Abstract
Background
People living with HIV (PLHIV) on effective antiretroviral therapy are living near-normal lives. Although they are less susceptible to AIDS-related complications, they remain highly vulnerable to non-communicable diseases. In this exploratory study of older PLHIV (OPLHIV) in Eswatini, we investigated whether epigenetic aging (i.e., the residual between regressing epigenetic age on chronological age) was associated with HIV-related parameters, and whether lifestyle factors modified these relationships. We calculated epigenetic aging focusing on the Horvath, Hannum, PhenoAge and GrimAge epigenetic clocks, and a pace of biological aging biomarker (DunedinPACE) among 44 OPLHIV in Eswatini.
Results
Age at HIV diagnosis was associated with Hannum epigenetic age acceleration (EAA) (β-coefficient [95% Confidence Interval]; 0.53 [0.05, 1.00], p = 0.03) and longer duration since HIV diagnosis was associated with slower Hannum EAA (− 0.53 [− 1.00, − 0.05], p = 0.03). The average daily dietary intake of fruits and vegetables was associated with DunedinPACE (0.12 [0.03, 0.22], p = 0.01). The associations of Hannum EAA with the age at HIV diagnosis and duration of time since HIV diagnosis were attenuated when the average daily intake of fruits and vegetables or physical activity were included in our models. Diet and self-perceived quality of life measures modified the relationship between CD4+ T cell counts at participant enrollment and Hannum EAA.
Conclusions
Epigenetic age is more advanced in OPLHIV in Eswatini in those diagnosed with HIV at an older age and slowed in those who have lived for a longer time with diagnosed HIV. Lifestyle and quality of life factors may differentially affect epigenetic aging in OPLHIV. To our knowledge, this is the first study to assess epigenetic aging in OPLHIV in Eswatini and one of the few in sub-Saharan Africa.
Similar content being viewed by others
Introduction
The advent of highly effective antiretroviral therapy (ART) has shifted the landscape of HIV from a deadly disease to a chronic condition, often associated with multi-morbidities [1]. Access to ART and viral load suppression is associated with decrease in morbidity and mortality [2, 3], which has resulted in an increase in life expectancy for people living with HIV (PLHIV), from 8 to 10 years from time of HIV diagnosis [4] to near-normal life expectancy compared to the general population [5, 6]. As a consequence of ART use, PLHIV have experienced decreases in AIDS-related complications, however, are now at a greater risk of developing age-related comorbidities, including cardiovascular diseases (CVDs) [7], type 2 diabetes (T2D) [8], neurocognitive disorders [9], and cancers [10]. This change is particularly relevant for sub-Saharan Africa, where HIV remains highly prevalent, particularly in Eswatini, the country with the highest prevalence in the world, at 27.2% among those age 15–49 years old [11]. With the expansion of HIV treatment in sub-Saharan Africa, PLHIV there are showing marked increases in life expectancy [12]. The number of older PLHIV (OPLHIV), defined as those ≥ 50 years of age, is growing worldwide [13] and with the number of OPLHIV increasing in sub-Saharan Africa at a more rapid pace compared to higher-income countries [14] has resulted in increased rates of non-AIDS age-related comorbidities [15]. This is particularly challenging for countries that have few programs for persons with non-communicable diseases [16].
At the interface between HIV and HIV-associated comorbidities are aging trajectories that may be dysregulated in OPLHIV. Although HIV-infected people are living longer lives, they are aging at a faster rate, which is a likely outcome of the physiological effects of HIV [17]. A significant portion of the ART-treated population display aging-associated phenotypes and morbidities at a higher rate and at a significantly younger age as compared to the general aging population [18], including frailty [19], reduced bone density [20], and age-related non-communicable diseases (CVDs, T2D, metabolic dysfunction, dementias, etc.) [21,22,23,24]. Although there is a paucity of data, with the increase in ART treatment access, similar trends have been observed in regions of sub-Saharan Africa [15, 25, 26]. These differences in the aging process and the earlier onset of age-related phenotypes among OPLHIV may arise from differences in biological aging, the accumulation of damage to tissues and cells throughout the body as the aging process proceeds across the life-course; a process that is affected by environmental influences (pollutants, chemicals, lifestyle behaviors, infections, etc.) [27, 28].
DNA methylation (DNAm), the addition of a methyl group to the 5′ position of cytosine at a cytosine-guanine dinucleotide (CpG), is an epigenetic modification involved in regulating cellular phenotype via gene regulation without changes to the genotype [29]. Epigenetic mechanisms, including DNAm, have gained considerable attention in biomedical research, given that these epigenetic markers are responsive to environmental influences (e.g., lifestyle factors, infections, chemical exposures, etc.) and can regulate underlying cellular functions via transcriptional regulation, and when dysregulated, may underlie disease pathogenesis. Importantly, microarray-based DNAm data have been used for the creation of epigenetic clocks, developed to predict chronological aging, such as the first-generation Horvath [30] and Hannum clocks [31] (i.e., epigenetic age), and the second-generation clocks, PhenoAge [32] and GrimAge [33], developed as biomarkers of biological age, and, finally, the pace of biological aging biomarker, DunedinPACE [34]. Higher epigenetic age, or biological age, as compared to chronological age, termed epigenetic age acceleration (EAA), has been associated with a wide range of age-related diseases and mortality [35,36,37]. Further, evidence suggests HIV infection may increase biological age by more than 5 years in untreated individuals [38], and despite ART and viral load suppression, which have been shown to slow biological aging, it remains relatively increased [39]. Although it is generally accepted that biological age is accelerated in OPLHIV, little is known about biological aging in OPLHIV in sub-Saharan Africa, a region disproportionately affected by HIV.
Additionally, in response to the growing epidemic of non-AIDS comorbidities in OPLHIV, strategies have been implemented that are targeted at mitigating, preventing, and managing non-communicable diseases in OPLHIV [40, 41]. However, in lower-middle-income countries, programs focused on the prevention of comorbidities in OPLHIV are limited for several reasons, including lack of resources, such as trained staff, integration of programs with health care systems, institutional support (e.g., government, department of health, facilities, etc.), and material resources (e.g., educational materials, equipment for HIV care, etc.) [42]. The limited programs to address non-communicable diseases in OPLHIV likely stem from—in addition to the lack of resources—HIV programs in sub-Saharan Africa that are primarily focused on getting individuals diagnosed, on ART treatment, and virally suppressed [43]. Even in uninfected populations, prevention of multimorbidity through interventions is inadequate due to economic constraints [44]. Investigating underlying features of HIV, and lifestyle and health behaviors that are associated with the aging process, will create better informed strategies to mitigate HIV-associated comorbidities in lower- and lower-middle-income countries.
In this study, we sought to investigate epigenetic aging among OPLHIV on ART in Eswatini by assessing whether HIV-related factors affect the aging process and to determine whether certain modifiable lifestyle and quality of life factors modify this relationship.
Methods
Study population
Participants for this study were recruited in an exploratory study of OPLHIV in Eswatini, detailed elsewhere [45]. In brief, a convenience sample of 50 PLHIV age ≥ 50 years receiving care at the Mankayane Government Hospital in Manzini region of Eswatini were recruited from October 2016 to January 2017. All participants provided written, informed consent. The study was approved by the Eswatini Directorate of Health Services/Public Health, the Eswatini Ethics Committee, and the Columbia University Irving Medical Center Institutional Review Board.
Data collection
Quantitative interviews were conducted, and clinical and sociodemographic information were self-reported and abstracted from paper and electronic-based medical records for each participant. Trained staff collected sociodemographic information (e.g., age, education, employment/occupation, financial and medical support, etc.), medical history, HIV and sexually transmitted disease information, lifestyle behaviors (e.g., physical activity, smoking, diet), psychosocial behavior (e.g., depressive symptoms), contraceptive use, non-communicable disease history, injuries, violence, social support, and acute quality of life measurements.
To assess quality of life, the Short Form Health Survey (SF-36) was used [46], which is a 36-item questionnaire that measures self-reported quality of life by assessing responses across several functional domains, including vitality, physical function, bodily pain, general health perception, physical functioning, emotional functioning, social functioning, and mental health/emotional well-being with each response scored from 0 (low health status) to 100 (high health status). Self-reported dietary intake was measured using the CORE physical activity and diet questions from the WHO STEPwise approach to surveillance (STEPS) survey [47]. Average daily dietary intake was calculated based on the number of fruits and/or vegetable servings eaten on a typical day divided by the number of servings eaten in a week; and weekly dietary intake was the total of fruits and/or vegetables, calculated as the number of servings in a typical day multiplied by the number of days fruits/vegetables are eaten in a week. Physical activity was also assessed using the STEPwise approach. Here, we included self-reported physical activity as cumulative physical activity in any given week, which included work-related physical activity (number of days of vigorous- and moderate-intensity work-related activities × number of minutes on any given day of vigorous- and moderate-intensity work-related activities), travel-related physical activity (i.e., number of days per week spent biking or walking to work × number of minutes spent biking or walking to work), and leisure time physical activity (number of days of vigorous- and moderate-intensity leisure physical activity × the number of minutes on any given day of vigorous- and moderate-intensity leisure physical activity). Similarly, self-reported recreational/leisure time physical activity minutes were calculated by the number of days in a week multiplied by the number of minutes of moderate and vigorous physical activity. HIV-related factors included data that were either self-reported (age at HIV diagnosis, age at ART initiation) or abstracted from clinical charts (WHO HIV clinical stage at the most recent visit, CD4+ T cell count [cells/µL] at enrollment and ART initiation, viral load at most recent visit). The number of years since HIV diagnosis and number of years on ART were calculated as the difference between age at HIV diagnosis and age at ART initiation with the participant’s chronological age at most recent visit.
DNA methylation preprocessing, normalization, and quantification
Each participant had dried blood spot (DBS) specimens obtained using a finger prick; five separate drops (75–80 µL each) of blood were collected on a Protein Saver card. DNAm was quantified from the DBS that were collected from 46 of the initially recruited 50 participants following standardized protocols; two participants had low-quality DNA collected, one participant was missing abstracted data, and one participant was missing key self-reported questionnaire data. Blood spots were purified for genomic DNA. DNA was bisulfite-converted and hybridized to Illumina’s Infinium MethylationEPIC BeadChip (850K; Illumina, Inc. San Diego, CA, USA), improving on the coverage, reproducibility, and accuracy from its predecessor the HumanMethylation450 BeadChip, and allowing for the quantification of DNAm at > 850,000 CpGs at single-nucleotide resolution across the genome, covering > 99% RefSeq genes. Raw DNAm IDAT files were generated and loaded into the R statistical environment (version 4.2.2) using the minfi package in R [48]. Initial quality control removed one sample whose mean detection p-value across all probes was unreliable (detection p-value ≥ 0.01). Next, normalization was performed using the ENmix package in R for background correction using a flexible exponential-normal mixture distribution along with a truncated normal distribution to reflect background noise, followed by a dye-bias correction method using RELIC, which corrects for dye-bias across the array between the two color channels of the 850K [49, 50]. Next, we performed inter-array normalization using the quantile normalization for methylation intensities between samples. One sample out of the 46 collected that lacked typical bimodal distribution was also excluded from downstream analyses. Next, we performed probe filtering to remove low-quality CpG probes across samples and removed CpG probes when detection p-values were not significant against the background (detection p-value > 0.01) in one or more individuals, resided on sex chromosomes, harbored known single-nucleotide polymorphisms (SNPs) in the probe sequence, and were cross-reactive. Finally, due to the differential abundance in type I and type II probes on the 850K, we performed probe-type bias adjustment using the regression on correlated probes (RCP) normalization method within the ENmix package, which recalibrates type II probes to nearby type I probes. From 45 participants, DNAm values (β-values) were thus generated for 763,250 CpGs, which is based on the ratio of the methylated CpG probe to the total methylation state of the probe (methylated + unmethylated probe) resulting in a DNAm proportion from 0.0 (unmethylated) to 1.0 (fully methylated). Due to potential confounding caused by differences in cell-type heterogeneity that underlies epigenetic variability in heterogeneous samples (e.g., blood spots, whole blood, etc.) [51, 52] coupled with changes in immune cell composition inherent to the aging process [53] and during HIV infection [54], we estimated cell-type proportion of each blood sample (CD4+ T cell, CD8+ T cell, NK cell, monocyte, B cell, neutrophil), using the Houseman’s projection method employed in the estimateCellCounts function in the minfi package [55] to include in our epigenetic aging analyses.
Epigenetic clock calculation
In order to estimate participants’ epigenetic/biological age, we included the two first-generation epigenetic clocks, the pan-tissue Horvath clock [30] and the blood immune cell Hannum clock [31], and two second-generation clocks that are shown to predict age-related phenotypes and lifespan (i.e., biological age), including PhenoAge [32] and GrimAge [33], these second-generation clocks differ from the first-generation epigenetic clocks that were trained to predict chronological age (i.e., epigenetic age). Due to one participant’s Hannum clock estimated epigenetic age deviating by more than 50 years compared to chronological age, we removed this participant from further analyses. Epigenetic aging trajectories were defined as the residuals between regressing estimated epigenetic/biological age on chronological age. When the resulting residual was positive, participants’ epigenetic/biological age was considered accelerated as compared to their chronological age (i.e., EAA) and negative values represented epigenetic age deceleration (EAD). Finally, we employed the DunedinPACE biomarker, which is intended as a DNAm-based biomarker that measures the rate of biological aging per calendar year [34].
Statistical analyses
Descriptive statistics for participants who had reliable DNAm data available (n = 44) were calculated as median (1st quartile, 3rd quartile) for continuous variables and counts (%) for categorical variables.
For all statistical analyses, we focused on the first-generation Horvath and Hannum clocks, the second-generation PhenoAge and GrimAge clocks, and the DunedinPACE biomarker of the pace of biological aging. Due to the missingness of data from those enrolled in this study, our analyses specifically focused on HIV-relevant clinical parameters, and lifestyle and perceived quality of life variables that were present in ≥ 70% of participants. To test whether HIV-related variables were associated with EAA in OPLHIV in Eswatini, in our primary analysis, we performed linear regression analyses between HIV-relevant variables as the predictor variables (number of years living with HIV since diagnosis [years], age at HIV diagnosis [years], number of years on ART [years], age at ART initiation [years], CD4+ T cell count at enrollment [cells/µL], CD4+ T cell count at ART initiation [cells/µL]) and epigenetic aging changes as the outcome variable (i.e., residuals of regressing estimated epigenetic metrics on chronological age), adjusting for relevant variables selected a priori, including age (years), sex (male or female), educational attainment (none, primary school, secondary school, high school, tertiary school), smoking status (never or ever), and estimated immune cell-type composition calculated from DNAm data (CD4+ cells, CD8+ cells, NK cells, B cells, monocytes, and neutrophils), which we considered in our primary analysis as Model 1. Although evidence suggests cell-type composition may underlie the variability in DNAm-based measures of biological aging, we have included regression models without further adjustment for DNAm estimations of cell-type composition in Additional file 1.
To investigate whether health and lifestyle factors modified the relationships between HIV-related variables and epigenetic aging, we first utilized linear regression analyses to test the association between health and lifestyle factors (i.e., average daily dietary intake of fruits and vegetables, weekly dietary intake of fruits and vegetables, total physical activity, and SF-36) as the predictor variable and epigenetic aging as the outcome. Next, we employed three separate models to determine whether lifestyle and perceived health factors modified the relationship between HIV-related characteristics and epigenetic aging, including, (1) a model that adjusted for a priori-selected covariates (the model used in the previous analysis [Model 1]) and average daily dietary intake fruits/vegetable servings (Model 2); (2) Model 1 with further adjustment for self-reported quality of life based on the SF-36 questionnaire (Model 3); and (3) Model 1 with further adjustment for total physical activity (total minutes of moderate and vigorous work, travel/commute, and recreational/leisure activity in a week) (Model 4). Statistical significance was defined as p < 0.05. Finally, similar to our other analyses, we have included regression models without DNAm-based estimation of cell-type composition in Additional file 1. Linear regression β-estimates (95% Confidence Intervals [CIs]) were calculated in R (version 4.2.2), and graphical figures were created using Prism (version 8; Dotmatics, Boston, MA).
Results
Participant characteristics
Descriptive statistics of participants included in this analysis (n = 44) are provided in Table 1. The median age (interquartile range [IQR]) was 59 [54, 66] years, slightly more than half of the participants were female (24 participants [54%]), the average weight was 75 (66, 87) kg, and the highest education attainment for most participants was primary school (grades 1–7; 23 participants [52%]). Participants had a median age of 53 (47, 58) years when diagnosed with HIV and had been living with HIV since diagnosis for 7 (5, 9) years until the most recent visit when DNAm data were collected. The median age at ART initiation was 53 (47, 59) years old, and participants had been on ART for 6 (4, 8) years. Most of the participants (40 [91%]) were categorized in WHO stage 1 (asymptomatic) at the most recent visit. CD4+ T cell counts at enrollment was 629 (457, 837) cell/µL, and at ART initiation, it was 202 (106, 289) cells/µL. At the most recent visit, the majority of participants were virally suppressed, including 40 participants that had ideal viral suppression (< 200 copies/mL), which included 27 participants with lower than detectable virus, 7 with < 20 copies/mL, and 6 with < 115 copies/mL, while the remaining 4 were missing (data not shown). Nine participants (21%) reported missing ART at least once in the last month. The median SF-36 quality of life score was 69 (53, 90). Participants reported consuming 0.8 (0.6, 1.3) fruits and/or vegetables per day, and no participants met the WHO standard for vegetable and fruit servings per week. Participants had 855 (390, 2220) minutes of total physical activity per week, with the majority through their work and commuting. Sixteen participants (36%) were past smokers, and three participants are current smokers (7%).
HIV status and epigenetic age acceleration
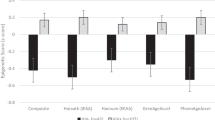
Descriptive statistics of epigenetic age calculated for each epigenetic clock and DunedinPACE are detailed in Table 1. Participants were generally biologically older when using the Horvath and PhenoAge clocks, having a median (IQR) age of 60 (56, 57) years and 68 (63, 77) years, respectively, as compared to a chronological age of 59 (54, 66) years. Participants were biologically younger using the Hannum (52 [45, 58] years) and GrimAge clocks (56 [50, 61] years) as compared to chronological age. Given the differences observed between epigenetic age and chronological age, we sought to determine whether HIV-related variables were associated with differences in epigenetic aging trajectories. Overall, we found no significant association between HIV-related variables with Horvath, PhenoAge, GrimAge or DunedinPACE (Additional file 1: Table S1; all p > 0.05). Using the Hannum epigenetic clock, we observed the age when participants were diagnosed with HIV was associated with a higher EAA (0.53 [0.05, 1.00], p = 0.03; Fig. 1; Additional file 1: Table S1) and the number of years since HIV diagnosis was associated with lower EAA (− 0.53 [− 1.00, − 0.05], p = 0.03; Fig. 1; Additional file 1: Table S1). Further, the age at which participants began ART treatments was approaching significance in its relationship with Hannum EAA (0.55 [-0.03, 1.12], p = 0.06) and the number of years on ARTs association with Hannum EAA was trending toward significance (− 0.55 [− 1.12, 0.03], p = 0.06).
Effect sizes for association between clinical variables of HIV status and epigenetic aging from older people living with HIV (OPLHIV) (N = 44), Eswatini, 2016–17. Effect estimates (95% Confidence Interval [CI]) for the change in epigenetic age acceleration (EAA) calculated from dried blood spots from OPLHIV using HIV status-related variables, including: age at HIV diagnosis (red), years since HIV diagnosis (green), age at antiretroviral therapy (ART) initiation (light blue), years on ART (pink), CD4+ T cell count at enrollment (navy) and CD4+ T cell count at ART initiation (tan). Figure depicts change in Hannum EAA. Plots are adjusted for age, sex, educational attainment, past smoking status, and estimated cell-type composition (CD4+ T cell, CD8+ T cell, NK cell, B cell, monocyte, and neutrophil). Significance taken at p < 0.05, represented by *; p < 0.05 *
Health and lifestyle factors and HIV-associated epigenetic age acceleration
We next investigated whether quality of life and lifestyle behaviors were associated with epigenetic aging (Additional file 1: Table S2). We observed a significant association between the average daily intake of fruits and vegetables and DunedinPACE (0.12 [0.03, 0.22]; p = 0.01). There were no significant associations for the first-generation or second-generation clocks with lifestyle and quality of life characteristics (all p > 0.05) (Additional file 1: Table S2).
Next, we sought to investigate whether health and lifestyle behaviors modified the relationship between HIV status and epigenetic aging trajectories (Fig. 2A–C; Additional file 1: Table S1). We found no notable effects of lifestyle factors on the associations between Horvath, GrimAge and DunedinPACE with clinical variables of HIV status (Additional file 1: Table S1). The association of age at HIV diagnosis with Hannum EAA in our primary model (Model 1) remained statistically significant after further adjustment for self-reported quality of life measurements derived from the SF-36 report (0.48 [0.04, 0.92], p = 0.03; Fig. 2A, Additional file 1: Table S1), which was similarly observed between Hannum EAA and the number of years since HIV diagnosis (− 0.48 [− 0.92, − 0.04], p = 0.03). However, when further adjusting our primary model for average daily intake of fruits and vegetables (Model 2) and physical activity (Model 4), we found the association between age at HIV diagnosis and Hannum EAA was attenuated and resulted in a non-significant association (Model 2: 0.47 [− 0.07, 1.00], p = 0.08; Model 4: 0.39 [− 0.08, 0.86], p = 0.10; Fig. 2A, Additional file 1: Table S1). Similarly, when adjusting for average daily intake of fruits and vegetables and physical activity, we found an attenuated association between the number of years since HIV diagnosis and Hannum EAA (Model 2: − 0.47 [− 1.06, 0.11], p = 0.10; Model 4: − 0.45 [− 0.99, 0.09], p = 0.10; Fig. 2B, Additional file 1: Table S1).
Effect sizes for the associations between HIV status variables and epigenetic aging adjusted for lifestyle and quality of life factors. Effect estimates (95% CI) for the change in A Hannum EAA by age at HIV diagnosis, B Hannum EAA by years since HIV diagnosis, and C Hannum EAA by CD4+ T cell counts at enrollment. Each plot represents effect estimates for four separate models: Model 1 (Green; covariates: age, sex, education level, past smoking status, and estimated cell-type composition); Model 2 (Orange; Model 1 + weekly dietary servings of fruits/vegetables); Model 3 (Navy; Model 1 + SF-36 questionnaire [perceived quality of life]); Model 4 (Green-Yellow; Model 1 + total physical activity [work-related moderate/vigorous + commuting + leisure/recreational moderate/vigorous activity] Significance taken at p < 0.05, represented by *; p < 0.05 *
We also observed that CD4+ T cell counts at enrollment were associated with higher Hannum EAA in the model that included the average daily intake of fruits and vegetables (0.02 [0.001, 0.03], p = 0.03; Fig. 2C, Additional file 1: Table S1) or self-reported quality of life measurements (0.01 [0.001, 0.02], p = 0.03; Fig. 2C, Additional file 1: Table S1). Lastly, we found a negative association that was approaching significance between CD4+ T cell counts after ART initiation with Horvath EAA (− 0.03 [− 0.07, 0.004], p = 0.07; Additional file 1: Table S1) and PhenoAge EAA (− 0.04 [− 0.08, 0.004], p = 0.07; Additional file 1: Table S1) in the model that was further adjusted for average daily intake of fruits and vegetables.
Discussion
Our results show that the age at HIV diagnosis and the duration since HIV diagnosis are associated with differences in epigenetic aging. HIV diagnosis at an older age was associated with higher epigenetic aging, and conversely, those living with HIV for a longer duration since HIV diagnosis displayed lower epigenetic aging. Additionally, our results showed that those who were diagnosed with HIV at an older age exhibited increased epigenetic aging. Prior to DNAm-based measures of epigenetic aging, several studies have reported increased incidence of age-related phenotypes and biological changes associated with aging in PLHIV, which occurred at a significantly younger age compared to non-infected counterparts [17, 56, 57]. This has been further established with the recent improvement in epigenetic clocks, with results showing that epigenetic and biological age is accelerated in PLHIV compared to HIV-uninfected individuals, although most of these studies were conducted in higher-income countries [38, 58]. In our study, we found epigenetic age was accelerated in OPLHIV in Eswatini as compared to their chronological age, which may reflect other reports suggesting higher epigenetic aging in PLHIV as compared to their non-infected counterparts [38, 58]. Whether epigenetic age in PLHIV is accelerated compared to non-infected individuals in Eswatini warrants further investigation. Finally, we found that lifestyle and self-reported measures of quality of life modified the relationship between HIV-related variables with epigenetic aging trajectories.
HIV infection has extensive effects on immune cellular activity and function, including, most notably, CD4+ cell depletion [59], and such changes in immune cell activity are characteristic of biological aging [60]. Even with early ART administration, immune cell functions are incompletely restored, and chronic adverse immune-related phenotypes persist [61, 62]. Low-grade, chronic inflammation is a significant feature of biological aging, termed by some as ‘inflammaging’, and it can have detrimental effects on physiology and cellular functions, and has been linked to age-related disease pathogenesis [63, 64]. Regarding the significant association between the age at HIV diagnosis and Hannum-derived epigenetic aging, having acquired HIV at an older age, when inflammaging is likely already running its course, may further exacerbate the inflammatory effects of HIV on an already inflammatory milieu observed in the elderly [65], which may underlie the rapid functional decline and vulnerability to non-communicable diseases observed in OPLHIV. Likewise, a recent meta-analysis suggests the time between HIV infection and diagnosis is around 3 years [66]; however, this has been performed in high- and upper-middle-income countries and whether it is similar in lower-middle income countries, such as Eswatini, is not known, and may, in fact, be longer. This is critical, given that the longer time intervals between infection and diagnosis—where diagnosis would come with immediate ART treatment—is indispensable not only for prevention of transmission but for long-term disease management and strategies to combat age-related morbidity and premature mortality. Left untreated, HIV can augment inflammation and has been shown to accelerate the aging process [67, 68]. We posit that the older age of diagnosis could encompass a long window between infection and treatment in which the virus can cause a myriad of detrimental effects. Contrary to higher epigenetic aging associating with being diagnosed with HIV at an older age, the slower epigenetic aging observed in those that have been living longer with HIV since diagnosis may be the result of the beneficial effects of early intervention and long-term adherence to ARTs on restoring the immune system and improving aging trajectories. Notably, epigenetic age was accelerated in PLHIV that have untreated HIV infection [68] and initiation of ART may decelerate epigenetic aging [69]. Taken together, it is possible that the older age at infection may augment an already pro-inflammatory milieu typical of the elderly or suggests a potentially longer duration of living with untreated HIV, which can also have effects on epigenetic aging trajectories through inducing chronic pro-inflammatory phenotypes, while timely initiation of ART may benefit aging trajectories by attenuating the effects of untreated HIV.
Despite the use of ART, the risk of developing comorbidities in OPLHIV remains relatively high, which is likely due to one or more factors (e.g., effects of HIV, ART, lifestyle behaviors, etc.) that influence the aging process. To address this, several interventions have been implemented to reduce HIV-associated comorbidities. For example, the PRECluDE Consortium, funded by the National Institute of Health, has modeled several interventions and clinical trials for PLHIV for CVD prevention [40] and the REPRIEVE phase 3 trial has shown marked reductions in CVD risk using pitavastatin calcium in PLHIV [70], although these efforts have yet to be implemented in sub-Saharan Africa. Mounting evidence suggests that health-promoting behaviors are linked to slower aging [71, 72]. Practical interventions have been developed for prevention and management of chronic diseases in OPLHIV, including physical activity and nutritional interventions [73, 74]. Thus, developing practical strategies to mitigate comorbidities in OPLHIV will be valuable in lower-middle-income countries where resources remain constrained. Here, we found lifestyle and quality of life measures may have a modifying role in the association between HIV-related factors and epigenetic aging trajectories.
In our study, we found the relationship between epigenetic age acceleration and the age at HIV diagnosis, and the number of years since diagnosis was modified by physical activity and diet. Prior evidence suggests moderate-intensity physical activity, including from occupational activity, can improve clinical indicators of immune function in HIV-infected patients and improve aging [75, 76], which is especially beneficial to older populations [77]. To our knowledge, there is no data on the association of dietary interventions and epigenetic aging trajectories in PLHIV. Previous evidence in HIV-uninfected individuals suggests that dietary interventions can have beneficial effects on biological aging, decelerating, and potentially reversing, biological aging, including interventions focused on higher quality foods and specific types of diets (e.g., Mediterranean Diet, DASH Diet, etc.) [78,79,80,81,82,83]. The reduction in epigenetic age acceleration in those that have been diagnosed at an older age could be derived from nutritional needs by OPLHIV to achieve proper immune function, reduce HIV-related complications and comorbidities associated with unhealthy eating patterns, which have all been linked to aging [13, 84]. The reduction in slower epigenetic aging that associated with the number of years since diagnosis when adjusting for diet we speculated could have been due to an interaction between long-term ART adherence and nutrition that could potentially affect drug metabolism, drug effectiveness, and the effects of ARTs in combination with certain foods on metabolic outcomes [85].
Next, we observed CD4+ T cell counts after enrollment into this study were significantly associated with higher Hannum-derived epigenetic age acceleration in models adjusted for dietary intake of fruits and vegetables or self-reported quality of life measurements derived from the SF-36. At the time of enrollment, participants were not yet treated for HIV, and untreated patients exhibit increased rates of epigenetic aging. Although higher levels of CD4+ T cell counts in HIV-infected individuals is an indicator of good health, in the early stages of HIV (i.e., acute infection) and prior to treatment, when patients may have CD4+ T cell counts that put them at a low risk for AIDS and AIDS-related complications, mortality still remains high compared to their uninfected counterparts [86], which potentially operates through excessive immune activation and chronic inflammation [67]. Evidence suggests nutrient quality both predicts CD4+ cell counts and is associated with a lower risk of mortality in PLHIV [87], and diet quality is associated with CD4+ T cell count [88, 89]. Finally, the positive relationship between CD4+ T cell counts at enrollment and Hannum epigenetic age acceleration when adjusted for self-reported quality of life, SF-36, could be due to those that perceive themselves as generally healthier having poor health-related behaviors. In one study, women who perceived their health as better also reported adverse alcohol drinking behaviors [90]. In HIV-infected participants, higher perceived health was associated with loss-to-follow for interventions and clinical care for HIV [91, 92]. Altogether, our findings suggest a differential role for lifestyle and quality of life measurements on epigenetic aging trajectories at different stages in HIV management and supports the utilization of practical interventions that would be beneficial for modifying biological aging trajectories and non-communicable disease risk for OPLHIV in lower- and lower-middle-income countries where costly interventions are not yet feasible.
While this study is, to our knowledge, the first to investigate epigenetic aging in OPLHIV in Eswatini, it has several limitations. First, this is an exploratory study and thus the findings are constrained by the limited sample of participants [45]. As our study used a cross-sectional design, we could not ascertain whether epigenetic aging trajectories changed over time, especially from early stages of infection (untreated) to viral load suppression. Likewise, these data were limited to the HIV-related factors included in the study. For example, the duration of time since HIV diagnosis is not reflective of time since HIV infection. Accumulating evidence suggests that the timeframe immediately after exposure until ART initiation may be important to biological aging trajectories and long-term health outcomes [68, 93]. Future research should examine changes in biological aging trajectories longitudinally, prior to ART initiation and potentially as early as when exposure occurred. Further, although Eswatini is a lower-middle income country, it is still constrained of resource, having high levels of poverty and unemployment, and collecting data from regions of the world that are still constrained of resources has its inherent problems, such as the robust collection of data across a range of variables over time, and future work should seek to recruit and retain participants in order to obtain a diversity of high-quality clinical, sociodemographic, and viral information over time. Finally, our study was performed exclusively in OPLHIV in Eswatini, thus, the findings may not be generalizable to other populations or regions in sub-Saharan Africa, to other lower-middle income countries, or, in general, to other OPLHIV. Although our study had these limitations, this exploratory study showed compelling evidence that may underlie aging trajectories in OPLHIV in a lower-middle-income country, which may provide a foundation for future studies to further explore the molecular mechanisms of aging in PLHIV in sub-Saharan Africa and other resource limited settings.
Conclusions
We found evidence that HIV-related variables are associated with epigenetic aging in OPLHIV on stable ART in Eswatini, a country with limited resources in sub-Saharan Africa. We also showed that modifiable risk factors may be important in influencing epigenetic aging trajectories in OPLHIV, which may unveil potential avenues for practical interventions that may be relevant to slowing aging trajectories and non-communicable disease risk in OPLHIV in regions with limited resources. With increased access to ART in Eswatini, and other regions of sub-Saharan Africa, life expectancy of PLHIV will continue to increase with an expanding older population living with HIV, with increasing risk of non-communicable diseases. Developing practical interventions that are aligned with “healthy aging” will be critically important.
Availability of data and materials
The data that supports findings from this study are available upon reasonable request from TGH. The data are part of an on-going collaboration between ICAP and the Eswatini Ministry of Health, and thus, is not currently publicly available.
References
Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet. 2013;382(9903):1525–33.
Krentz HB, Kliewer G, Gill MJ. Changing mortality rates and causes of death for HIV-infected individuals living in Southern Alberta, Canada from 1984 to 2003. HIV Med. 2005;6(2):99–106.
Quinn TC. HIV epidemiology and the effects of antiviral therapy on long-term consequences. AIDS. 2008;22(Suppl 3):S7-12.
Porter K, Johnson AM, Phillips AN, Darbyshire JH. The practical significance of potential biases in estimates of the AIDS incubation period distribution in the UK register of HIV seroconverters. AIDS. 1999;13(14):1943–51.
Marcus JL, Leyden WA, Alexeeff SE, Anderson AN, Hechter RC, Hu H, et al. Comparison of overall and comorbidity-free life expectancy between insured adults with and without HIV infection, 2000–2016. JAMA Netw Open. 2020;3(6): e207954.
Trickey A, Sabin CA, Burkholder G, Crane H, d’Arminio Monforte A, Egger M, et al. Life expectancy after 2015 of adults with HIV on long-term antiretroviral therapy in Europe and North America: a collaborative analysis of cohort studies. Lancet HIV. 2023;10(5):e295–307.
Triant VA. Cardiovascular disease and HIV infection. Curr HIV/AIDS Rep. 2013;10(3):199–206.
Kalra S, Kalra B, Agrawal N, Unnikrishnan A. Understanding diabetes in patients with HIV/AIDS. Diabetol Metab Syndr. 2011;3(1):2.
Valcour V, Shikuma C, Shiramizu B, Watters M, Poff P, Selnes O, et al. Higher frequency of dementia in older HIV-1 individuals: the Hawaii Aging with HIV-1 Cohort. Neurology. 2004;63(5):822–7.
Chen CH, Chung CY, Wang LH, Lin C, Lin HL, Lin HC. Risk of cancer among HIV-infected patients from a population-based nested case–control study: implications for cancer prevention. BMC Cancer. 2015;15:133.
MacKellar D, Williams D, Bhembe B, Dlamini M, Byrd J, Dube L, et al. Peer-delivered linkage case management and same-day ART initiation for men and young persons with HIV infection-Eswatini, 2015–2017. MMWR Morb Mortal Wkly Rep. 2018;67(23):663–7.
Reniers G, Blom S, Calvert C, Martin-Onraet A, Herbst AJ, Eaton JW, et al. Trends in the burden of HIV mortality after roll-out of antiretroviral therapy in KwaZulu-Natal, South Africa: an observational community cohort study. Lancet HIV. 2017;4(3):e113–21.
Wing EJ. HIV and aging. Int J Infect Dis. 2016;53:61–8.
Negin J, Cumming RG. HIV infection in older adults in sub-Saharan Africa: extrapolating prevalence from existing data. Bull World Health Organ. 2010;88(11):847–53.
Roomaney RA, van Wyk B, Pillay-van Wyk V. Aging with HIV: increased risk of HIV comorbidities in older adults. Int J Environ Res Public Health. 2022;19(4):2359.
Neugut AI, El-Sadr WM, Ruff P. The looming threat: cancer in Sub-Saharan Africa. Oncologist. 2021;26(12):e2099–101.
Meir-Shafrir K, Pollack S. Accelerated aging in HIV patients. Rambam Maimonides Med J. 2012;3(4): e0025.
McMillan JM, Krentz H, Gill MJ, Hogan DB. Managing HIV infection in patients older than 50 years. CMAJ. 2018;190(42):E1253–8.
Bloch M. Frailty in people living with HIV. AIDS Res Ther. 2018;15(1):19.
Kruger MJ, Nell TA. Bone mineral density in people living with HIV: a narrative review of the literature. AIDS Res Ther. 2017;14(1):35.
Aberg JA. Aging and HIV infection: focus on cardiovascular disease risk. Top Antivir Med. 2020;27(4):102–5.
Pao V, Lee GA, Grunfeld C. HIV therapy, metabolic syndrome, and cardiovascular risk. Curr Atheroscler Rep. 2008;10(1):61–70.
Lam JO, Hou CE, Hojilla JC, Anderson AN, Gilsanz P, Alexeeff SE, et al. Comparison of dementia risk after age 50 between individuals with and without HIV infection. AIDS. 2021;35(5):821–8.
Capeau J. Premature aging and premature age-related comorbidities in HIV-infected patients: facts and hypotheses. Clin Infect Dis. 2011;53(11):1127–9.
Coetzee L, Bogler L, De Neve JW, Bärnighausen T, Geldsetzer P, Vollmer S. HIV, antiretroviral therapy and non-communicable diseases in sub-Saharan Africa: empirical evidence from 44 countries over the period 2000 to 2016. J Int AIDS Soc. 2019;22(7): e25364.
Yuyun MF, Sliwa K, Kengne AP, Mocumbi AO, Bukhman G. Cardiovascular diseases in Sub-Saharan Africa compared to high-income countries: an epidemiological perspective. Glob Heart. 2020;15(1):15.
Oblak L, van der Zaag J, Higgins-Chen AT, Levine ME, Boks MP. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res Rev. 2021;69: 101348.
Gavazzi G, Krause KH. Ageing and infection. Lancet Infect Dis. 2002;2(11):659–66.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67.
Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–91.
Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11(2):303–27.
Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022;11:e73420.
Salameh Y, Bejaoui Y, El Hajj N. DNA methylation biomarkers in aging and age-related diseases. Front Genet. 2020;11:171.
Joyce BT, Gao T, Zheng Y, Ma J, Hwang SJ, Liu L, et al. Epigenetic age acceleration reflects long-term cardiovascular health. Circ Res. 2021;129(8):770–81.
Faul JD, Kim JK, Levine ME, Thyagarajan B, Weir DR, Crimmins EM. Epigenetic-based age acceleration in a representative sample of older Americans: Associations with aging-related morbidity and mortality. Proc Natl Acad Sci USA. 2023;120(9): e2215840120.
Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212(10):1563–73.
Schoepf IC, Esteban-Cantos A, Thorball CW, Rodés B, Reiss P, Rodríguez-Centeno J, et al. Epigenetic ageing accelerates before antiretroviral therapy and decelerates after viral suppression in people with HIV in Switzerland: a longitudinal study over 17 years. Lancet Healthy Longev. 2023;4(5):e211–8.
Gamble-George JC, Longenecker CT, Webel AR, Au DH, Brown AF, Bosworth H, et al. ImPlementation REsearCh to DEvelop Interventions for People Living with HIV (the PRECluDE consortium): combatting chronic disease comorbidities in HIV populations through implementation research. Prog Cardiovasc Dis. 2020;63(2):79–91.
Lagrange RD, Mitchell SJ, Lewis M, Abramowitz S, D'Angelo LJ. Health protective behaviors among young people living with HIV/AIDS. J AIDS Clin Res. 2012;01(S1).
Ngcobo S, Scheepers S, Mbatha N, Grobler E, Rossouw T. Roles, barriers, and recommendations for community health workers providing community-based HIV care in Sub-Saharan Africa: a review. AIDS Patient Care STDS. 2022;36(4):130–44.
Harris TG, Rabkin M, El-Sadr WM. Achieving the fourth 90: healthy aging for people living with HIV. AIDS. 2018;32(12):1563–9.
Alkhatib A, Nnyanzi LA, Mujuni B, Amanya G, Ibingira C. Preventing multimorbidity with lifestyle interventions in Sub-Saharan Africa: a new challenge for public health in low and middle-income countries. Int J Environ Res Public Health. 2021;18(23):12449.
Harris TG, Floren S, Mantell JE, Nkambule R, Lukhele NG, Malinga BP, et al. HIV and aging among adults aged 50 years and older on antiretroviral therapy in Eswatini. Afr J AIDS Res. 2021;20(1):107–15.
Ware JE, Sherbourne CD. The MOS 36-item short-form health survey (SF-36). I. Conceptual framework and item selection. Med Care. 1992;30(6):473–83.
Riley L, Guthold R, Cowan M, Savin S, Bhatti L, Armstrong T, et al. The World Health Organization STEPwise approach to noncommunicable disease risk-factor surveillance: methods, challenges, and opportunities. Am J Public Health. 2016;106(1):74–8.
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9.
Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44(3): e20.
Xu Z, Langie SA, De Boever P, Taylor JA, Niu L. RELIC: a novel dye-bias correction method for illumina methylation BeadChip. BMC Genom. 2017;18(1):4.
Lowe R, Rakyan VK. Correcting for cell-type composition bias in epigenome-wide association studies. Genome Med. 2014;6(3):23.
Houseman EA, Kelsey KT, Wiencke JK, Marsit CJ. Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC Bioinform. 2015;16:95.
Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8(9):1844–65.
Wang S, Zhang Q, Hui H, Agrawal K, Karris MAY, Rana TM. An atlas of immune cell exhaustion in HIV-infected individuals revealed by single-cell transcriptomics. Emerg Microbes Infect. 2020;9(1):2333–47.
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86.
Akusjarvi SS, Neogi U. Biological aging in people living with HIV on successful antiretroviral therapy: do they age faster? Curr HIV/AIDS Rep. 2023;20(2):42–50.
Smith RL, de Boer R, Brul S, Budovskaya Y, van Spek H. Premature and accelerated aging: HIV or HAART? Front Genet. 2012;3:328.
Horvath S, Lin DTS, Kobor MS, Zoller JA, Said JW, Morgello S, et al. HIV, pathology and epigenetic age acceleration in different human tissues. Geroscience. 2022;44(3):1609–20.
Balasubramaniam M, Pandhare J, Dash C. Immune control of HIV. J Life Sci (Westlake Village). 2019;1(1):4–37.
Salam N, Rane S, Das R, Faulkner M, Gund R, Kandpal U, et al. T cell ageing: effects of age on development, survival & function. Indian J Med Res. 2013;138(5):595–608.
Wilson EM, Sereti I. Immune restoration after antiretroviral therapy: the pitfalls of hasty or incomplete repairs. Immunol Rev. 2013;254(1):343–54.
Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39(4):633–45.
Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9(6):7204–18.
Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4-9.
Nasi M, De Biasi S, Gibellini L, Bianchini E, Pecorini S, Bacca V, et al. Ageing and inflammation in patients with HIV infection. Clin Exp Immunol. 2017;187(1):44–52.
Gbadamosi SO, Trepka MJ, Dawit R, Jebai R, Sheehan DM. A systematic review and meta-analysis to estimate the time from HIV infection to diagnosis for people with HIV. AIDS Rev. 2022;24(1):32–40.
Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–55.
Sehl ME, Breen EC, Shih R, Chen L, Wang R, Horvath S, et al. Increased rate of epigenetic aging in men living with HIV prior to treatment. Front Genet. 2021;12: 796547.
Sehl ME, Rickabaugh TM, Shih R, Martinez-Maza O, Horvath S, Ramirez CM, et al. The effects of anti-retroviral therapy on epigenetic age acceleration observed in HIV-1-infected adults. Pathog Immun. 2020;5(1):291–311.
Grinspoon SK, Fitch KV, Zanni MV, Fichtenbaum CJ, Umbleja T, Aberg JA, et al. Pitavastatin to prevent cardiovascular disease in HIV infection. N Engl J Med. 2023;389(8):687–99.
Thomas A, Belsky DW, Gu Y. Healthy lifestyle behaviors and biological aging in the US National Health and Nutrition Examination Surveys 1999–2018. J Gerontol A Biol Sci Med Sci. Series A. 2023; 78(9):1535–42.
Sabia S, Singh-Manoux A, Hagger-Johnson G, Cambois E, Brunner EJ, Kivimaki M. Influence of individual and combined healthy behaviours on successful aging. CMAJ. 2012;184(18):1985–92.
Botros D, Somarriba G, Neri D, Miller TL. Interventions to address chronic disease and HIV: strategies to promote exercise and nutrition among HIV-infected individuals. Curr HIV/AIDS Rep. 2012;9(4):351–63.
Cade WT, Reeds DN, Mondy KE, Overton ET, Grassino J, Tucker S, et al. Yoga lifestyle intervention reduces blood pressure in HIV-infected adults with cardiovascular disease risk factors. HIV Med. 2010;11(6):379–88.
Dempsey PC, Musicha C, Rowlands AV, Davies M, Khunti K, Razieh C, et al. Investigation of a UK biobank cohort reveals causal associations of self-reported walking pace with telomere length. Commun Biol. 2022;5(1):381.
Dalene KE, Tarp J, Selmer RM, Ariansen IKH, Nystad W, Coenen P, et al. Occupational physical activity and longevity in working men and women in Norway: a prospective cohort study. Lancet Public Health. 2021;6(6):e386–95.
Shim MS, Noh D. Effects of physical activity interventions on health outcomes among older adults living with HIV: a systematic review and meta-analysis. Int J Environ Res Public Health. 2022;19(14):8439.
Simpson RJ, Campbell JP, Gleeson M, Krüger K, Nieman DC, Pyne DB, et al. Can exercise affect immune function to increase susceptibility to infection? Exerc Immunol Rev. 2020;26:8–22.
Fitzgerald KN, Hodges R, Hanes D, Stack E, Cheishvili D, Szyf M, et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging (Albany NY). 2021;13(7):9419–32.
Kim Y, Huan T, Joehanes R, McKeown NM, Horvath S, Levy D, et al. Higher diet quality relates to decelerated epigenetic aging. Am J Clin Nutr. 2022;115(1):163–70.
Kresovich JK, Park YM, Keller JA, Sandler DP, Taylor JA. Healthy eating patterns and epigenetic measures of biological age. Am J Clin Nutr. 2022;115(1):171–9.
Gensous N, Garagnani P, Santoro A, Giuliani C, Ostan R, Fabbri C, et al. One-year Mediterranean diet promotes epigenetic rejuvenation with country- and sex-specific effects: a pilot study from the NU-AGE project. Geroscience. 2020;42(2):687–701.
Fiorito G, Caini S, Palli D, Bendinelli B, Saieva C, Ermini I, et al. DNA methylation-based biomarkers of aging were slowed down in a two-year diet and physical activity intervention trial: the DAMA study. Aging Cell. 2021;20(10): e13439.
Martínez de Toda I, Maté I, Vida C, Cruces J, De la Fuente M. Immune function parameters as markers of biological age and predictors of longevity. Aging. 2016;8(11):3110–9.
Raiten DJ. Nutrition and pharmacology: general principles and implications for HIV. Am J Clin Nutr. 2011;94(6):1697S-S1702.
Bassett IV, Sax PE. Untreated HIV: harmful even at high CD4 cell counts. Lancet. 2010;376(9738):306–8.
Rawat R, McCoy SI, Kadiyala S. Poor diet quality is associated with low CD4 count and anemia and predicts mortality among antiretroviral therapy-naive HIV-positive adults in Uganda. J Acquir Immune Defic Syndr. 2013;62(2):246–53.
Keshani P, Sarihi S, Parsaie N, Joulaei H. Dietary pattern association with CD4 cells count in patients living with human immunodeficiency virus: a cross-sectional study. J Public Health Res. 2023;12(2):22799036231181200.
Somarriba G, Neri D, Schaefer N, Miller TL. The effect of aging, nutrition, and exercise during HIV infection. HIV AIDS (Auckl). 2010;2:191–201.
Riediger ND, Bombak AE, Mudryj AN. Health-related behaviours and their relationship with self-rated health among Canadian adults. BMC Public Health. 2019;19(1):960.
Tran N, Nishi A, Young LE, Endo A, Cumberland WG, Young SD. The role of perceived health in retention disparity: a HIV-testing-related behavioral intervention among African American and Latinx men who have sex with men in the United States. Prev Med Rep. 2023;33: 102195.
Fuente-Soro L, López-Varela E, Augusto O, Bernardo EL, Sacoor C, Nhacolo A, et al. Loss to follow-up and opportunities for reengagement in HIV care in rural Mozambique: a prospective cohort study. Medicine (Baltimore). 2020;99(20): e20236.
Schoepf IC, Esteban-Cantos A, Thorball CW, Rodes B, Reiss P, Rodriguez-Centeno J, et al. Epigenetic ageing accelerates before antiretroviral therapy and decelerates after viral suppression in people with HIV in Switzerland: a longitudinal study over 17 years. Lancet Healthy Longev. 2023;4(5):e211–8.
Acknowledgements
We would like to thank the Eswatini Ministry of Health leadership, the Mankayane Government Hospital staff, the Eswatini Health Laboratory Services, Wilson Ramos, Nicholas Kisyeri, Samkelo Simelane, Nomcebo Dlamini, Nomenzile Mamba, Sarah Floren, Joanne Mantell, Rejoice Nkambule, Nomthandazo Lukhele, Bongiwe Malinga, Rhinos Chekenyere, Gugu Maphalala, all the participants from Eswatini who took part in this study, and everyone else who helped with conducting the exploratory study, which our study builds from, without your help, none of this could have been made possible.
Author Disclaimer
The content of this study is solely the responsibility of the authors and does not represent the official views of any of the funding agencies.
Funding
This study was supported by the National Institute of Environmental Health Sciences Grant: T32ES007322 and R01ES032638-03S1 (CKD). This project was also supported by the Columbia University Mailman School of Public Health Dean’s Pilot Fund and Innovation Fund awards.
Author information
Authors and Affiliations
Contributions
CKD involved in manuscript authorship (writing, reviewing, editing, figures/tables), data processing (DNA methylation data), and data analyses. HW involved in manuscript authorship (reviewing, editing), data analysis interpretation, and study coordination. GLJ involved in sample processing and coordination. AK involved in data collection and management and technical supervision. RN involved in administrator for study participation. NGL involved in clinical specialist overseeing participant data collection. BPM involved in medical specialist administration of study. RC involved in study site coordination. WE-S involved in cohort oversight and supervision, HIV analysis and interpretation, and manuscript review and editing. AAB involved in intellectual resources of DNA methylation analyses, supervision, and manuscript authorship (reviewing, editing). TGH involved in conceptualization, supervision, interpretation, and manuscript authorship (writing, reviewing, and editing).
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Eswatini Directorate of Health Services/Public Health, the Eswatini Ethics Committee, and the Columbia University Irving Medical Center Institutional Review Board. Informed consent was obtained from all participants included in this study.
Consent to publish
Not applicable.
Competing interests
The authors declare they have no known competing financial or personal interests that could appear to influence the findings reported in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplementary Tables.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Dye, C.K., Wu, H., Jackson, G.L. et al. Epigenetic aging in older people living with HIV in Eswatini: a pilot study of HIV and lifestyle factors and epigenetic aging. Clin Epigenet 16, 32 (2024). https://doi.org/10.1186/s13148-024-01629-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-024-01629-7