Abstract
Background
Psoriasis (Ps) is a common chronic inflammatory skin disease. The keratinocytes of psoriatic skin defy normal apoptosis and exhibit active cell proliferation. Aberrant DNA methylation (DNAm) has been suggested relevant through regulating the expression of Ps susceptibility genes. However, it is unclear whether the biological age inferred from DNA methylome is affected.
Results
To address the above issue, we applied a recently developed methylation clock model to our Chinese Han population dataset, which includes DNAm data of 114 involved psoriatic skin tissues (PP) and 41 uninvolved psoriatic skin tissues (PN) from Ps patients, and 62 normal skin tissues (NN) from health controls. We first confirmed the applicability of the clock in PN and NN. We then showed that PP samples have largely unchanged DNAm age, and that no association was observed between available clinical features and DNAm age acceleration. Examination of genome-wide CpGs yielded age-associated CpGs with concordant age-association coefficients among the three groups, which was also supported by an external dataset. We also interestingly observed two clock CpGs differentially methylated between PP and PN.
Conclusions
Overall, our results suggest no significant alteration in DNAm age in PN and PP. Therefore, the increase in keratinocyte proliferation and alteration in DNAm caused by Ps may not affect the biological age of psoriatic skin tissue.
Similar content being viewed by others
Introduction
Psoriasis (Ps) is a chronic autoimmune inflammatory skin disease and affected by a complex interplay between the immunologic factors, Ps-associated susceptibility loci and various environmental factors [1,2,3]. This disease is characterized by abnormal growth, differentiation, and apoptosis of keratinocytes [4]. In normal skin apoptosis offsets keratinocyte proliferation and regulates the formation of stratum corneum. However, psoriatic keratinocytes exhibit an enhanced ability to resist apoptosis. This is a key potential step during the pathogenesis of Ps [5]. Whereas such resistance leads to the abnormal and rapid proliferation of keratinocytes, it remains unclear whether it causes acceleration or deceleration in the biological aging process of skin tissue.
Importantly, the age of onset is a key clinical character for classifying Ps subtypes [6]: patients are classified as early stage (or type I) if their onset ages are < 40 years old or late stage (or type II) if otherwise. In a UK cohort, the onset age of Ps incidence is bimodally distributed, with a clear separation between early and late stage patients [7]. The two subtypes have differences in certain aspects. For example, only early but not late stage patients are strongly associated with the HLA-C*06:02 allele in a case-control study [8]. Moreover, patients from the two stages are unevenly distributed across the three Ps subgroups classified based on DNA methylation (DNAm) [9]. These findings indicate the existence of an age effect during the occurrence and progression of Ps.
Ps is associated with change in DNAm levels at various CpG sites, which regulate the expression of genes at key processes related to Ps progression [10]. On the other hand, DNAm can record the biological age of an organism. Numerous CpGs throughout the genome can increase or decrease their methylation levels during the aging process, a pattern that can be observed ubiquitously across tissues and species [11,12,13,14,15,16,17,18,19]. The prevalence of age-associated CpGs has led to the development of epigenetic aging markers (or DNAm clocks) that use the combination of tens to a few hundreds of CpGs to predict age [17, 18, 20,21,22,23,24,25]. These DNAm clocks have been proven competent in predicting age in various cell/tissue types with reasonable accuracy [18, 22, 23]. Beyond accurately predicting age, DNAm clocks can identify the change in lifespan that are caused by genetic and environmental interventions [20,21,22] and indicate the acceleration of biological age caused by pathological conditions [26,27,28]. These suggest the usefulness of these clocks in understanding the impact of chronic diseases on lifespan.
Given the involvement of age and DNAm in Ps progression, and the active proliferation of keratinocytes, here we asked whether Ps skin tissues have anomalous DNAm ages in the Chinese Han population. To test this, we applied a DNAm clock model to a dataset from a previous epigenome-wide association study [29], which includes 114 involved psoriatic skin tissues (PP) and 41 uninvolved psoriatic skin tissues (PN) from Ps patients, and 62 normal skin tissues (NN) from health controls (Table 1).
Results and discussion
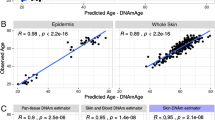
Here, we focused on the multi-tissue DNAm age clock developed by Horvath [18, 25]. We first examined normal skin tissues (NN and PN) to address the accuracy of this age predictor on our methylation dataset. Indeed, we observed a strong positive correlation between the estimated DNAm age and chronological age for both NN and PN (Fig. 1, Spearman’s ρ ≥ 0.86, P < 2.2E-16). Moreover, the age acceleration residuals, which measure the increase/decrease in DNAm age relative to chronological age, are near zero for both NN and PN (Fig. 2, t test P > 0.15). Therefore, these suggest that the methylation age clock model works reasonably well in NN and that the DNAm age of PN is unchanged. We then examined the DNAm data of PP. A strong correlation between DNAm age and chronological age was also observed (Fig. 1, ρ = 0.78, P < 2.2E-16), although somewhat lower than that from NN and PN. Moreover, the age acceleration residual of PP also showed no significant difference from zero (Fig. 2, t test, P = 0.78), or compared to NN and PN (P > 0.3). The different correlations between DNAm age and chronological age in the three groups may be affected by some external reasons, for example, previous studies have shown that numerous conditions such as obesity [26], Huntington’s disease [30], race [31], and calorie restriction [21] could somehow affect DNAm age. We further specifically examined the 41 paired PP and PN samples and again did not observed significant difference in age acceleration residual (paired t test, P = 0.51). These data suggest that Ps may not alter the DNAm age of PP.
To explore the potential impact of clinical features on DNAm age acceleration across samples, we applied a multi-linear model to associate age acceleration residual with available clinical features. These include age, body mass index (BMI), gender, smoking status (current, former, and never), and Ps area and severity index (PASI) score. As a result, we observed no significant association between clinical features and age acceleration residual in all skin groups (Table 2).
We observed 100, 588, and 8032 CpGs, respectively, in NN, PN, and PP that are significantly associated with age (false discovery rate (FDR) < 0.05, Additional file 1: Table S1, Additional file 2: Table S2, and Additional file 3: Table S3). Interestingly, almost all of these significant CpGs (95, 585, and 7765 CpGs in NN, PN, and PP, respectively) undergo hypermethylation during aging. Also, almost all of these CpGs in NN (94.0%) and PN (93.7%) remain age-associated in PP, while 51 CpGs are shared across the three groups (Fig. 3a). All these overlaps are significantly higher than by chance (hypergeometric test P < 2.2E-16). Moreover, the shared CpGs have largely consistent age-association coefficients across comparisons (ρ > = 0.75, Fig. 3b, Additional file 4: Figure S1A and S1B). Despite the consistence in coefficients, the discrepancy in numbers of age-associated CpGs across groups seems intriguing. To address whether this is genuine or caused by factors such as sample size and the range of age, we considered specifically the 41 individuals with paired PN and PP samples appears suitable to account for these effects. As a result, we observed 588 and 325 CpGs (Additional file 5: Table S4) associated with age (FDR < 0.05) in these paired PN and PP samples, respectively, which are rather comparable numbers. Therefore, the discrepancies in numbers of age-associated CpGs across groups are rather a reflection of difference in statistical power.
Comparison of age-associated CpGs among groups and with external dataset. a The overlap of age-associated CpGs across the NN, PN, and PP groups. b For the 94 overlapped age-associated CpGs between NN and PP, their coefficients are strongly positively correlated (Spearman’s ρ = 0.84, P < 2.2E-16). c For the overlapped age-associated CpGs between an additional epidermis dataset and PP, their coefficients are also significantly positively correlated (Spearman’s ρ = 0.45, P < 2.2E-16)
To further assess these age-associated CpGs, we re-analyzed a study of 108 healthy epidermis methylomes measured under the same platform [32]. We observed that the vast majority of age-associated CpGs (> 77.5%) are also significant in this dataset, and that the age-association coefficients are significantly positively correlated (ρ ≥ 0.45, Fig. 3c, Additional file 4: Figure S1C and S1D). These results support the consistency across the three categories of skin tissue samples and with external skin data.
Our previous analyses identified 426 and 1514 CpGs differently methylated between PP and NN, and between PP and PN, respectively, with 264 shared between the two comparisons [29]. We then asked how many Ps-specific CpGs are also age-associated. Among the 8032 age-associated CpGs observed in PP, we identified 39 (Additional file 6: Table S5) and 1 (cg11426075) CpGs also differentially methylated between PP and PN, and between PP and NN, respectively. The age- and Ps-associated shared CpGs are uniquely mapped to 23 genes and we found these genes are significantly enriched in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway “Regulation of actin cytoskeleton” (hypergeometric test P < 0.05). This observation suggests that the actin cytoskeleton pathway might be affected in both psoriatic and aged skins, which appears compatible with previous findings that aberrant actin cytoskeleton organization may induce Ps [33], and that cytoskeletal integrity declines as a function of age [34]. However, no age-associated CpG is shared between the two comparisons. Likewise, among the 588 age-associated CpGs in PN, 4 CpGs (cg04154027, cg09655403, cg15197065, and cg23005885) were differentially methylated between PP and PN. The difference in the overlaps is again likely due to different sample sizes. By further comparing with the 353 clock CpGs defined by Horvath [18, 25], we interestingly observed 2 clock CpGs (cg10266490 and cg22171829) also differentially methylated between PP and PN.
We finally asked which CpGs have changed their age-association patterns upon developing Ps (or Ps-dependent age-associated CpGs), such as from age-associated to unassociated, or vice versa. To address this, we used a method that resembles Fisher’s transformation (details are described in the “Materials and methods” section) to identify possible Ps-dependent age-associated CpGs. By contrasting PP with PN and NN, respectively, we obtained 301 and 881 CpGs (P < 0.01; Additional file 7: Table S6 and Additional file 8: Table S7), but none of these sites passed the threshold of FDR < 0.05. Overall, this data suggests that the age-associated pattern of DNAm in the skin might be largely independent of Ps.
This study is the first to explore whether psoriatic skin tissues suffer from an accelerated DNAm age. Our study is partially motivated by the fact that keratinocytes of Ps involved skins usually undergo abnormal and rapid proliferation. This is a feature that resembles aged individuals and tumors where cells have experienced excessive mitotic cell division. Such excessive division in soma cells could cause widespread passive DNAm change, especially in late DNA replicating domains [35, 36]. This provides us a tempting hypothesis that the involved skins may have an affected DNA methylome and methylation age that resembles the pattern in aged cells or tumor cells. However, our observations, the DNAm age and the age-association pattern of the majority of CpGs appear unaffected by Ps, contradict this hypothesis. This suggests a phenomenon where the methylome of Ps skin cells is affected in a manner that is generally independent to the usual pattern of passive epigenetic drift. As a comparison, the exposure to sun/UV is linked to change in methylome that resembles the aging process [37]. Further investigation into the interactions between Ps and environmental stimuli that are likely important in skin aging might be meaningful, such as UV radiation.
From the technical perspective, DNAm age is calculated through averaging the information of 353 CpGs scattered across the genome, each CpG has very limited weight in the algorithm. Therefore, it is possible that changing the methylation of only a few CpGs, e.g., two clock CpGs are differentially methylated between PP and PN, may not have perceptible impact on the outcome. It may also be that the effect of Ps on aging is relatively small for detection. If this is the case, a larger sample size might be needed to further verify our observation, since our analyses were based on a relatively small set of Ps patients that were exclusively from the Chinese Han population.
Overall, we showed that the multi-tissue DNAm clock model works well in skin tissue of the Chinese Han population. No significant alteration in DNAm age was observed in the uninvolved and involved psoriatic skin tissues, and there is a lack of association between PASI score and age acceleration residual. Therefore, the increase in keratinocyte proliferation and alteration in DNAm caused by Ps may not affect the biological age of psoriatic skin tissue. Our findings expand the understanding of Ps in terms of its impact on skin aging.
Materials and methods
Study samples and dataset description
Our previous epigenome-wide association study collected 114 PP and 41 PN from Ps patients (41 patients with paired PP and PN), and 62 NN from health controls from the outpatient at Department of Dermatology, the First Affiliated Hospital, Anhui Medical University, Anhui, China. The detailed inclusion and exclusion criteria of Ps patients and normal controls, the requirement of skin tissues, and methylation experiment were described in our previous study [29]. Here, we analyzed the DNAm dataset of the tissue samples. The clinical features of NN, PN, and PP samples were matched, and no significant difference was detected (P > 0.01, Table 1).
Calculation of DNA methylation age
Horvath [18] developed an epigenetic clock that incorporates the methylation of 353 CpGs, which were obtained through an elastic-net regression, to estimate age. The approach has shown to be robust across a range of human tissues. Here, we took advantage of the corresponding online tool “DNA methylation age calculator” (https://dnamage.genetics.ucla.edu) to estimate the DNAm age of Ps and non-Ps skin samples. Specifically, we prepared and uploaded a compressed text file that include methylation beta values which were measured from all available samples, as well as a file with brief annotation information. After online analysis, an output file with calculated methylation age and other estimates was obtained. The output contains DNAm age and age acceleration residual for each sample. The age acceleration residual is defined as the residual of a linear model where the independent variable is chronological age and the response is DNAm age. As a measure of increase/decrease of DNAm age, the advantage of the residual over the difference between DNAm age and chronological is that the latter could be naturally negatively correlated with chronological age.
Identification of Ps-dependent age-associated CpG sites
Ps-dependent age-associated CpG sites are considered those age-associated in NN/PN but becoming unassociated in Ps, or vice versa. To obtain them, we first applied a linear regression to test the association between age and CpG methylation for NN, PN, and PP, separately. For the linear model, the independent variable is age and the response is the logit of methylation level M, or \( \log \frac{M}{1-M} \). For each site, the regression model yielded a coefficient value as well as a corresponding standard error (i.e., b and se) of the independent variable age. Then, to test the difference of the coefficient between Ps and NN/PN groups, we calculated a Z score by using the following equation:
Here, b1 and se1 denote the coefficient and the corresponding standard error in Ps, while b2 and se2 denote those in NN/PN. The Z score was considered normally distributed, and a P value could be obtained for each CpG accordingly.
KEGG pathway analysis
KEGG pathway analysis was conducted by R package clusterProfiler [38]. For age-associated CpGs, we dropped those located in intergenic regions. We then mapped CpGs to genomic context according to Illumina 450 K methylation array annotation file and CpGs within 5′UTR to 3′UTR for one gene were selected. These genes were searched against the human pathway background. Enrichment test was based on hypergeometric distribution.
Statistical analysis
Statistical analyses were performed in R software (version 3.3.2) and SPSS (version 16.0). Quantitative data are presented as mean ± standard error. Differences between two groups were analyzed by the Student’s t test or Wilcoxon rank sum test. Differences among multiple groups were analyzed by the analysis of variance (ANOVA). P values were determined by ANOVA for all quantifications. P values < 0.05 were regarded as statistically significant.
Abbreviations
- ANOVA:
-
Analysis of variance
- BMI:
-
Body mass index
- DNAm:
-
DNA methylation
- FDR:
-
False discovery rate
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- NN:
-
Normal skin tissue of health control
- PASI:
-
Psoriasis area and severity index
- PN:
-
Uninvolved psoriatic skin tissue
- PP:
-
Involved psoriatic skin tissue
- Ps:
-
Psoriasis
- UV:
-
Ultraviolet
References
Shen C, Gao J, Yin X, Sheng Y, Sun L, Cui Y, et al. Association of the late cornified envelope-3 genes with psoriasis and psoriatic arthritis: a systematic review. J Genet Genomics. 2015;42:49–56.
Dou J, Zhang L, Xie X, Ye L, Yang C, Wen L, et al. Integrative analyses reveal biological pathways and key genes in psoriasis. Br J Dermatol. 2017;177:1349–57.
Hawkes JE, Chan TC, Krueger JG. Psoriasis pathogenesis and the development of novel targeted immune therapies. J Allergy Clin Immunol. 2017;140:645–53.
Nograles KE, Davidovici B, Krueger JG. New insights in the immunologic basis of psoriasis. Semin Cutan Med Surg. 2010;29:3–9.
Zhang Y, Tu C, Zhang D, Zheng Y, Peng Z, Feng Y, et al. Wnt/beta-catenin and Wnt5a/ca pathways regulate proliferation and apoptosis of keratinocytes in psoriasis lesions. Cell Physiol Biochem. 2015;36:1890–902.
Queiro R, Tejon P, Alonso S, Coto P. Age at disease onset: a key factor for understanding psoriatic disease. Rheumatology (Oxford). 2014;53:1178–85.
Springate DA, Parisi R, Kontopantelis E, Reeves D, Griffiths CE, Ashcroft DM. Incidence, prevalence and mortality of patients with psoriasis: a U.K. population-based cohort study. Br J Dermatol. 2017;176:650–8.
Wisniewski A, Matusiak L, Szczerkowska-Dobosz A, Nowak I, Luszczek W, Kusnierczyk P. The association of ERAP1 and ERAP2 single nucleotide polymorphisms and their haplotypes with psoriasis vulgaris is dependent on the presence or absence of the HLA-C*06:02 allele and age at disease onset. Hum Immunol. 2018;79:109–16.
Zhou F, Shen C, Hsu YH, Gao J, Dou J, Ko R, et al. DNA methylation-based subclassification of psoriasis in the Chinese Han population. Front Med. 2018. https://doi.org/10.1007/511684-017-0588-6. [Epub ahead of print]
Pollock RA, Abji F, Gladman DD. Epigenetics of psoriatic disease: a systematic review and critical appraisal. J Autoimmun. 2017;78:29–38.
Swisshelm K, Disteche CM, Thorvaldsen J, Nelson A, Salk D. Age-related increase in methylation of ribosomal genes and inactivation of chromosome-specific rRNA gene clusters in mouse. Mutat Res. 1990;237:131–46.
Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–8.
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602.
Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–9.
Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–6.
Garagnani P, Bacalini MG, Pirazzini C, Gori D, Giuliani C, Mari D, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–4.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115.
Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15:R24.
Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, et al. Caloric restriction delays age-related methylation drift. Nat Commun. 2017;8:539.
Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, Gladyshev VN. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 2017;25:954–960 e956.
Stubbs TM, Bonder MJ, Stark AK, Krueger F, Team BIAC, von Meyenn F, et al. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 2017;18:68.
Wagner W. Epigenetic aging clocks in mice and men. Genome Biol. 2017;18:107.
Wang T, Tsui B, Kreisberg JF, Robertson NA, Gross AM, Yu MK, et al. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 2017;18:57.
Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371-384
Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111:15538–43.
Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, et al. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–5.
Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212:1563–73.
Zhou F, Wang W, Shen C, Li H, Zuo X, Zheng X, et al. Epigenome-wide association analysis identified nine skin DNA methylation loci for psoriasis. J Invest Dermatol. 2016;136:779–87.
Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, et al. Huntington's disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY). 2016;8:1485–512.
Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171.
Bormann F, Rodriguez-Paredes M, Hagemann S, Manchanda H, Kristof B, Gutekunst J, et al. Reduced DNA methylation patterning and transcriptional connectivity define human skin aging. Aging Cell. 2016;15:563–71.
Choi JH, Choi DK, Sohn KC, Kwak SS, Suk J, Lim JS, et al. Absence of a human DnaJ protein hTid-1S correlates with aberrant actin cytoskeleton organization in lesional psoriatic skin. J Biol Chem. 2012;287:25954–63.
Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, et al. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 2014;346:360–3.
Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44:40–6.
Zhou W, Dinh HQ, Ramjan Z, Weisenberger DJ, Nicolet CM, Shen H, et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet. 2018;50:591–602.
Vandiver AR, Irizarry RA, Hansen KD, Garza LA, Runarsson A, Li X, et al. Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol. 2015;16:80.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Acknowledgements
We are grateful to the participants and their families involved in this study. We also thank the editors and reviewers for their important comments and useful suggestions to this manuscript.
Funding
This project was mainly supported from the National Natural Science Foundation (Grant Nos.: 81502713, 81573033, 81872516) of China, Grant for Scientific Research of BSKY (XJ201634) from Anhui Medical University, the Key Program of Outstanding Talents of Anhui Province (gxyqZD2017028), the Fundamental Research Funds for the Central Universities (3332018182), China, and Innovation Fund for Graduate Students (2018-1002-01-26), Peking Union Medical College, China.
Availability of data and materials
The epigenetic dataset was analyzed in this study are available from the corresponding author on reasonable request. The raw data had been submitted to GEO under accession GSE73894 (title: genome-wide DNA methylation analysis of psoriatic and normal skin tissues).
Author information
Authors and Affiliations
Contributions
CBS, MW, YC, XJZ, and FSZ participated in the design of the study. CBS, FSZ, LLW, JG, and XS carried out the samples collection, and methylation array interpretation. CBS, MW, LLW, JG, and FSZ drafted the manuscript. MW, CBS, FSZ, LDS, and XBZ performed the bioinformatics analyses of the data. YHH, XJZ, YC, LDS, and RK reviewed and revised this manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from all of the subjects, under an Anhui Medical University approved protocol, and the study was performed according to the Declaration of Helsinki principles.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. One hundred CpGs significantly associated with age in NN. (XLSX 14 kb)
Additional file 2:
Table S2. Five hundred and eighty-eight CpGs significantly associated with age in PN. (XLSX 46 kb)
Additional file 3:
Table S3. Eight thousand and thirty-two CpGs significantly associated with age in PP. (XLSX 534 kb)
Additional file 4:
Figure S1. Extra comparisons in addition to Fig. 3b, c. (TIFF 903 kb)
Additional file 5:
Table S4. CpGs associated with age (FDR < 0.05) in paired PN and PP samples. (XLSX 90 kb)
Additional file 6:
Table S5. Thirty nine CpGs that are both age-associated in PP and differently methylated between PP and PN. (XLSX 10 kb)
Additional file 7:
Table S6. Three hundred and one candidates Ps-dependent age-associated CpGs contrasted by PN and PP using a loose cut-off (P < 0.01). (XLSX 79 kb)
Additional file 8:
Table S7. Eight hundred and eighty-one candidates Ps-dependent age-associated CpGs contrasted by NN and PP using a loose cut-off (P < 0.01). (XLSX 208 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shen, C., Wen, L., Ko, R. et al. DNA methylation age is not affected in psoriatic skin tissue. Clin Epigenet 10, 160 (2018). https://doi.org/10.1186/s13148-018-0584-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-018-0584-y