Abstract
Background
Maternal smoking during pregnancy (MSDP) affects development of multiple organ systems including the placenta, lung, brain, and vasculature. In particular, children exposed to MSDP show lifelong deficits in pulmonary function and increased risk of asthma and wheeze. Our laboratory has previously shown that vitamin C supplementation during pregnancy prevents some of the adverse effects of MSDP on offspring respiratory outcomes. Epigenetic modifications, including DNA methylation (DNAm), are a likely link between in utero exposures and adverse health outcomes, and MSDP has previously been associated with DNAm changes in blood, placenta, and buccal epithelium. Analysis of placental DNAm may reveal critical targets of MSDP and vitamin C relevant to respiratory health outcomes.
Results
DNAm was measured in placentas obtained from 72 smokers enrolled in the VCSIP RCT: NCT03203603 (37 supplemented with vitamin C, 35 with placebo) and 24 never-smokers for reference. Methylation at one CpG, cg20790161, reached Bonferroni significance and was hypomethylated in vitamin C supplemented smokers versus placebo. Analysis of spatially related CpGs identified 93 candidate differentially methylated regions (DMRs) between treatment groups, including loci known to be associated with lung function, oxidative stress, fetal development and growth, and angiogenesis. Overlap of nominally significant differentially methylated CpGs (DMCs) in never-smokers versus placebo with nominally significant DMCs in vitamin C versus placebo identified 9059 candidate “restored CpGs” for association with placental transcript expression and respiratory outcomes. Methylation at 274 restored candidate CpG sites was associated with expression of 259 genes (FDR < 0.05). We further identified candidate CpGs associated with infant lung function (34 CpGs) and composite wheeze (1 CpG) at 12 months of age (FDR < 0.05). Increased methylation in the DIP2C, APOH/PRKCA, and additional candidate gene regions was associated with improved lung function and decreased wheeze in offspring of vitamin C-treated smokers.
Conclusions
Vitamin C supplementation to pregnant smokers ameliorates changes associated with maternal smoking in placental DNA methylation and gene expression in pathways potentially linked to improved placental function and offspring respiratory health. Further work is necessary to validate candidate loci and elucidate the causal pathway between placental methylation changes and outcomes of offspring exposed to MSDP.
Clinical trial registration ClinicalTrials.gov, NCT01723696. Registered November 6, 2012. https://clinicaltrials.gov/ct2/show/record/NCT01723696.
Similar content being viewed by others
Background
Maternal smoking during pregnancy (MSDP) is the leading preventable cause of prematurity, intrauterine growth restriction (IUGR), and perinatal death [1,2,3,4]; however, despite smoking cessation efforts, over 50% of smokers will continue to smoke during pregnancy [5, 6]. MSDP affects development of multiple organ systems including placenta, lung, brain, and vasculature [7,8,9,10,11,12] and is associated with altered DNA methylation in placenta, blood, and buccal epithelium [13,14,15,16,17,18,19,20,21,22,23,24]. Perhaps the best-characterized effects of MSDP on long-term offspring health are respiratory outcomes. Offspring exposed to MSDP exhibit lifetime decreases in airway function and increased risk of wheeze and asthma [12, 25, 26]. Mechanisms of MSDP on fetal lung development include both direct effects of nicotine, nicotine metabolites, and other harmful components in cigarette smoke on the developing fetus [12], and indirect effects on the feto-placental unit [27].
The placenta plays a key role in the overall health and development of the fetus through the supply of oxygen and nutrients, gas and waste exchange, and regulation of fetal growth in response to fetal endocrine signals [28]. Compromised placental function is associated with increased risk of cardiometabolic diseases in later life [29, 30], and smoking during pregnancy is specifically associated with impairment in both placental structure and function. For instance, multiple Doppler-ultrasound (Doppler-US) studies have shown that smoking during pregnancy decreases placental blood flow and increases umbilical artery pulsatility index [11, 31, 32]. Placental pathologies in smokers include increased thickness of the villous membrane, increased collagen deposition, decreased vascularization of the placental bed, as well as reduced intervillous area and capillary volume [27, 33].
DNA methylation (DNAm) is the covalent addition of a methyl group, found primarily at cytosine–guanine (CpG) dinucleotides, and can act to regulate gene expression at local and distant loci through modification of chromatin state and transcription factor binding [34]. DNA methylation regulates processes critical to placental development [35] and may provide a mechanistic link between in utero exposures and future health outcomes [36,37,38,39]. Placental DNAm is dysregulated with in utero exposure to heavy metals, alcohol, and air pollution [40,41,42], with some of the largest effect sizes reported in association with MSDP [15, 43,44,45,46,47,48,49]. Previous studies have reported global and gene-specific changes in placental DNAm in response to MSDP [19, 42, 50], and some of these changes are proposed to mediate the effect of MSDP on health outcomes, such as infant birth weight and psychiatric morbidity [13, 23, 51].
We have previously demonstrated that vitamin C supplementation (500 mg/day) to pregnant smokers unable to quit smoking significantly increased newborn lung function and infant airway function through one year of age in two double-blind, randomized control trials [52,53,54]. In our pilot clinical trial population, we demonstrated using targeted bisulfite sequencing that vitamin C supplementation during pregnancy could restore levels of DNAm in candidate genes in placenta, in parallel with improved lung function, as well as in cord blood and childhood buccal DNA [55]. We also recently demonstrated that vitamin C supplementation improved placental hemodynamics in an ancillary study of Doppler-US in a subset of participants in the second VCSIP RCT [56]. Therefore, improved lung function in offspring from pregnancies supplemented by vitamin C may parallel improved placental structure and function. We hypothesize that epigenome-wide analysis of placental DNAm in this cohort may reveal loci dysregulated with MSDP that are improved or protected by vitamin C supplementation, and that these changes may also relate to improved infant lung function.
To test this hypothesis, we measured placental DNAm genome-wide using the Illumina MethylationEPIC array platform in a subset of placentas collected at delivery from participants in the VCSIP RCT. We used a concept-driven analysis approach (Additional file 1: Figure S1) to identify candidate differentially methylated CpGs (DMCs) potentially relevant to lung function. We also performed differentially methylated region (DMR) analysis, which may provide greater evidence of a functional impact on gene expression than individual CpGs [57]. We conducted enrichment analyses within candidate overlapping (i.e., “normalized”) DMCs and DMRs to identify biological pathways and processes which vitamin C supplementation may protect, and expression quantitative trait methylation (eQTM) analysis to identify genes with mRNA correlated with methylation changes. Lastly, we examined association of candidate CpGs with infant lung function and composite wheeze measured at 12 months of age.
Results
Baseline characteristics
We measured epigenome-wide DNA methylation in placentas obtained at delivery from 72 smoking participants from the “Vitamin C to Decrease the Effects of Smoking in Pregnancy on Infant Lung Function” (VCSIP) multi-center, double-blind RCT (35 placebo and 37 vitamin C supplemented) and from 24 never-smokers for reference [53, 54, 58]. Additional file 2: Table S1 presents the maternal and infant demographics and birth statistics per group (never-smokers, placebo smokers, vitamin C smokers) for the 96 subjects with placental epigenome-wide DNAm measurements. There were no significant differences in baseline characteristics between the vitamin C-supplemented smokers and placebo-supplemented smokers included on the MethylationEPIC arrays. Gene expression (total RNA-sequencing) was also available in 71 of the placental samples (26 placebo, 27 vitamin C, and 18 never-smokers; Additional file 1: Figure S2) [56].
Differentially methylated loci between treatment groups
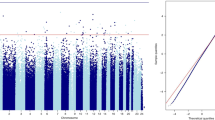
We performed epigenome-wide robust linear regression analysis to measure differential methylation in placentas from smokers randomized to placebo versus vitamin C. We also recruited pregnant never-smokers, as a reference group, to identify loci differentially methylated with MSDP and potentially normalized with vitamin C. All models were adjusted for cellular heterogeneity, infant sex, and gestational age at delivery. Comparison of vitamin C supplemented smokers and placebo smokers identified a single Bonferroni significant CpG (cg20790161; chr2:204553666), located in an intergenic region 17.5 Kbp upstream of CD28 (Fig. 1a; Table 1), which was also the only FDR-DMC between RCT groups. The majority (61%) of nominally significant CpGs between RCT groups had increased methylation in placentas from vitamin C smokers versus placebo. Comparison of never-smokers to placebo supplemented smokers identified 17 CpGs at Bonferroni significance (Fig. 1b; Table 1), 7 of which have been previously reported as differentially methylated with MSDP in placenta with consistent direction and magnitude of effect size and 726 CpGs with FDR adjusted p < 0.05. Results from sensitivity analyses adding ethnicity (self-reported), or removing adjustments for cell heterogeneity, infant sex, or gestational age, were not observably different from the original model (Additional File 1: Figure S3). The top CpG associated with maternal smoking status was cg27402634 (chr3:156536860), with a striking decrease of 31.4% in DNAm in placentas from placebo smokers versus never-smokers (beta scale SE = 0.018; p = 3.88E−31), and was not restored with vitamin C. The majority (82%) of MSDP associated loci (placebo-smokers compared with never-smokers) were hypomethylated. Novel CpGs differentially methylated in placebo versus never-smokers at Bonferroni significance mapped to 10 unique genes (MIR100HG, SSPO, ST3GAL6-AS1, MARK2, BDP1, SLC9A8, NECTIN3-AS1, PPP1R3G, PRDM2, and SRC) (Fig. 2).
Manhattan plots of differential placental DNAm between a placebo and vitamin C smokers and b placebo smokers and never-smokers. The horizontal red line marks Bonferroni adjusted significance (unadjusted p < 0.05/714666 CpGs = 6.99 e−08). The top 3 Bonferroni significant CpGs per comparison are annotated to the nearest proximal gene
Selection of candidate CpG sites used for functional enrichment analysis, eQTM analysis, and association with FEF75 and wheeze at 12 months of age. a Venn diagram showing overlap of nominally significant DMCs between groups (n = 9541 CpGs overlapping). b Heatmap showing the magnitude and direction of methylation change (delta beta) between sample groups (P–N: placebo vs never-smoker; V–P: vitamin C vs placebo). Only the CpGs with a reversal in the direction of methylation change with vitamin C supplementation (n = 9059 CpGs “partially restored”) were considered in downstream analyses
We next used comb-p to identify differentially methylated regions (DMRs) [59], a method that combines adjacent p values in sliding windows, and required a p value of 0.05 to start a region and extended the region if another significant p value was within 500 base pairs [60]. We identified 93 DMRs between randomized treatment groups (vitamin C-smokers vs placebo-smokers; VP-DMRs; Fig. 3a; Additional file 2: Table S3) that spanned 548 individual CpGs (278 hypomethylated in placebo/ 270 hypermethylated) and 82 unique genes. Visual inspection of individual DMR beta values and genomic locations confirmed consistent patterns of DNAm within loci and proximity to critical regulatory features (i.e., CpG islands, transcription factor binding sites, DNAse hypersensitivity sites, etc.) for the majority of DMRs (Additional file 3). The top significant PV-DMR (mean ∆β P–V = 9.85%; Šidák adjusted p = 3.45E−10) covered a CpG island and shore across the transcription start site (TSS) for ANKDD1B (chr5:74907152–74908171). Further, we identified 3 PV-DMR loci with a mean DNA methylation difference greater than 10% (PLIN1, HSPA1A/1L, and XXYTL1). In the comparison of never-smokers versus placebo-supplemented smokers, we identified 1359 Šidák significant NP-DMRs (Fig. 3b; Additional file 2: Table S4), with the top 3 significant DMRs annotated to LRP1, LINC00886, and GPR20. Out of the 93 PV-DMR loci between randomized treatment groups, 25 overlapped with NP-DMRs, proximal to 16 unique genes (Table 2). The top significant restored DMR (mean ∆β V–P = 4.8%; mean ∆β P–N = −2.3%; Šidák adjusted p = 2.52E−7) was located in an intergenic region (chr5:72596701–72597716) located between TMEM174 and FOXD1. Additionally, multiple restored DMRs mapped to PRKCA (3 DMRs) and DIP2C (2 DMRs).
Volcano plots of differentially methylated regions (DMRs) in placenta between a placebo and vitamin C smokers and b placebo smokers and never-smokers. DMRs with a Šidák adjusted p value < 0.05 are shown in red. The top 20 DMRs per comparison are annotated with the nearest gene and number of CpGs. The x-axis denotes the average delta-beta between sample groups across the DMR region
For downstream analyses, we focused on “candidate restored DMCs” based on the overlap of nominally significant CpGs between the two comparisons (PN: placebo vs never-smoker; VP: vitamin C vs placebo; n = 9541 CpGs; Fig. 2a) and showing any restoration with vitamin C treatment of the methylation change caused by maternal smoking. In 9059 out of 9541 overlapping CpGs (95%), the average mean difference in methylation between vitamin C smokers and placebo smokers was in the opposite direction of the effect between placebo smokers and never-smokers (Fig. 2b; Additional file 2: Table S2), consistent with partial restoration.
Association of candidate DMCs with infant lung function and wheeze
We next examined the relationship between DNAm at candidate CpGs with infant lung function (FEF75; the measurement of forced expiratory flows (FEF) at 75% of the expired volume) and wheeze assessed at 12 months of age. Due to a smaller sample size, we considered nominal p < 0.05 significant within the candidate CpGs tested for association with outcome. Out of 9059 candidates partially “normalized” with vitamin C supplementation, 1584 CpGs (annotated to 1208 unique genes) were nominally associated (and 34 FDR significant; Additional file 1: Figures S6–S7) with infant lung function after adjustment for infant length at PFT, infant sex, and GA at delivery. Of note, 52 candidate CpGs associated with lung function annotated to PRKCA, 18 CpGs annotated to ADAMTS2, and 10 CpGs annotated to FOXP4 were positively associated with FEF75 (Additional file 2: Table S2). We also identified 30 genes annotated to CpGs associated with FEF75 with known roles in lung development, including 6 genes (ROR2, SOX9, GATA6, RUNX2, RUNX3, and ACTN4) previously associated with adult lung function (Fig. 4; Additional file 2: Table S2) [61]. The top CpG among candidates associated with FEF75 was cg03172077, annotated to TOP3B (Fig. 5a).
Overlap of genes containing restored DMRs and candidate loci nominally associated with respiratory outcomes (12 month FEF75 and/or composite wheeze score) and candidate lung development genes [61]
We also examined association of candidate restored CpGs with composite wheeze as defined by any of the following: parental report of wheeze, healthcare provider diagnosis of wheeze, any bronchodilator or steroids use. A total of 620 candidate CpGs (annotated to 530 unique genes) were nominally associated with wheeze after adjustment for infant length at 12 months of age, infant sex, and GA at delivery (Additional file 2: Table S2). Eighteen wheeze associated CpGs in placenta annotated to 14 unique genes with known roles in lung development (Fig. 4; Additional file 2: Table S2) [61]. The top (and only FDR significant) CpG associated with wheeze was cg26433839, annotated to PRKCA, in addition to 47 PRKCA CpGs and 6 CpGs annotated to APOH (directly upstream of PRKCA) found to have nominally significant lower methylation associated with wheeze (Fig. 5b). The majority (42/53, 79%) of wheeze associated CpGs annotated to APOH/ PRKCA were also positively associated with FEF75 at 12 months of age.
Replication look-up
Prenatal exposure to MSDP (never-smokers vs placebo-smokers: NP) was associated with placental DNAm at 17 Bonferroni significant CpGs and 726 FDR-significant DMCs (Table 1; Fig. 1b). We compared our results with those of two previous EWAS of MSDP in placenta [13, 62]. In lookup study 1, we focused on the overlap of our results with DMCs associated with sustained MSDP (Everson et al.: Table S6 [62]), since the majority of smokers in our study were persistent smokers throughout pregnancy. Out of all 19,219 CpGs sustained-smoking associated CpGs (FDR < 0.05) identified by Everson et al. [62], 17,039 were represented by probes on the EPIC platform and 4034 were also nominally significant in our comparison of placebo smokers to never-smokers on the beta scale (Additional file 2: Table S5). Within those 4034 CpGs, 3964 (98%) showed a consistent direction of change between smokers and non-smokers in the two datasets (overall correlation = 0.88; p < 2.2e−16) and 131 CpGs were nominally restored with vitamin C treatment. Out of 726 FDR NP-DMCs in our study, 105 were FDR significant in the PACE study [62] (correlation = 0.95; p < 2.2e−16; Additional file 1: Figure S4A). The second lookup study was from the Gen3G cohort which used the same Illumina MethylationEPIC platform as this study [13]. We replicated 62/71 Gen3G DMCs at nominal significance and 20/71 at FDR significance between placebo smokers and never-smokers with consistent direction and magnitude of effect sizes (overall correlation = 0.99; p < 2.2e−16; Additional file 1: Figure S4B).
Functional enrichment within candidate loci
We next performed enrichment analyses in the 9059 candidate restored CpGs (Fig. 2) to identify biological functions and processes dysregulated with MSDP and improved with vitamin C supplementation. Analysis for enrichment using ConsensusPathDB of the 5613 unique genes annotated nearest to candidate restored CpGs identified enrichment of 156 pathways (q value < 0.05; Additional file 2: Table S6) and 461 GO-terms (q value < 0.05; Additional file 2: Table S7). The top enriched pathway and GO-terms were “neuronal system” (q value = 2.3E−08) and “nervous system development” (q value = 6.8E−36), respectively. Other significant pathways were related to cell signaling (i.e., PI3K-AKT-mTOR, Wnt, VEGFA-VEGFR2, Hippo, GPCR, ERBB2, MAPK1/MAPK3), differentiation (ectoderm, neural crest), insulin secretion and Type II diabetes, growth factors (i.e., VEGFs, FGFs, EGFR1), calcium regulation in the cardiac cell, extracellular matrix, and many others. Overrepresented GO-terms were related to development, morphogenesis, and embryogenesis, as well as growth factor signaling. For context, we also performed CPDB pathway analysis using the “Top 9059 NP DMCs,” regardless of whether they were restored with vitamin C. The top smoking associated pathways in this study included “nervous system development,” “axon guidance,” and “neuronal system” (Additional file 2: Table S6). We next used IPA software to identify upstream regulators enriched among candidate CpGs associated with MSDP and blunted by vitamin C supplementation. Top upstream regulators of candidate restored CpGs included ESR1 (p = 1.1E−11; 447 target molecules in dataset), CREBBP (p = 3.85E−10; 150 targets), and TGFB1 (p = 1.35E−09; 567 downstream targets; Additional file 2: Table S8).
Expression quantitative trait methylation (eQTM) at differentially methylated loci
We investigated the possible impact of DNA methylation changes on the expression of mRNA using RNA-seq data available from 71 placentas used in EPIC methylation analysis. To this end, we calculated the residuals for both the CpG beta-values and logcpm mRNA expression regression analyses adjusted for infant sex, cell type composition, gestational age at delivery, and RNA batch. We then tested the association between methylation residuals of candidate CpGs (described in methods) with flanking (± 250 kb) mRNA residuals. Out of 10,010 candidate CpGs tested, we identified 432 total significant associations with at least one mRNA transcript (at FDR < 0.05). The 432 FDR significant eQTM associations included 357 unique CpGs within 250 kb of transcripts annotated to 268 unique gene regions (Additional file 2: Table S9). The top significant eQTM in candidate restored CpGs was cg02283691 at chr19:33182526, annotated to NUDT19, and was negatively associated with the expression of NUDT19 (p = 1.34E−19; β = − 3.41; SE = 0.27; Additional file 1: Figure S5A). Notably, we identified several non-DMR genes from our candidate list of restored CpGs with multiple significant eQTM CpGs including 8 CpGs annotated to LOXL2 and positively associated with ENTPD4 mRNA. Within the 93 VP-DMRs we identified 89 eQTM CpGs, annotated to 21 unique gene regions (FDR p < 0.05; Table 3). Nine of the VP-DMRs contained more than one significant eQTM CpG inversely associated with one or more mRNA transcripts including PCK2 (13 CpGs), IRF7 (11 CpGs), IVD (11 CpGs), ZNF214/ZNF215 (11 CpGs), LINC00526/LINC00667 (7 CPGs), LRRC4 (5 CpGs), ZNF85 (4 CpGs), AL161785.1 (3 CpGs), and ETF1P1 (2 CpGs). Further, four PV-DMR regions contained more than one positive eQTM association located nearest COL21A1 (8 CpGs), ZFP57 (4 CpGs), RNF39 (2 CpGs), and TMEM105 (2 CpGs).
Among the 17 CpGs associated with MSDP at FDR significance in placebo versus never-smokers, 3 were eQTMs after FDR multiple testing correction. Notably, the top eQTM associated with MSDP was at cg03313447, annotated to CCDC97, and was strongly associated with decreased expression of nearby TGFB1 (β = − 11.0; FDR adjusted p = 6.94E−08; Additional file 1: Figure S5B). The top CpG associated with MSDP, cg27402634, was not significantly associated with expression of flanking genes including LEKR1 (β = − 0.41; FDR adjusted p = 0.65; Additional file 1: Figure S5C); however, cg20385913 located in the body of CYP1A2 was associated with expression of CYP1A1 mRNA (β = 35.7; FDR adjusted p < 0.01); Additional file 1: Figure S5D).
Discussion
This study of placental DNA methylation nested within the VCSIP RCT [53] provides suggestive evidence for partial or full restoration of 9059 CpGs associated with MSDP in placentas from smokers randomized to vitamin C versus placebo. We observed consistently lower placental DNAm among smokers supplemented to placebo versus never-smokers and versus vitamin C supplemented smokers (6691 CpGs hypomethylated, 2368 CpGs hypermethylated) and identified 21 candidate restored DMRs in addition to 268 unique DMC genes associated with mRNA expression using eQTM analysis. Importantly, a subset of candidate CpGs dysregulated with MSDP and normalized with vitamin C were associated with FEF75 and/or composite wheeze, assessed only in offspring born to pregnant smokers at 12 months of age.
Only one CpG, cg20790161, located in an intergenic region 17.5 Kbp upstream of CD28 reached epigenome-wide significance in the comparison of randomized treatment groups. However, there was no significant difference between placebo smokers and non-smokers, and we found no previous reports for differential methylation or functional relevance at this locus. Therefore, we have focused our discussion on candidate restored DMCs and DMRs nearest genes with multiple significant associations in downstream analyses (Fig. 4; Additional file 2: Table S2), with known association with lung development and function [61], and/or literature connections to biological processes and pathways enriched among candidate loci. The first gene candidate supported by multiple lines of evidence is DIP2C (disco-interacting protein 2 homolog C). We identified 2 DMRs hypomethylated in placebo vs never-smokers and restored with vitamin C located across the intergenic CpG island and shore regions upstream of DIP2C. Additionally, we identified 6 restored candidate DMCs nominally associated with lung function, and 1 restored DMC (cg27315601) associated with both wheeze and lung function. Furthermore, 4 DMCs located in the body of DIP2C were significantly associated with DIP2C expression (Additional file 1: Figure S8).
The DIP2 family members (DIP2A, DIP2B, and DIP2C) are highly conserved, and DIP2B and DIP2C are both expressed in human lung and placenta (Human Protein Atlas available from http://www.proteinatlas.org) [63]. Transcriptome profiling in lungs from Dip2a−/− versus wild-type mice revealed dysregulation of genes critical to vasculogenesis, alveologenesis, and branching morphogenesis [64], while loss of Dip2b in mice results in embryonic lethality due to abnormal lung development [65, 66], suggesting a likely role for DIP2 members in human lung development. Further, an EWAS of lung function in a Korean COPD cohort identified one significant DMC in DIP2C (cg03559389) associated with FEV1/FVC ratio, strengthening the potential relevance of this gene in lung development and function [67].
We also identified six DMRs restored with vitamin C in gene regions associated with both lung function and wheeze that were not associated with changes in mRNA expression in the placenta (3 DMRs in PRKCA, 1 DMR in APOH, TMEM174/FOXD1, and SSPO). Within PRKCA (Protein kinase C alpha type) we identified 3 restored DMRs and 41 candidate CpGs that were nominally associated with both lung function and wheeze. PRKCA is involved in the regulation of critical pulmonary and cardiovascular processes including angiogenesis [68], vascular endothelial barrier function [69], platelet function [70], arterial blood flow [71], cardiac hypertrophy [72], and endothelial cell migration and adhesion [73]. Increased expression of PRKCA in pulmonary artery smooth muscle cells from smokers is associated with increased pulmonary artery wall thickness [74]. Moreover, PRKCA has been suggested as a positional candidate for the shared genetic predisposition to asthma and obesity [75], and in utero exposure to polycyclic aromatic hydrocarbons from both ambient sources and MSDP has been previously reported as a risk factor for both asthma and obesity in early life [76].
Directly upstream of PRKCA is the APOH (Apolipoprotein H) gene, also known as beta-2-glycoprotein I (β2GPI). We identified one DMR (6 CpGs) annotated to APOH, similarly hypomethylated in placentas from placebo-smokers compared to placentas from both never-smokers and vitamin C-smokers. Interestingly, β2GPI is expressed in placental syncytiotrophoblasts and extravillous trophoblasts, and one of the key targets for antiphospholipid antibodies (aPL) that are associated with adverse pregnancy outcomes such as intrauterine growth restriction (IUGR), preeclampsia, and recurrent miscarriage [77]. Additionally, β2GPI is associated with hypoxia in endothelial cells, and β2GPI-derived peptides have been tested for therapeutic potential in limiting tumor growth by regulating angiogenesis [78].
FOXD1 (forkhead box D1) belongs to the forkhead family of transcription factors and regulates gene expression in a wide variety of biological processes including kidney morphogenesis and retinal development. Moreover, Foxd1 expressing progenitor cells play a role in lung development and lung fibrosis [79]. FOXD1 mutations have been implicated in obstetric complications including preeclampsia, IUGR, repeated implantation failure, and recurrent pregnancy loss through regulation of endometrial and placental genes [80]. We identified a restored DMR located in the intergenic region between TMEM174 and FOXD1 that spanned ENCODE regulatory motifs, in addition to candidate restored DMCs outside the DMR positively associated with respiratory outcomes (Additional file 3; Additional file 1: Table S2). One CpG was correlated with FOXD1 mRNA expression (r = 0.244; p = 0.039) in crude analysis but did not reach FDR significance in the adjusted eQTM analysis. In all, these findings suggest that DNA methylation loci dysregulated by MSDP in placenta and restored with vitamin C supplementation are involved in biological processes critical to angiogenesis and embryonic morphogenesis that are highly relevant to both placental function and lung development.
We examined the consistency of our results with those from two previous EWAS of MSDP in placenta (Additional file 2: Table S5). The top CpG associated with MSDP in our study, cg27402634, was also the top DMC in previous reports [13, 21] and we observed a similar large magnitude of difference between placebo-smokers and never-smokers (− 31 ± 2%). Contrary to previous reports, we did not observe increased expression of LEKR1 mRNA associated with cg27402634 (r = − 0.19; p = 0.09). The absolute value of effect sizes for MSDP were, on average, greater in our cohort than in a meta-analysis of sustained MSDP in placental DNAm [62], suggesting potentially increased duration and magnitude of exposure in our population. This is not surprising given that our randomized clinical trial consisted of pregnant smokers unable to quit smoking, compared with prospective birth cohorts with lower prevalence of MSDP and potentially a lower proportion of heavy smokers. Other possible explanations for differences in effect size include differences in sample size, probe efficiency between the MethylationEPIC and 450 K platforms, maternal demographics (i.e., race and age), and overall health status.
We further identified novel loci in the comparison of placebo-treated smokers to never-smokers and in the candidate restored CpGs, possibly due to differences in overall health status and prevalence of smoking in our clinical trial population compared to population-based studies, with limited information on smoke exposure level and vitamin C status. Across both novel and replicated CpGs, the majority were hypomethylated in smokers versus non-smokers. A large proportion of replicated loci associated with MSDP and not reversed by vitamin C were dose-dependently associated with level of exposure, based on maternal cotinine measurements performed throughout pregnancy [81].
Enrichment analysis of genes nearest candidate “normalized CpGs” identified many of the same pathways previously reported to be associated with sustained MSDP [62] (Additional file 2: Table S6). The top three pathways associated with sustained MSDP [62] and normalized or improved with vitamin C included “calcium regulation in the cardiac cell,” “VEGFA-VEGFR2 signaling,” and “Wnt signaling”. Placental vasculogenesis is regulated by a number of growth factors and aberrations in this process are a common theme identified in pregnancy complications such as intrauterine growth restriction and preeclampsia [82]. Dysregulated angiogenesis is also associated with pulmonary complications such as broncho-pulmonary dysplasia, pulmonary hypertension, and COPD [83]. Therefore, vitamin C supplementation to pregnant smokers may restore the balance of angiogenic factors in the placenta, in parallel with changes in the developing lung, in order to elicit the measured effects on lung function in our RCT.
To confirm the potential importance of these methylation changes on lung development and function, we examined our results for genes with known roles in lung development, based on a list of 391 genes compiled by Portas et al. Out of 391 known lung development genes, 126 were listed among our candidate restored DMCs (Additional file 2: Table S2), and 36 were associated with infant lung function and/or wheeze within RCT participants (Fig. 4) [61]. We speculate that although DNA methylation profiles are largely tissue specific at the CpG level, maternal smoking during pregnancy disrupts an overlapping set of critical developmental and homeostatic pathways across fetal and placental tissues through epigenetic mechanisms. This is supported by a previous study showing overlap of nicotine associated methylation changes between placenta and fetal lung collected in early development [84]. Future studies are necessary to validate these findings in additional populations and in animal models, and to identify the specific mechanism(s) whereby altered DNA methylation by vitamin C in the placenta influences infant lung growth and development.
Our findings in this study, combined with a previous study of placental hemodynamics and transcriptome analysis in this cohort [56], suggest that altered placental methylation and gene expression may mediate changes in vasculature and angiogenic signaling in response to MSDP and that these changes may be blunted by supplemental vitamin C during pregnancy. Our results are supported by the overall consistency of our findings with previous EWAS studies of MSDP and by histological studies in placentas from smokers which report increased collagen deposition, increased thickness of the villous membrane, vascular remodeling, as well as reduced intervillous area and capillary volume [27]. Importantly, as these reported histological changes in the placenta are similar to changes observed in blood vessels from offspring of smokers [85, 86], the mechanism by which vitamin C improves placental blood flow in smokers may parallel the widespread effects of MSDP on pulmonary and cardiovascular development.
Our study is the first to use the MethylationEPIC array (over 850,000 CpGs) for association with placental DNAm in a majority smoking (and RCT) population with extensive exposure measurements collected at multiple time-points throughout pregnancy. To our knowledge, the only previous EWAS of placental DNAm with MSDP measured on this platform was from the Genetics of Glucose Regulation in Gestation and Growth Study (Gen3G), which included 403 non-smokers and 38 participants with self-reported smoking during pregnancy [13]. We are also the first to combine epigenome-wide placental DNAm association with MSDP and direct measurements of childhood lung function.
The primary limitation to our study is our available sample size, which was underpowered to detect small differences in methylation at the majority of loci [87]. As we anticipated that our sample size may be insufficient to detect epigenome-wide significance between placebo and vitamin C treated smokers, we used a concept-driven analysis approach with the overall goal to identify candidate loci that may be “normalized” with vitamin C supplementation toward the level of never-smokers. We also performed DMR analysis using comb-p for consistency with previous studies [55]. However, comb-p has been recently reported to have greater Type 1 error, also known as “higher false positives,” than other DMR calling methods, and therefore, these candidate regions require further validation. An additional limitation is that these results represent only a subset of placentas available from the parent RCT. In the parent RCT, we collected 210 placentas from pregnancies of smokers at delivery and selected a subset of available placentas using a blocked- randomization design after exclusion of placentas from pregnancies complicated by preeclampsia, preterm delivery, gestational hypertension, and placentas collected outside the 3-h window post-delivery (Additional file 1: Figure S2). However, our previous transcriptome-wide analysis of 80 placentas (60 RCT smokers and 20 never-smokers for reference) also suggested activation of vasculogenesis, endothelial tissue development, and response to growth factors and we confirmed expression of genes critical to these processes by RT-qPCR in the larger RCT population [56]. Although these findings are suggestive and require replication, given that we focused on the overlap of nominally significant CpGs and a less stringent DMR method, our results are supported by consistency with prior studies of smoking-associated placental DNA methylation and downstream analyses demonstrating association of DNAm with placental gene expression and infant lung function.
Conclusions
This epigenome-wide analysis of placental DNA methylation within a randomized clinical trial population of pregnant smokers identified candidate loci associated with vitamin C supplementation. Critically, some of the treatment associated CpGs were also associated with infant lung function and wheeze measured at 12 months of age. These findings suggest the potential for vitamin C supplementation to mitigate negative consequences of MSDP on placental gene expression through epigenetic mechanisms.
Methods
Study design
This study was nested within a multi-center, double-blind RCT that demonstrated improved airway function at 3 and 12 months of age in offspring whose mothers were randomized to supplemental vitamin C (500 mg/day) versus placebo [53, 54, 58]. For the current study, we analyzed 96 placentas and prioritized samples used previously in transcriptome analysis [56]. We excluded placentas from subjects with gestational hypertension, preeclampsia, and preterm delivery (< 37 weeks), and placentas sampled more than 3 h after delivery (Additional file 1: Figure S2).
Study population
The parent RCT recruited women with singleton pregnancies (≥ 15 years old; < 23 weeks gestation) with a history of current smoking and documented refusal/inability to quit. Women were randomized to receive vitamin C versus placebo after a successful run-in trial for medication compliance that required 75% adherence and return for follow-up within 7–21 days. Randomization to vitamin C or placebo was blocked in rotations of two and four subjects, and stratified by gestational age at randomization (≤ 18 vs > 18 weeks) and site (Oregon Health & Science University [OHSU], Portland, Oregon; PeaceHealth Southwest Washington Medical Center [SWW], Vancouver, Washington; Indiana University [IU], Indianapolis, Indiana). A total of 252 pregnant smokers were randomized and 243 infants were available for study at delivery. The RCT was approved by each site’s Institutional Review Board and monitored by an NIH appointed Data Safety Monitoring Board. A group of 33 pregnant never-smokers were enrolled toward the end of the RCT as a reference group for an ancillary study of placental blood flow, histology, and molecular biomarkers (Additional File 1: Figure S2). We obtained written informed consent from all subjects prior to enrollment [58].
Statistical analysis of patient demographics
Normally distributed variables are expressed as mean and standard error and compared for group differences using an unadjusted F-test. Non-normally distributed variables are expressed as median and interquartile range (25th–75th percentile), and the Wilcoxon rank sum test was used to compare groups. Chi-square test was used to compare categorical variables. Significance was defined as p < 0.05.
Placental DNA methylation (DNAm) acquisition and pre-processing
Epigenome-wide placental DNAm was measured with the Infinium MethylationEPIC BeadChip (Illumina, San Diego, California) at the Fred Hutchinson Cancer Genomics Resource (Seattle, WA). See Supplemental Methods for details of placental collection, DNA extraction and DNA methylation acquisition (Additional file 1). Data normalization and QC were performed using ChAMP: non-CpG probes, probes with a beadcount < 3 in at least 5% of samples, probes annotated to SNPs [88], probes with a detection p value > 0.01 in one or more samples, cross-hybridizing probes [89], and probes on X/Y chromosomes were removed and remaining probes (n = 714,666) were normalized via functional normalization.
Estimate of placental cellular heterogeneity
We used the RefFreeEWAS package to estimate proportions of cell types in placental samples. We used the top 10,000 most variable CpGs from our dataset in the bootstrap to determine the optimal number of cell types. The optimal k for this dataset (k = 2) was selected based on the minimal deviance metric. The entire set of filtered CpGs (n = 714,666) was then used to estimate the proportions of each cell type.
CpG annotation to nearest gene
For downstream enrichment analysis we annotated all intergenic CpGs to the nearest proximal gene using the chromosome and positions (GrCh37/hg19) provided for each probe in the Illumina HumanMethylationEPIC annotation file, matched to the nearest gene symbol using the GenomicRanges package in R.
Analysis of differentially methylated CpGs (DMCs) and regions (DMRs)
We performed differential methylation analysis using the lmFit (method = “robust”), contrasts.fit and eBayes functions in limma with methylation for each CpG site as the response variable on the M-scale (logit2 beta) and randomization group as the predictor. Covariates were selected from a list of potential a priori confounders assessed for variance contribution using champ.SVD. We adjusted models for infant sex, gestational age at delivery, and estimated cellular heterogeneity. We used quantile–quantile plots of p values to visualize genomic inflation between unadjusted and adjusted models (Additional file 1: Figure S3). We computed estimated coefficients and standard errors for each contrast of interest (never-smoker vs placebo smoker) and (placebo smoker vs vitamin C smoker). We also calculated the coefficients and standard error from β value regression for more intuitive biological interpretation and comparison to previous studies. To identify differentially methylated regions (DMRs) we used comb-p [59] using the results from each fully adjusted limma model as input and the following parameters to initiate and extend a region: –seed 0.05 –dist 500. A DMR was considered significant when it included at least 2 probes within a window of 500 bp and the Šidák corrected p value was < 0.05.
Association of candidate DMCs with infant lung function and wheeze
We measured airway function in infants born to smoking participants in the RCT as described previously [53]. Briefly, FEFs were obtained from forced expiratory flow volume curves using the raised volume rapid thoracic compression technique following the American Thoracic Society/ European Respiratory Society criteria for performance and acceptance [90]. The measurement of FEF at 75% of the expired volume (FEF75) was defined a priori as the primary outcome in our RCT and was measured at 3 and 12 months of age. A modified form of the International Study of Asthma and Allergies in Childhood (ISAAC) respiratory questionnaire [91] was administered at least quarterly to the infant’s caretaker. Composite wheeze was defined as a positive response to any of the following questions: parental report of wheeze, healthcare provider diagnosis of wheeze or any bronchodilator or steroids use. We used robust linear regression analysis adjusted for infant length at PFT, infant sex, and gestational age (GA) at birth to check for association between candidate loci and lung function or wheeze assessed in infants at 12 months of age. Due to our small sample size, CpGs with unadjusted p < 0.05 were considered significant for discussion relevant to lung function or wheeze.
Replication look-up in previous EWAS
We compared our results with previous findings from two previous EWAS of MSDP associated methylation changes in placenta: (1) a meta-analysis for the association between sustained MSDP and placental DNAm measured on the Illumina HumanMethylation450 BeadChip (Additional file 2: Table S6 [62]), and (2) results from the Gen3G study (Web Table 3 [13]) measured using the same Illumina MethylationEPIC platform as this study. For our comparisons, we utilized the nominally significant results from the contrast of never-smokers versus smokers randomized to placebo and compared beta-value scale coefficients to the beta-coefficients reported previously for MSDP.
Enrichment analysis of biological pathways and gene ontology (GO) terms
Functional enrichment analyses were performed at the gene level using ConsensusPathDB [92] and Ingenuity Pathway Analysis (Qiagen Inc., MD, USA) [93]. We focused our enrichment analysis on genes annotated nearest to candidate CpGs partially restored in the overlap of nominal p values (Fig. 2). ConsensusPathDB performs enrichment analysis using a hypergeometric test, and we report significant pathway and GO_term results after multiple testing correction with FDR < 5%. In IPA, we used the “Core Analysis” pipeline with default settings to test for enriched canonical pathways, upstream regulators, diseases, and functions.
Expression quantitative trait methylation (eQTM) loci
We performed expression quantitative trait methylation (eQTM) analysis using the MEAL package [94] for correlation of expression and methylation. Genome-wide RNA-sequencing was available for 71 placentas (26 placebo, 27 vitamin C, and 18 never-smokers) with MethylationEPIC data (Additional file 1: Figure S2; Additional file 1: Supplemental Methods). We focused our eQTM analysis on CpGs partially restored in the overlap of nominal p-values (Fig. 2), CpGs located in DMRs between placebo and vitamin C (Additional file 2: Table S3), and FDR significant CpGs between placebo and never-smokers (Table 1). Combined, these three sets of candidate CpGs included 10,010 unique CpGs for eQTM analysis. We used the default flanking parameter to identify mRNA transcripts with a transcription start site (TSS) located within 250 kb of each candidate CpG. We calculated the residuals for both the CpG beta-values and logcpm mRNA expression regression analyses adjusted for infant sex, cell type composition, and gestational age at delivery. The association between methylation and expression residuals was performed in 55825 CpG-mRNA pairs and we report statistically significant eQTM with a FDR < 5% (Additional file 2: Table S9).
Availability of data and material
The raw and processed DNAm data are publically accessible through NCBI Gene Expression Omnibus (GEO) via accession series GSE169598.
Abbreviations
- MSDP:
-
Maternal smoking during pregnancy
- CpG:
-
Cytosine–guanine dinucleotide
- DNAm:
-
DNA methylation
- DMC:
-
Differentially methylated CpG
- DMR:
-
Differentially methylated region
- Doppler-US:
-
Doppler ultrasound
- EWAS:
-
Epigenome-wide association study
- eQTM:
-
Expression quantitative trait methylation
- RCT:
-
Randomized clinical trial
- IUGR:
-
Intra-uterine growth restriction
- TSS:
-
Transcription start site
- FDR:
-
False discovery rate
- FEF:
-
Forced expiratory flow
- FEF75 :
-
Forced expiratory flow at 75% of vital capacity
- PN:
-
Placebo versus non-smoker
- VP:
-
Vitamin C versus placebo
- Gen3G:
-
Genetics of Glucose regulation in Gestation and Growth study
- PACE:
-
Pregnancy And Childhood Epigenetics
References
Anderson TM, Lavista Ferres JM, Ren SY, Moon RY, Goldstein RD, Ramirez JM, et al. Maternal smoking before and during pregnancy and the risk of sudden unexpected infant death. Pediatrics. 2019;143(4).
Berlanga Mdel R, Salazar G, Garcia C, Hernandez J. Maternal smoking effects on infant growth. Food Nutr Bull. 2002;23(3 Suppl):142–5.
Kalinka J, Hanke W. Tobacco smoking—a risk factor for intrauterine growth retardation, preterm delivery and low birth weight. Ginekol Pol. 1996;67(2):75–81.
Horta BL, Victora CG, Menezes AM, Halpern R, Barros FC. Low birthweight, preterm births and intrauterine growth retardation in relation to maternal smoking. Paediatr Perinat Epidemiol. 1997;11(2):140–51.
Allen AM, Dietz PM, Tong VT, England L, Prince CB. Prenatal smoking prevalence ascertained from two population-based data sources: birth certificates and PRAMS questionnaires, 2004. Public Health Rep. 2008;123(5):586–92.
Tong VT, Jones JR, Dietz PM, D’Angelo D, Bombard JM, et al. Trends in smoking before, during, and after pregnancy—Pregnancy Risk Assessment Monitoring System (PRAMS), United States, 31 sites, 2000–2005. MMWR Surveill Summ. 2009;58(4):1–29.
Albuquerque CA, Smith KR, Johnson C, Chao R, Harding R. Influence of maternal tobacco smoking during pregnancy on uterine, umbilical and fetal cerebral artery blood flows. Early Hum Dev. 2004;80(1):31–42.
Gishti O, Jaddoe VW, Felix JF, Reiss I, Steegers E, Hofman A, et al. Impact of maternal smoking during pregnancy on microvasculature in childhood: the generation R study. Early Hum Dev. 2015;91(10):607–11.
Slotkin TA, Seidler FJ, Spindel ER. Prenatal nicotine exposure in rhesus monkeys compromises development of brainstem and cardiac monoamine pathways involved in perinatal adaptation and sudden infant death syndrome: amelioration by vitamin C. Neurotoxicol Teratol. 2011;33(3):431–4.
Suzuki K, Nakai K, Hosokawa T, Oka T, Okamura K, Sakai T, et al. Association of maternal smoking during pregnancy and infant neurobehavioral status. Psychol Rep. 2006;99(1):97–106.
Rizzo G, Capponi A, Pietrolucci ME, Arduini D. Effects of maternal cigarette smoking on placental volume and vascularization measured by 3-dimensional power Doppler ultrasonography at 11+0 to 13+6 weeks of gestation. Am J Obstet Gynecol. 2009;200(4):415 e – 5.
McEvoy CT, Spindel ER. Pulmonary effects of maternal smoking on the fetus and child: effects on lung development, respiratory morbidities, and life long lung health. Paediatr Respir Rev. 2017;21:27–33.
Cardenas A, Lutz SM, Everson TM, Perron P, Bouchard L, Hivert MF. Mediation by placental DNA methylation of the association of prenatal maternal smoking and birth weight. Am J Epidemiol. 2019;188(11):1878–86.
Hannon E, Schendel D, Ladd-Acosta C, Grove J, Hansen CS, Hougaard DM, et al. Variable DNA methylation in neonates mediates the association between prenatal smoking and birth weight. Philos Trans R Soc Lond B Biol Sci. 2019;374(1770):20180120.
Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–96.
Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 450K Epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012.
Kheirkhah Rahimabad P, Anthony TM, Jones AD, Eslamimehr S, Mukherjee N, Ewart S, et al. Nicotine and its downstream metabolites in maternal and cord sera: biomarkers of prenatal smoking exposure associated with offspring DNA methylation. Int J Environ Res Public Health. 2020;17(24).
Kupers LK, Xu X, Jankipersadsing SA, Vaez A, la Bastide-van GS, Scholtens S, et al. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int J Epidemiol. 2015;44(4):1224–37.
Maccani JZ, Koestler DC, Houseman EA, Marsit CJ, Kelsey KT. Placental DNA methylation alterations associated with maternal tobacco smoking at the RUNX3 gene are also associated with gestational age. Epigenomics. 2013;5(6):619–30.
Markunas CA, Xu Z, Harlid S, Wade PA, Lie RT, Taylor JA, et al. Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2014;122(10):1147–53.
Morales E, Vilahur N, Salas LA, Motta V, Fernandez MF, Murcia M, et al. Genome-wide DNA methylation study in human placenta identifies novel loci associated with maternal smoking during pregnancy. Int J Epidemiol. 2016;45(5):1644–55.
Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet. 2015;24(8):2201–17.
Witt SH, Frank J, Gilles M, Lang M, Treutlein J, Streit F, et al. Impact on birth weight of maternal smoking throughout pregnancy mediated by DNA methylation. BMC Genomics. 2018;19(1):290.
Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462–7.
Elliot J, Vullermin P, Robinson P. Maternal cigarette smoking is associated with increased inner airway wall thickness in children who die from sudden infant death syndrome. Am J Respir Crit Care Med. 1998;158(3):802–6.
Gilliland FD, Berhane K, McConnell R, Gauderman WJ, Vora H, Rappaport EB, et al. Maternal smoking during pregnancy, environmental tobacco smoke exposure and childhood lung function. Thorax. 2000;55(4):271–6.
Jauniaux E, Burton GJ. Morphological and biological effects of maternal exposure to tobacco smoke on the feto-placental unit. Early Hum Dev. 2007;83(11):699–706.
Murphy VE, Smith R, Giles WB, Clifton VL. Endocrine regulation of human fetal growth: the role of the mother, placenta, and fetus. Endocr Rev. 2006;27(2):141–69.
Thornburg KL, O’Tierney PF, Louey S. Review: The placenta is a programming agent for cardiovascular disease. Placenta. 2010;31(Suppl):S54–9.
Barker DJ, Thornburg KL, Osmond C, Kajantie E, Eriksson JG. The surface area of the placenta and hypertension in the offspring in later life. Int J Dev Biol. 2010;54(2–3):525–30.
Newnham JP, Patterson L, James I, Reid SE. Effects of maternal cigarette smoking on ultrasonic measurements of fetal growth and on Doppler flow velocity waveforms. Early Hum Dev. 1990;24(1):23–36.
Pintican D, Poienar AA, Strilciuc S, Mihu D. Effects of maternal smoking on human placental vascularization: a systematic review. Taiwan J Obstet Gynecol. 2019;58(4):454–9.
Zdravkovic T, Genbacev O, McMaster MT, Fisher SJ. The adverse effects of maternal smoking on the human placenta: a review. Placenta. 2005;26(Suppl A):S81–6.
Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb). 2010;105(1):105–12.
Bianco-Miotto T, Mayne BT, Buckberry S, Breen J, Rodriguez Lopez CM, Roberts CT. Recent progress towards understanding the role of DNA methylation in human placental development. Reproduction. 2016;152(1):R23-30.
Tekola-Ayele F, Zeng X, Ouidir M, Workalemahu T, Zhang C, Delahaye F, et al. DNA methylation loci in placenta associated with birthweight and expression of genes relevant for early development and adult diseases. Clin Epigenetics. 2020;12(1):78.
Tian FY, Everson TM, Lester B, Punshon T, Jackson BP, Hao K, et al. Selenium-associated DNA methylation modifications in placenta and neurobehavioral development of newborns: an epigenome-wide study of two US birth cohorts. Environ Int. 2020;137:105508.
Liu J, Zhang Z, Xu J, Song X, Yuan W, Miao M, et al. Genome-wide DNA methylation changes in placenta tissues associated with small for gestational age newborns; cohort study in the Chinese population. Epigenomics. 2019;11(12):1399–412.
Paquette AG, Houseman EA, Green BB, Lesseur C, Armstrong DA, Lester B, et al. Regions of variable DNA methylation in human placenta associated with newborn neurobehavior. Epigenetics. 2016;11(8):603–13.
Loke YJ, Muggli E, Nguyen L, Ryan J, Saffery R, Elliott EJ, et al. Time- and sex-dependent associations between prenatal alcohol exposure and placental global DNA methylation. Epigenomics. 2018;10(7):981–91.
Cai J, Zhao Y, Liu P, Xia B, Zhu Q, Wang X, et al. Exposure to particulate air pollution during early pregnancy is associated with placental DNA methylation. Sci Total Environ. 2017;607–608:1103–8.
Rousseaux S, Seyve E, Chuffart F, Bourova-Flin E, Benmerad M, Charles MA, et al. Immediate and durable effects of maternal tobacco consumption alter placental DNA methylation in enhancer and imprinted gene-containing regions. BMC Med. 2020;18(1):306.
Rauschert S, Melton PE, Heiskala A, Karhunen V, Burdge G, Craig JM, et al. Machine learning-based DNA methylation score for fetal exposure to maternal smoking: development and validation in samples collected from adolescents and adults. Environ Health Perspect. 2020;128(9):97003.
Nielsen CH, Larsen A, Nielsen AL. DNA methylation alterations in response to prenatal exposure of maternal cigarette smoking: a persistent epigenetic impact on health from maternal lifestyle? Arch Toxicol. 2016;90(2):231–45.
Bozack AK, Cardenas A, Quamruzzaman Q, Rahman M, Mostofa G, Christiani DC, et al. DNA methylation in cord blood as mediator of the association between prenatal arsenic exposure and gestational age. Epigenetics. 2018;13(9):923–40.
Gona S, Sanders AP, Miranda ML, Fry RC. Prenatal exposure to cadmium and cotinine and CpG island DNA methylation in Mother-Infant pairs. Genom Data. 2015;5:378–80.
Xu P, Wu Z, Yang W, Wang L. Dysregulation of DNA methylation and expression of imprinted genes in mouse placentas of fetal growth restriction induced by maternal cadmium exposure. Toxicology. 2017;390:109–16.
Sharp GC, Arathimos R, Reese SE, Page CM, Felix J, Kupers LK, et al. Maternal alcohol consumption and offspring DNA methylation: findings from six general population-based birth cohorts. Epigenomics. 2018;10(1):27–42.
Gruzieva O, Xu CJ, Yousefi P, Relton C, Merid SK, Breton CV, et al. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ Health Perspect. 2019;127(5):57012.
Suter M, Ma J, Harris A, Patterson L, Brown KA, Shope C, et al. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6(11):1284–94.
Wiklund P, Karhunen V, Richmond RC, Parmar P, Rodriguez A, De Silva M, et al. DNA methylation links prenatal smoking exposure to later life health outcomes in offspring. Clin Epigenetics. 2019;11(1):97.
McEvoy CT, Schilling D, Clay N, Jackson K, Go MD, Spitale P, et al. Vitamin C supplementation for pregnant smoking women and pulmonary function in their newborn infants: a randomized clinical trial. JAMA. 2014;311(20):2074–82.
McEvoy CT, Shorey-Kendrick LE, Milner K, Schilling D, Tiller C, Vuylsteke B, et al. Vitamin C to pregnant smokers persistently improves infant airway function to 12 months of age: a randomised trial. Eur Respir J. 2020.
McEvoy CT, Shorey-Kendrick LE, Milner K, Schilling D, Tiller C, Vuylsteke B, et al. Oral Vitamin C (500 mg/d) to pregnant smokers improves infant airway function at 3 months (VCSIP): a randomized trial. Am J Respir Crit Care Med. 2019;199(9):1139–47.
Shorey-Kendrick LE, McEvoy CT, Ferguson B, Burchard J, Park BS, Gao L, et al. Vitamin C prevents offspring DNA methylation changes associated with maternal smoking in pregnancy. Am J Respir Crit Care Med. 2017;196(6):745–55.
McEvoy CTSK, Lyndsey E., Tepper R, Morris CD, Haas D, Frias A, Spindel E. Vitamin C supplementation to pregnant smokers is associated with improved placental circulation and gene expression. Pediatric Academic Societies; 2020.
Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41(1):200–9.
McEvoy CT, Milner KF, Scherman AJ, Schilling DG, Tiller CJ, Vuylsteke B, et al. Vitamin C to decrease the effects of smoking in pregnancy on infant lung function (VCSIP): rationale, design, and methods of a randomized, controlled trial of vitamin C supplementation in pregnancy for the primary prevention of effects of in utero tobacco smoke exposure on infant lung function and respiratory health. Contemp Clin Trials. 2017;58:66–77.
Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28(22):2986–8.
Li D, Xie Z, Pape ML, Dye T. An evaluation of statistical methods for DNA methylation microarray data analysis. BMC Bioinform. 2015;16:217.
Portas L, Pereira M, Shaheen SO, Wyss AB, London SJ, Burney PGJ, et al. Lung development genes and adult lung function. Am J Respir Crit Care Med. 2020;202(6):853–65.
Everson TM, Vives-Usano M, Seyve E, Cardenas A, Lacasaña M, Craig JM, et al. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. bioRxiv. 2019:663567.
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics tissue-based map of the human proteome. Science. 2015;347(6220):1260419.
Sah RK, Yang A, Bah FB, Adlat S, Bohio AA, Oo ZM, et al. Transcriptome profiling of mouse brain and lung under Dip2a regulation using RNA-sequencing. PLoS ONE. 2019;14(7):e0213702.
Sah RK, Ma J, Bah FB, Xing Z, Adlat S, Oo ZM, et al. Targeted disruption of mouse Dip2B leads to abnormal lung development and prenatal lethality. Int J Mol Sci. 2020;21(21).
Adlat S, Sah RK, Hayel F, Chen Y, Bah FB, Al-Azab M, et al. Global transcriptome study of Dip2B-deficient mouse embryonic lung fibroblast reveals its important roles in cell proliferation and development. Comput Struct Biotechnol J. 2020;18:2381–90.
Lee MK, Hong Y, Kim SY, Kim WJ, London SJ. Epigenome-wide association study of chronic obstructive pulmonary disease and lung function in Koreans. Epigenomics. 2017;9(7):971–84.
Xu H, Czerwinski P, Hortmann M, Sohn HY, Forstermann U, Li H. Protein kinase C alpha promotes angiogenic activity of human endothelial cells via induction of vascular endothelial growth factor. Cardiovasc Res. 2008;78(2):349–55.
Vandenbroucke Amant E, Tauseef M, Vogel SM, Gao XP, Mehta D, Komarova YA, et al. PKCalpha activation of p120-catenin serine 879 phospho-switch disassembles VE-cadherin junctions and disrupts vascular integrity. Circ Res. 2012;111(6):739–49.
Konopatskaya O, Gilio K, Harper MT, Zhao Y, Cosemans JM, Karim ZA, et al. PKCalpha regulates platelet granule secretion and thrombus formation in mice. J Clin Invest. 2009;119(2):399–407.
Partovian C, Zhuang Z, Moodie K, Lin M, Ouchi N, Sessa WC, et al. PKCalpha activates eNOS and increases arterial blood flow in vivo. Circ Res. 2005;97(5):482–7.
Muth JN, Bodi I, Lewis W, Varadi G, Schwartz A. A Ca(2+)-dependent transgenic model of cardiac hypertrophy: a role for protein kinase Calpha. Circulation. 2001;103(1):140–7.
Wang A, Nomura M, Patan S, Ware JA. Inhibition of protein kinase Calpha prevents endothelial cell migration and vascular tube formation in vitro and myocardial neovascularization in vivo. Circ Res. 2002;90(5):609–16.
Xaing M, Liu X, Zeng D, Wang R, Xu Y. Changes of protein kinase Calpha and cyclin D1 expressions in pulmonary arteries from smokers with and without chronic obstructive pulmonary disease. J Huazhong Univ Sci Technol Med Sci. 2010;30(2):159–64.
Murphy A, Tantisira KG, Soto-Quiros ME, Avila L, Klanderman BJ, Lake S, et al. PRKCA: a positional candidate gene for body mass index and asthma. Am J Hum Genet. 2009;85(1):87–96.
Jung KH, Perzanowski M, Rundle A, Moors K, Yan B, Chillrud SN, et al. Polycyclic aromatic hydrocarbon exposure, obesity and childhood asthma in an urban cohort. Environ Res. 2014;128:35–41.
Tong M, Tsai BW, Chamley LW. Antiphospholipid antibodies and extracellular vesicles in pregnancy. Am J Reprod Immunol. 2020:e13312.
Smalley H, Rowe JM, Nieto F, Zeledon J, Pollard K, Tomich JM, et al. Beta2 glycoprotein I-derived therapeutic peptides induce sFlt-1 secretion to reduce melanoma vascularity and growth. Cancer Lett. 2020;495:66–75.
Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;188(7):820–30.
Quintero-Ronderos P, Jimenez KM, Esteban-Perez C, Ojeda DA, Bello S, Fonseca DJ, et al. FOXD1 mutations are related to repeated implantation failure, intra-uterine growth restriction and preeclampsia. Mol Med. 2019;25(1):37.
Shorey-Kendrick LE. Epigenome-wide analysis of placental DNA methylation associated with objective measures of maternal smoking. Pediatric Academic Societies; 2021; Virtual.
Rana S, Burke SD, Karumanchi SA. Imbalances in circulating angiogenic factors in the pathophysiology of preeclampsia and related disorders. Am J Obstet Gynecol. 2020.
Laddha AP, Kulkarni YA. VEGF and FGF-2: promising targets for the treatment of respiratory disorders. Respir Med. 2019;156:33–46.
Chhabra D, Sharma S, Kho AT, Gaedigk R, Vyhlidal CA, Leeder JS, et al. Fetal lung and placental methylation is associated with in utero nicotine exposure. Epigenetics. 2014;9(11):1473–84.
Kyrklund-Blomberg NB, Hu J, Gennser G. Chronic effects of maternal smoking on pulse waves in the fetal aorta. J Matern Fetal Neonatal Med. 2006;19(8):495–501.
Geelhoed JJ, El Marroun H, Verburg BO, van Osch-Gevers L, Hofman A, Huizink AC, et al. Maternal smoking during pregnancy, fetal arterial resistance adaptations and cardiovascular function in childhood. BJOG. 2011;118(6):755–62.
Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics. 2019;20(1):366.
Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45(4):22.
Nordlund J, Backlin CL, Wahlberg P, Busche S, Berglund EC, Eloranta ML, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14(9):r105.
Stoller JK, Snider GL, Brantly ML, Fallat RJ, Stockley RA, Turino GM, et al. American thoracic society/european respiratory society statement: standards for the diagnosis and management of individuals with alpha-1 antitrypsin deficiency. Pneumologie. 2005;59(1):36–68.
Asher MI, Keil U, Anderson HR, Beasley R, Crane J, Martinez F, et al. International study of asthma and allergies in childhood (ISAAC): rationale and methods. Eur Respir J. 1995;8(3):483–91.
Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database issue):793–800.
Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–30.
Ruiz-Arenas CGJ. MEAL: perform methylation analysis. R package version 1.16.0. (2019).
Acknowledgements
We would like to thank the women and their infants who participated in the VCSIP RCT to make this study possible. We also thank all VCSIP team members who contributed to clinical data and sample collection.
Funding
Supported by the NHLBI (R01 HL105447 and R01 HL 105460) with co-funding from the Office of Dietary Supplements (ODS) and by P51 OD011092565 and NIH UH3 OD023288. Additional support from the Oregon Clinical Translational Research Institute funded by the National Center for Advancing Translational Sciences (UL1TR000128).
Author information
Authors and Affiliations
Contributions
CTM, RST, and ERS initiated, designed, and acquired the funding for this study. LES, ERS, and SMO coordinated all laboratory analysis. CTM, RST, DH, KM, and BV coordinated all clinical data collection. CDM and AV oversaw data collection and data integrity. LG and BP provided statistical consultations and LES performed the analysis. LES, CTM, RST, and ERS drafted the manuscript. All authors contributed to the interpretation of results and revisions to the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The trial was approved by each hospital’s institutional review board and monitored by a National Institutes of Health-appointed data safety monitoring board. Written informed consent was obtained from pregnant smokers before enrollment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Figures:
Figure S1. Flowchart of the analysis steps; Figure S2. Consort diagram of samples used in analysis; Figure S3. QQ-plot for unadjusted vs adjusted models; Figure S4. Comparison of FDR significant smoking DMCs with previous studies; Figure S5. Top eQTMs associated with vitamin C vs placebo DMRs; Figure S6. Top DMCs associated with m12 FEF75; Figure S7. Heatmap of FDR DMCs associated with m12 FEF75; Figure S8. Visual summary of DIP2C findings. Supplemental Methods.
Additional file 2. Tables:
Table S1. Population characteristics; Table S2. Candidate restored DMCs and association with outcomes; Table S3. VP-DMRs; Table S4. PN-DMRs; Table S5. Lookup analysis; Table S6. CPDB Pathway; Table S7. CPDB GO terms; Table S8. IPA Top regulators; Table S9. eQTMs.
Additional file 3.
Gviz plots of significant DMRs between vitamin-C supplemented smokers and placebo. All 93 DMRs are plotted and arranged in alphabetical order based on the nearest annotated gene. Each page represents a single DMR and is organized into multiple tracks showing genomic ranges, CpG island locations, DMR location, and CpG level data tracks for each comparison (placebo vs non-smoker and vitamin C vs placebo).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shorey-Kendrick, L.E., McEvoy, C.T., O’Sullivan, S.M. et al. Impact of vitamin C supplementation on placental DNA methylation changes related to maternal smoking: association with gene expression and respiratory outcomes. Clin Epigenet 13, 177 (2021). https://doi.org/10.1186/s13148-021-01161-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-021-01161-y