fig. 3a). Contrastingly, the obvious hemolysis rings were shown on the blood plates with S. gallolyticus ATCC9809 (Fig. 3b). From the VF and phenotypic predictions at the gene level, it could be concluded that S. alactolyticus strain FGM belonged to γ-streptococcus.
Acid tolerance
Gastric acid tolerance (pH 2–3) is one of the barriers that probiotic bacteria must overcome to survive (Tulumoglu et al. 2018). Normally, the liquid food stays in the stomach of monogastric animals for 1~2 h (Shen et al. 2007). As shown in Fig. 4, the bacterial concentration in the control group increased with time (0 h~4 h). Compared to the control group (CG), the growth of FGM cells in the acid-treated group was extremely inhibited at 3 h and 4 h (P < 0.01). The survival rate of live cells rates in pH 2 group were 91.18% (1 h), 86.51% (2 h), 58.85% (3 h), and 17.73% (4 h). However, it should be noted that there were still about 3.10 × 108 CFU/mL alive cells in pH 2 broth at 4 h.
The bacterial concentration of FGM cells in different acid-treated groups. Results are expressed as means ± SD (n = 3) of viable cells. The pH value of the control group (CG) was 6.0. The data of treated groups were compared with the data of CG at the same time point. Mean values with a letter (a) differ significantly (P < 0.05), and with different letters (ab) differ extremely significantly (P < 0.01).
Bile salt tolerance
Bile plays a fundamental role in the specific and nonspecific defense mechanisms of the gut. The magnitude of its inhibitory effects is determined primarily by the concentration of bile salts (Charteris et al. 1998). Normally, relevant physiological concentrations of animal bile range from 0.03 to 0.30% (Chang et al. 2001). As shown in Fig. 5, compared with the control group, the growth of FGM cells in the treated groups were both inhibited obviously at 1 h~4 h (P < 0.05). The germ survival rates in 0.15% BS group were 77.44% (1 h), 77.08% (2 h), 71.04% (3 h), and 26.93% (4 h). The germ survival rates in 0.30% BS group were 65.10% (1 h), 60.06% (2 h), 46.06% (3 h), and 16.35% (4 h). The status of bacterial proliferation in the 0.30% BS group was worse than in the 0.15% BS group. However, at 4 h, there were still 4.67 × 108 CFU/mL alive cells in the broth with 0.15% bile salts and 2.83 × 108 CFU/mL alive cells in the broth with 0.30% bile salts.
The bacterial concentration of FGM cells in different bile treated groups with bile salts. Results are expressed as means ± SD (n = 3) of viable cells. The data of treated groups were compared with the data of CG at the same time point. Mean values with a letter (a) differ significantly (P < 0.05), and with different letters (ab) differ extremely significantly (P < 0.01)
Adherence assays to Caco-2 cells
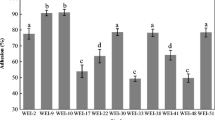
As shown in Fig. 6b, a certain amount of gram-positive, oval-shaped, paired or chained strain FGM cells adhered to caco-2 cells, and attached mainly at the outside of the cell membrane. On the contrary, no such observation was made in Caco-2 cells in Fig. 6a. To evaluate the adherence ability of bacteria to Caco-2 cells easily and accurately, an improved fluorescence tagging assay and a designed formula were used to calculate the bacterial adhesion rate. Figure 7 showed that S. alactolyticus strain FGM was capable of adhering to the intestinal epithelial cells. There was a positive correlation between the concentration of germ and the adherence rate. The adherence rate of FGM cells with 1.0 × 109 CFU/mL was 37.51 ± 3.18% and was the highest among groups which (P < 0.05).
Adherence rate of different concentrations of strain FGM on Caco-2 cell by fluorescence detection. The concentrations of strain FGM in group 1, group 2, group 3 were 3.0 × 108 CFU/mL, 6.0 × 108 CFU/mL and 1.0 × 109 CFU/mL, respectively. Results are expressed as means ± SD (n = 4) of viable cells. Mean values with a letter (a) differ significantly (P < 0.05)
Pathogens inhibitory activity
In our test, two methods, anti-E. coli growth test and competition test with E. coli were used to compare the pathogens inhibitory ability of S. alactolyticus strain FGM with other 3 recognized probiotics, and E. coli O78 was selected as the indicator bacteria. The diameter of the inhibition zone against E. coli O78 in 4 strains is shown in Fig. 8. The inhibition spectrum of strain FGM was significantly bigger than those of L. acidophilus, L. reuteri, and L. casei (P < 0.05). This indicated that FGM pellets have a strong feature to inhibit the growth of E. coli O78.
A pathogenic bacterium first attaches to the intestinal epithelium when it enters into the host intestines. Hence, inhibiting its adherence to the intestinal epithelium can decrease the infection rate in animals. As shown in Fig. 9, S. alactolyticus strain FGM and other regarded probiotics (L. acidophilus, L. casei, and L. reuteri), all prevented E. coli O78 from adhering to Caco-2 cells. The adherence loss rate of E. coli O78 in the strain FGM group was significantly higher than in the group of L. acidophilus (P < 0.05), and significantly lower than in the group of L. reuteri (P < 0.05). There was no obvious difference in the loss of adherence rate of E. coli O78 between the strain FGM group and the L. casei group (P > 0.05). The result proved that S. alactolyticus strain FGM can inhibit the adhesion of E. coli O78 to the epithelium.
Discussion
With increasing knowledge about their essential role in the host health, the gut microbiota is now considered an important ally, interacting with most cells (Cani 2018). Recent studies showed that probiotics in the gut microbiome play a very important role in the health of humans and animals (Vitetta et al. 2017). S. alactolyticus strain FGM isolated from the chicken cecum has a good ability to ferment A. membranaceus, which improves the concentration of polysaccharides, and the fermented products are more beneficial to the health of broilers. It is reported that the health benefits of fermented food is very associated with the probiotic lactic acid bacteria (Chen et al. 2020). To indicate the safety and benefits of application in animal feed in future, this study sequenced the genome of S. alactolyticus strain FGM and subjected it to genomic analyses and probiotic properties analysis.
Undoubtedly, genome sequencing and analysis of potential probiotic candidates have become mandatory in the last years, to gain information on their safety aspects and functional properties (Reid et al. 2019; Esther et al. 2020). However, no literature had reported on the genome information of S. alactolyticus so far. The sequencing analysis showed that the GC content of strain FGM is 40.38% which is similar to the other species of the genus Streptococcus (Lifu et al. 2013). Based on the assembly sequence of genes, 2655 genes were annotated according to the GO database. It is noticeable that the strain FGM had a high number (151) of genes annotated to biological adhesion. The capability to adhere to animal intestinal cells is an important characteristic for probiotic bacteria to favor their colonization of the host gut. Using 1.0 × 108 CFU/mL bacterial concentration incubated with Caco-2 cells, Li et al. found that the highest adherence rate among S. alactolyticus strains isolated from swine was 9.713% (Li et al. 2016). In this study, we used image observation and fluorescent quantitation to evaluate the capability of S. alactolyticus strain FGM to adhere to intestinal epithelial cells, and the adherence rate of 1.0 × 109 CFU/mL FGM cells was 37.51 ± 3.18%. The result indicates the strain FGM has capability to adhere to intestinal epithelial cell.
Because of the increasing concern on the diffusion of antibiotic-resistant bacteria, probiotic bacteria must be checked for the presence of antibiotic resistance genes that could be transferred to other bacteria, in particular to pathogens (Curragh and Collins 1992). Contrary to transmissible antibiotic resistance, intrinsic resistance could be quietly favorable, giving a chance to probiotics to survive an antibiotic treatment on the host (Charteris et al. 1998). The susceptibility result of S. alactolyticus strain FGM was not strictly consistent with the detection of antibiotic resistance genes conducted using CARD, and just showed the resistance to trimoxazole and oxytetracycline. S. alactolyticus strains from the respiratory and genitourinary of porcine clinical specimens in Brazil were reported to have high resistance rates to tetracyclines and macrolides (Moreno et al. 2016). The resistance to kanamycin is also considered intrinsic in all streptococci strains (Farrow et al. 1984). However, S. alactolyticus strain FGM did not show resistance to kanamycin (Aminoglycosides) and Erythromycin (Macrolides) in the test. β-Hemolytic activity is one of the main safety concerns besides antibiotics susceptibility and should therefore be evaluated even for strains belonging to bacterial species that possess the QPS or GRAS status (Armin et al. 2019). In our study, S. alactolyticus strain FGM did not show the presence of hemolytic gene and activity which reflected its safety. This was similar to the previous findings (Farrow et al. 1984)
As potential probiotics for animals, the bacteria have to adapt to the environments with acid and bile salt to survive in the gastrointestinal tract. It was also reported that chyme can decrease the damage degree of probiotics by acid when passing through the gastrointestinal tracts. Once the bacteria pass through the stomach and duodenum, they enter into the ileum and cecum with the chyme and increase drastically (Kos et al. 2000). Our results indicated that S. alactolyticus strain FGM could remain a certain amount cells in pH 2 broth and 0.15~0.30% bile salt broth in 1~4 h. It showed a similar property to S. alactolyticus strains (FB027, FB034, FB018) for bile salt tolerance and acid tolerance (Li et al. 2016). Besides, the functions of amino acids predicted by VFanalyzer and CARD such as bile-salt hydrolase, capsule, MOMP, and cfrA contribute to the survival and colonization of the strain in the harsh gastrointestinal environment.
Research has shown that lactic acid bacteria with high adhesion can compete with pathogenic bacteria for adhesion receptors of the intestinal epithelium and get priority for colonization, to effectively inhibit colonization of pathogenic bacteria and eliminate them (Sun et al. 2007). Meanwhile, competition from lactic acid bacteria for nutrition also limits the excessive multiplication of pathogenic bacteria and maintains the balance of intestinal microecology (Tallon et al. 2007). Lactobacillus known as probiotics showed strong adherence and inhibition to pathogens adhesion and growth (Ma et al. 2006; Fernández et al. 2018). In this paper, we compared S. alactolyticus strain FGM with three lactobacillus strains on pathogens inhibition. The result indicated that S. alactolyticus strain FGM was similar with the Lactobacillus strains, not only inhibited significantly the growth of E. coli O78 on adapted medium but also induced competitively the loss of adhering E. coli O78 amount on Caco-2 cells. This probiotic property provides a reasonable basis for why the strain FGM could improve the health of broilers infected by E. coli O78 (Wu et al. 2020).
Conclusions
The genomic and phenotypic assessments of safety and probiotic features were studied for Streptococcus alactolyticus strain FGM isolated from the chicken cecum in China that already had indicated some technological potential on herb fermentation. Our results demonstrated the absence of adverse effects in the strain regarding hemolytic activity, hemolytic gene, susceptibility to most commonly used antibiotics, putative resistance-related genes and survive-related genes. We also got promising results regarding probiotic potential, such as resistance to acid and bile salts, adhesion ability to Caco-2 cells and inhibition activity to E. coli, thus contributing to increase our knowledge of S. alactolyticus. However, further studies are needed in order to exclude any possible safety concern and supplement other benefits for animal health prior to use the strain FGM in animal feed.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
22 September 2021
A Correction to this paper has been published: https://doi.org/10.1186/s13213-021-01651-7
References
Aponte M, Murru N, Shoukat M (2020) Therapeutic, prophylactic, and functional use of probiotics: a current perspective. Front Microbiol 11:562048. https://doi.org/10.3389/fmicb.2020.562048
Armin T, Vinícius SD, Shadi P, Viviana C, Alessio G (2019) Genomic and phenotypic assessments of safety and probiotic properties of Streptococcus macedonicus strains of dairy origin. Food Res Int 130:108931. https://doi.org/10.1016/j.foodres.2019.108931
Baele M, Devriese LA, Butaye P, Haesebrouck F (2002) Composition of enterococcal and streptococcal flora from pigeon intestines. J Appl Microbiol 92(2):348–351. https://doi.org/10.1046/j.1365-2672.2002.01537.x
Bianchi MA, Rio DD, Pellegrini N, Sansebastiano G, Brighenti F (2010) Fluorescence-based method for the detection of adhesive properties of lactic acid bacteria to Caco-2 cells. Lett Appl Microbiol 39:301–305
Cani PD (2018) Human gut microbiome: hopes, threats and promises. Gut 67(9):1716–1725. https://doi.org/10.1136/gutjnl-2018-316723
Chang YH, Kim JK, Kim HJ, Kim WY, Kim YB, Park YH (2001) Selection of a probiotic lactobacillus strain and subsequent invivo studies. Antonie Van Leeuwenhoek 80(2):193–199. https://doi.org/10.1023/A:1012213728917
Charteris WP, Kelly PM, Morelli L, Collins JK (1998) Antibiotic susceptibility of potentially probiotic Lactobacillus species. J Food Prot 61(12):1636–1643. https://doi.org/10.4315/0362-028x-61.12.1636
Chen ZD, Kang JY, Zhang Y, Yi XX, Gao X (2020) Differences in the bacterial profiles and physicochemical between natural and inoculated fermentation of vegetables from Shanxi Province. Ann Microbiol 70(1):66. https://doi.org/10.1186/s13213-020-01605-5
Curragh HJ, Collins MA (1992) High levels of spontaneous drug resistance in Lactobacillus. J Appl Microbiol 73(1):31–36
Czerwinski J, Hojberg O, Smulikowska S, Engberg RM, Mieczkowska A (2010) Influence of dietary peas and organic acids and probiotic supplementation on performance and caecal microbial ecology of broiler chickens. Br Poultry Sci 51(2):258–269. https://doi.org/10.1080/00071661003777003
Esther M, Jose DF, Martha HRB, Jose MI, Paula GF, Alvaro P, Encana V (2020) Genome analysis of Endobacterium cerealis, a Novel Genus and Species Isolated from Zea mays Roots in North Spain. Microorganisms 8:939
Farrow JAE, Kruze J, Phillips BA, Bramley AJ, Collins MD (1984) Taxonomic studies on Streptococcus bovis and Streptococcus equinus: description of Streptococcus alactolyticus sp. nov. and Streptococcus saccharolyticus sp. nov. Syst Appl Microbiol 5(4):467–482. https://doi.org/10.1016/S0723-2020(84)80004-1
Fernández S, Fraga M, Silveyra E, Trombert AN, Ravaza A, Pla M, Zunino P (2018) Probiotic properties of native Lactobacillus spp. strains for dairy calves. Benef Microbes 9(4):613–624. https://doi.org/10.3920/BM2017.0131
Hao GJ, Zhang K, Zhang JY, Wang XR, Qin Z, Wang XZ, Wang L, Meng JR, Yang ZQ, Li JX (2013) RT-qPCR analysis of dexB and galE gene expression of Streptococcus alactolyticus in Astragalus membranaceus fermentation. Appl Microbiol Biotechnol 97(13):6009–6018. https://doi.org/10.1007/s00253-013-4873-2
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, McArthur AG (2017) Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573. https://doi.org/10.1093/nar/gkw1004
Jozefiak D, Rutkowski A, Jensen BB, Engberg RM (2006) The effect of beta-glucanase supplementation of barley- and oat-based diets on growth performance and fermentation in broiler chicken gastrointestinal tract. Br Poultry Sci 47(1):57–64. https://doi.org/10.1080/00071660500475145
Kaushik JK, Kumar K, Duary RK, Mohanty AK, Grover S, Batish VK (2009) Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS One 4(12):e8099. https://doi.org/10.1371/journal.pone.0008099
Kos B, Suskovic J, Goreta J, Matosic S (2000) Effect of protectors on the viability of Lactobacillus acidophilus M92 in simulated gastrointestinal conditions. Food Technol Biotechnol 38:121–127
Li LL, Wang YT, Yang X, Zhou LX, Wang LH, Du EQ (2016) Isolation and characterizeation of lactic acid bacteria from swine. J Northwest A&F Univ 44(02):1–7
Lifu S, Wei W, Georg C, Anke R, Helena S, Michael R, Irene WD, An-ping Z (2013) Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics 14:430
Liu B, Zheng D, Jin Q, Chen L, Yang J (2018) A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 36:D539–D542
Lossa A, Pollice RI (2019) Bacillus megaterium SF185 spores exert protective effects against oxidative stress in vivo and in vitro. Sci Rep 9:12082
Ma YL, Guo T, Xu ZR, You P, Ma JF (2006) Effect of Lactobacillus isolates on the adhesion of pathogens to chicken intestinal mucus in vitro. Lett Appl Microbiol 42(4):369–374. https://doi.org/10.1111/j.1472-765X.2006.01844.x
Maria LP, Laurie O, Shiau PT, Peter M, Helen H, Montserrat G, Jonathan AL, Rita MH, Peadar GL, Gillian EG (2014) In vitro assessment of marine bacillus for use as livestock probiotics. Mar Drugs 12(5):2422–2445
Mazzoli A, Donadio G, Lanzilli M, Saggese A, Guarino AM, Rivetti M, Crescenzo R, Ricca E, Ferrandino I, Pieniz S, Andreazza R, Anghinoni T, Camargo F, Brandelli A (2014) Probiotic potential, antimicrobial and anti-oxidant activities of Enterococcus durans strain LAB18s. Food Control 37:251–256
Moreno LZ, Matajira CEC, Gomes VTM, Silva APS, Mesquita RE, Christ APG, Sato MIZ, Moreno AM (2016) Molecular and antimicrobial susceptibility profiling of atypical Streptococcus species from porcine clinical specimens. Infect Genet Evol 44:376–381. https://doi.org/10.1016/j.meegid.2016.07.045
Qin Z, Li JX, Yang ZQ, Zhang K, Zhang JY, Meng JR, Wang L, Wang L (2012) Effects of Fermentation Astragalus Polysaccharides on Experimental Hepatic Fibrosis. J Anim Vet Adv 11:1195–1203
Reid G, Gadir AA, Dhir R (2019) Probiotics: reiterating what they are and what they are not. Front Microbiol 10:242. https://doi.org/10.3389/fmicb.2019.00424
Rinkinen ML, Koort JMK, Ouwehand AC, Westermarck E, Björkroth KJ (2004) Streptococcus alactolyticus is the dominating culturable lactic acid bacterium species in canine jejunum and feces of four fistulated dogs. FEMS Microbiol Lett 1:35–39
Shen ZY, Zhao SM, Liang YX (2007) Screening of lactic acid bacteria strains for pig additives and the study of its probiotic properties in vitro. J Huazhong Agric Univ 26(3):348–352
Shin D, Chang SY, Bogere P, Won K, Choi JY, Choi YJ, Lee HK, Hur J, Park BY, Kim Y, Heo J (2019) Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS One 14(8):e0220843. https://doi.org/10.1371/journal.pone.0220843
Sun J, Le GW, Shi YH, Su GW (2007) Factors involved in binding of Lactobacillus plantarum Lp6 to rat small intestinal mucus. Lett Appl Microbiol 44(1):79–85. https://doi.org/10.1111/j.1472-765X.2006.02031.x
Tallon R, Arias S, Bressollier P, Urdaci MC (2007) Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J Appl Microbiol 102(2):442–451. https://doi.org/10.1111/j.1365-2672.2006.03086.x
Todorov SD (2009) Bacteriocins from Lactobacillus Plantarum-Production, Genetic Organization and Mode of Action. Braz J Microbiol 40(2):209–221
Tulumoglu S, Erdem B, Simsek O (2018) The effects of inulin and fructo-oligosaccharide on the probiotic properties of Lactobacillus spp. isolated from human milk. Zeitschrift fur Naturforschung C 73(9-10):367–373
Vitetta L, Coulson S, Thomsen M, Nguyen T, Hall S (2017) Probiotics, D-Lactic acidosis, oxidative stress and strain specificity. Gut Microbes 8(4):311–322. https://doi.org/10.1080/19490976.2017.1279379
Wang H, Zhang H, Zhang K, Wang L, Zhang K, Ma YJ, Li JX, Yang XP, Zhang JY (2020) Preliminary safety evaluation of the fermented astragalus liquid by FGM strain. Chin J Vet Drug 54:49–56
Wang L (2012) Research on the biological characteristics of Streptococcus alactolyticus strain FGM for fermenting Astragalus polysaccharides. Dissertation, Chinese Academy of Agricultural Sciences, Beijing.
Wu KK, Zhang K, Wang L, Zhang K, Qiu ZY, Zhang LJ, Zhang JY, Li JX (2020) The effects of Streptococcus alactolyticus FGM on intestinal health in broilers infected with E. coli. Chin Vet Sci 50:1–7
Zhang K, Yang ZQ, Wang XZ, Meng JR, Zhang JY, Qin Z, Wang L, Wang L, Li JX (2011) Study on effects of fermented Radix Astragalus extraction on growth, immunoglobulin of broiler chicken. Hubei Agric Sci 50:1216–1218
Zhang W M (2014) Analysis of the colonization and probiotic effect of Lactobacilli with high adhesive ability isolated from piglet intestine and its surface proteins. Dissertation, Zhe Jiang University, Hangzhou.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China, grant number 31602101; the Key Tasks of Science and Technology Innovation Project of CAAS, grant number CAAS-LM-02; the National Key Research and Development Program of China, grant number 2017YFE0114400.
Author information
Authors and Affiliations
Contributions
Jingyan Zhang conducted research on correlation analysis and drafted a manuscript. Hong Zhang and Kang Zhang participated in the experiments. Lei Wang participated in its design, coordination, and modification. Zhengying Qiu participated in the data analysis. Kai Zhang and Yong Zhang improved the writing of the manuscript. Cong Yue participated in the modification of manuscript. Jianxi Li and Xingxu Zhao designed the experimental scheme, and carried out the overall planning and improvement of the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: The order of the corresponding authors in the author group has been corrected.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, J., Zhang, H., Wang, L. et al. The safety and potential probiotic properties analysis of Streptococcus alactolyticus strain FGM isolated from the chicken cecum. Ann Microbiol 71, 19 (2021). https://doi.org/10.1186/s13213-021-01630-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13213-021-01630-y