Abstract
Background
Louse flies (Diptera, Hippoboscidae) are important blood-sucking parasites of birds and mammals with a worldwide distribution. The aim of our study was to collect louse flies from birds across multiple sites in Hungary and evaluate the effects of avian traits on louse fly–host relationships.
Methods
Between 2015 and 2022, 237 louse flies were collected from birds at multiple locations in Hungary. The louse flies were identified to species level by morphological and molecular methods. Louse fly species and their seasonal dynamics were analyzed.
Results
Six louse fly species were identified: Ornithomya avicularia, Ornithomya fringillina, Ornithomya biloba, Ornithomya chloropus, Ornithoica turdi and Ornithoctona laticornis. Results of statistical analyses indicated that habitat, migration habits and the feeding places of birds have significant effects on their possible role as hosts of O. avicularia, O. fringillina and O. turdi. Analysis of the temporal distribution of avian louse flies showed different seasonal patterns according to species. Phylogenetic analyses highlighted that O. turdi clustered separately from other members of the subfamily Ornithomyinae which thus did not form a monophyletic group.
Conclusions
This study presents one of the longest continuous collections of ornithophilic louse fly species in Europe so far. Avian traits were shown to influence louse-fly infestation. To our best knowledge, this is the first report on O. laticornis in Europe. The ability of this African louse fly species to survive in Europe, as demonstrated in the present study, may be an indication of its future establishment. Our findings, in accordance with previous reports, also indicated that the subfamily Ornithomyinae should be taxonomically revised.
Graphical Abstract

Similar content being viewed by others
Background
The role of birds in the “transportation” of arthropods with vector potential is long-known [1]. This aspect of bird life as parasite hosts is suspected to become more and more important due to the changes in ecological conditions and which may become a consequence of the currently ongoing climate change [2].
Louse flies (Diptera, Hippoboscidae) are blood-sucking parasites of birds and mammals, with 213 known species worldwide [3]. The family Hippoboscidae contains three subfamilies: Hippoboscinae, Lipopteninae and Ornithomyinae. Ornithophilic louse flies generally belong to the Ornithomyinae subfamily [4]; however, members of the Hippoboscinae subfamily (e.g. Hippobosca equina and Hippobosca longipennis) can also parasitize birds [5]. Bird lice can negatively affect the health and condition of birds when present and they also play an important role in the ecology of other parasites, possibly contributing to their evolution by phoresis [6]. They can carry a multitude of different pathogens with high veterinary-medical significance, as exemplified by the West-Nile virus [7] and Babesia species [8], although their vector role is not yet clear. Research on hippoboscids is flourishing nowadays, i.e. with investigations focusing on their ecology, evolution, and potential role in the transmission of pathogens [3, 9,10,11,12,13,14].
Although studies on louse flies originating from Central [3] Northern [14], Southern [10], Western [15] and Eastern Europe [16] have also been conducted, only a few of these report long-term evaluations with continuous sample collection. The number of studies conducted on avian ectoparasites steadily increased in Hungary and in other Central European countries during the previous decade [2, 3, 12, 17, 18], but studies on ornithophilic hippoboscids appear to have been neglected compared to other arthropod vectors that are generally considered epidemiologically more important (i.e. ticks and mosquitoes) [19, 20]. Previous Hungarian studies on hippoboscids date back to the previous century and around the turn of the millenium [21, 22]. In light of the above, research on these insects has both regional and international importance.
The aim of the present study was to monitor the louse fly fauna of foraging birds in Hungary, elaborate on the effects of the hosts' attributes (migration habit, habitat, feeding place) on louse fly burden and identify potential trends in the temporal distribution of hippoboscids in Hungary.
Methods
Sample collection
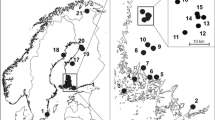
Hippoboscids were collected from passeriform, strigiform and piciform hosts by professional bird-ringers at multiple locations in Hungary (Fig. 1): Ócsa (47°19′ N, 19°13′ E), Fenékpuszta (46°44′N, 17°14′E), Dávod (46°0′N, 18°51′E), Izsák (46°46′N, 19°19′E), Szalonna (48°27′N, 20°42′E) and Tömörd (47°21′N, 16°39′E). Birds were mist-netted for ringing by standard ornithological (Ecotone, Gdynia, Poland) mist-nets (length 12 m, mesh size 16 × 16 mm). During the ringing procedure, birds were examined for the presence of ectoparasites. Samples were also collected at Barbacs (47°38′N, 17°18′E) and Lajta-Hanság (47°50′N, 17°14′E) by licensed hunters during the thinning of Hooded Crow (Corvus cornix) population.
Map of the Carpathian Basin showing sample collection sites (stars with numbers). The yellow star (marked with 1) denotes the ringing station at Ócsa where 8-year-long sampling activities took place. White stars mark other locations where sample collection occurred only in 2022: (2) Lajta-Hanság, (3) Barbacs, (4) Tömörd, (5) Fenékpuszta, (6) Szalonna, (7) Izsák, (8) Dávod
Hippoboscids were also caught from the environment of the bird-ringing facilities. As many flies can fall off or escape from the bird during the capturing and the ringing procedure [10], some avian louse flies were found in and around the buildings where the birds were ringed. In these cases, it was impossible to identify the original hosts and, therefore, the term “environment” was used.
In Ócsa, collection activities started in 2015 and continued in all consecutive years until 2022, with collections from March to November. At the other sample collection sites, sample collection started in March 2022 and ended in November 2022.
Ornithophilic louse flies were collected with fine tweezers and stored in 2-ml screwcap tubes filled with 96% ethanol. Data on the collection (date, location, avian host species) were recorded (Additional file 1: Table S1).
Categorization of birds according to their habitat, migration habit and feeding place
HURING codes were used instead of the birds’ full English and/or binomial names in all figures and tables presented in this study. These abbreviations are explained in Table 1 and Additional file 2: Table S2. English bird species names are capitalized, following international recommendations (https://bou.org.uk/britishlist/bird-names/).
All louse fly-bird associations can be found in Additional file 1: Table S1. Birds were categorized according to their feeding place, migration habits and habitats according to ornithological data and previous reports [17, 19, 23] as well as the expertise of two of the authors (TCS and DK) (Additional file 2: Table S2). In order to categorize birds according to their feeding places, an “Above ground” category was created. Birds belonging to this group feed on, for example, reed trunks, bushes or branches that do not touch the ground directly but are not far from it either.
Morphological identification of louse fly species
Louse fly species were identified based on standard taxonomic keys: [3, 15, 24].
DNA extraction
Seven hippoboscids were chosen for molecular identification: two Ornithomya avicularia, and one each from the other louse fly species identified (Ornithomya fringillina, Ornithomya biloba, Ornithomya chloropus, Ornithoica turdi, Ornithoctona laticornis). The surface of louse flies was disinfected by sequential washing for 15 s in 10% NaClO, tap water and distilled water. For the DNA extraction, one leg of each specimen was cut off. DNA was extracted with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction, including an overnight digestion in tissue lysis buffer and Proteinase-K at 56 °C. Extraction controls (tissue lysis buffer) were also processed with the hippoboscid samples to monitor cross-contamination.
Molecular identification of Hippoboscidae species
The cytochrome c oxidase subunit I (cox 1) encoding gene was chosen as the target for molecular analysis. The PCR was modified from Folmer et al. [25] and amplifies an ~710-bp-long fragment of the gene. The primers HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) and LCO1490 (5’-GGT CAACAA ATC ATA AAG ATA TTG G-3’) were used in a reacti on volume of 25 μl, containing 1 U (stock 5 U/μl)HotStarTaq Plus DNA Polymerase (Qiagen, Hilden, Germany), 2.5 μl 10 × CoralLoad Reacti on buffer (including 15 mM MgCl2), 0.5 μl PCR nucleotide Mix (Qiagen, Hilden, Germany) (stock 10 mM), 0.5 μl of each primer (stock 50 μM), 15.8 μl ddH2O and 5 μl template DNA. For amplification, an initial denaturation step at 95°C for 5 min was followed by 40 cycles of denaturation at 94°C for 40 s, annealing at 48°C for 1 min and extension at 72°C for 1 min. Final extension was performed at 72°C for 10 min.
Sequencing
A non-template reaction mixture served as the negative control in all PCR analyses. Extraction controls and negative controls remained PCR negative in all tests. The PCR products were purified and sequenced by Eurofins Biomi Ltd. (Gödöllő, Hungary). The BioEdit program was used for quality control and trimming of sequences. The analyses of assembled sequences were performed with BLASTN via GenBank (https://blast.ncbi.nlm.nih.gov). The sequences obtained in the current study were deposited in the GenBank database and are available under accession numbers PP111350–PP111356.
Statistical analyses
All data from the sample collectors were recorded in Microsoft Office Excel (Microsoft Corp., Redmond, WA, USA), and all calculations, with the except of Fisher’s exact tests and calculations for the phylogenetic tree, were performed in this program. The mean and median rates of infestation were calculated according to Reiczigel et al. [26]. For the comparison of host attributes (habitat, migratory habit, feeding place), Fisher’s exact tests were used (R program 4.3.0) [27]; R program 4.3.0 was also used to generate Fig. 2 (bipartite package) [28]) and Fig. 3 (igraph [29], ggraph [30] and RColorBrewer [31] packages).
Phylogenetic analysis
In order to construct the tree, cox1 nucleotide sequences of hippoboscids were chosen from the GenBank database (https://www.ncbi.nlm.nih.gov/nucleotide/). Sequences were aligned with MUSCLE (MEGA 11 program [32]). To root the tree, we chose three species from the Calyptratae subsection.
The evolutionary history was inferred by using the maximum likelihood method and general time reversible (GTR) model [33]. The percentage of trees in which the associated taxa clustered together is shown below the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 46 nucleotide sequences. There were a total of 589 positions in the final dataset. Evolutionary analyses were conducted in MEGA 11 [32].
Results
Species and numbers of louse flies
During the 8-year-long sample collection period, 237 louse flies were collected from 175 birds (nflies = 219) of 32 species, and from the environment of the ringing facilities (nflies = 18) at multiple locations across Hungary. Mean and median intensity of infestation was 1.13 and 1, respectively. According to the morphological identification, the louse flies found belonged to six species: Ornithomya avicularia (n = 168), Ornithomya biloba (n = 23), Ornithomya fringillina (n = 17), Ornithomya chloropus (n = 3), Ornithoica turdi (n = 24) and Ornithoctona laticornis (n = 1). The parasite–host associations are visualized in Fig. 2 and listed in Table 1.
The maximum number of different louse fly species infesting the same bird was two. This only happened for two birds: a Song Thrush (Turdus philomelos) and a Common Reed Bunting (Emberiza schoeniclus). The co-infestation detected on these birds consisted of O. avicularia and O. turdi (one–one specimens) (Additional file 1: Table S1). The species and number of louse flies identified and their bird hosts are as follows:
-
Ornithoctona laticornis (n = 1). Only one specimen of this species was found. It was removed from a Eurasian Blue Tit (Cyanistes caeruleus) (Table 1; Figs. 3, 4; Additional file 1: Table S1).
-
Ornithomya aviculara (n = 168). This species of louse fly was the most abundant during the study period, with 71% of the detected louse flies belonging to this species. The most common host of this species was Savi’s Warbler (Locustella luscinioides) (nbirds = 29, nflies = 36) (Table 1). Ornithomya avicularia specimens were collected from 26 different bird species of three orders (Passeriformes, Strigiformes, Piciformes) (Figs. 5, 6).
-
Ornithomya biloba (n = 23). Specimens of this species were collected from Barn Swallow (Hirundo rustica) (nbirds = 20, nflies = 21) and from Sand Martin (Riparia riparia) (nbirds = 1, nflies = 1); one from the environment (Table 1; Fig. 7).
-
Ornithomya fringillina (n = 17). Fifteen specimens of this louse fly species were removed from nine different passeriform bird species among which the most abundant were the Eurasian Blackcap (Sylvia atricapilla) (nbirds = 3, nflies = 3) and the Tree Pipit (Anthus trivialis) (nbirds = 3, nflies = 3). Two specimens were collected from the environment (Table 1; Figs. 5, 7).
-
Ornithomya chloropus (n = 3). In total, three specimens of this louse fly species were collected: one from a Dunnock (Prunella modularis) (nbird = 1, nfly = 1), one from a Bearded Reedling (Panurus biarmicus) (nbird = 1, nfly = 1) and one from the environment (Table 1, Fig. 8).
-
Ornithoica turdi (n = 25). This louse fly species was the second most abundant species, even though they accounted for only 10.5% of all louse flies. The most common host of this species was the Red-backed Shrike (Lanius collurio) (nbirds = 5, nflies = 7). Ornithoica turdi were found feeding on 10 different bird species during the study period (Table 1; Figs. 5, 7).
Key morphological characters of the louse fly Ornithoctona laticornis. A Dorsal view of the head (1 denotes the anterior ocellus situated slightly above the level of posterior eye margins). B Ventral view of the head, mesosternal processes (1 denotes the antennae, which were twice as long as broad; 2, the length of the mesosternal process). C Scutellum with four prominent hairs (1 denotes the medial hairs, which were twice as long as the lateral ones), D Dorsal view of the abdomen (1 denotes three median tergal plates; 2, two log hairs on both plates of tergite six; 3, antero-lateral area of abdomen with long hair; 4, caudal area of abdomen with long hair)
Molecular identification and phylogenetic analyses
In general, based on the cox1 gene of the representative louse fly specimens, the identity of all six louse fly species was confirmed (O. avicularia, n = 2 specimens tested; O. biloba, n = 1; O. fringillina, n = 1; O. chloropus, n = 1; O. turdi, n = 1; O. laticornis, n = 1).
More specifically, according to their cox1 sequences:
-
Ornithoctona laticornis (PP111350) showed 99.83% identity to an Ornithoctona sp. (EF531223) from the collection of the North Carolina State University (host and collection site are unknown).
-
The two O. avicularia specimens (PP111351 and PP111352) belonged to different haplotypes (98.28% identity). PP111351 showed 99.69% identity to O. avicularia (OR064832) from Russia and 99.53% to O. avicularia (MZ261702) from Canada. PP111352 showed 99.69% identity to O. avicularia (OP035933) from Slovakia and the same percent of identity to multiple O. avicularia specimens from Canada (MZ261701, MZ261708, MZ261714) (Fig. 9).
-
Ornithomya biloba (PP111353) was 99.84% identical to O. biloba specimens from the Czech Republic (MF496010), Slovakia (OP036771) and from Portugal (OL505728) (Fig. 9).
-
Ornithoica turdi (PP111354) showed 100% identity to O. turdi (OR064834) from Moldova, and 99.69% to O. turdi (MK234697) from Austria (Fig. 9).
-
Ornithomya chloropus (PP111355) showed 99.84% identity to O. chloropus specimens from Finland (MW590960, MW590969) and from Russia (OR054225) (Fig. 9).
-
Ornithomya fringillina (PP111356) showed 100% identity to O. fringillina specimens from Finland (MW590981, MW590974, MW590963) (Fig. 9).
Statistical analyses
Due to the low numbers of O. chloropus and O. laticornis, no statistical probes were performed involving these species. Ornithomya biloba was also excluded from the analysis as its host specificity towards the Hirundinidae family would have led to biased (or obvious) results.
According to our analyses, there were significant differences between the host habitats (reed, forest, meadow, forest/meadow) of O. avicularia and O. turdi (P < 0.0001). Ornithomya avicularia was the most abundant louse fly found on reed-associated birds, and O. turdi was the most commonly found louse fly on forest- and meadow-associated hosts. In the case of O. avicularia and O. fringillina, the result of the same comparison was non-significant (P = 0.2958). The difference between O. fringillina and O. turdi was also non-significant (P = 0.1985) (Table 2).
Comparisons were made based on the migration habits (resident, resident/short-distance migrants, short-distance migrants and long-distance migrants) of the hosts of O. avicularia, O. fringillina and O. turdi. The difference between the hosts of O. avicularia and O. fringillina was significant (P = 0.0036) as we found no O. fringillina on short-distance migrants; however the difference was non-significant in the comparison of the hosts of O. avicularia and O. turdi (P = 0.4033). A significant difference was also visible for the hosts of O. fringillina and O. turdi, as the latter was frequently found on short-distance migrants (P = 0.0343) (Table 3).
The avian hosts were also categorized according to their feeding place (Ground, Above ground, Ground/Above ground). There was a significant difference between the hosts of O. avicularia and O. fringillina (P = 0.0024) as O. fringillina was not found feeding on birds belonging to the “Ground” category during the study period; in contrast, 36% of O. avicularia-parasitized birds belonged to the “Ground” category. The difference was also significant for the hosts of O. avicularia and O. turdi (P = 0.0068), with 64% of the O. turdi specimens found on birds belonging to the “Ground” category of feeding. The difference between O. fringillina and O. turdi according to feeding place was also significant (P < 0.0001) (Table 4).
Temporal distribution of louse flies
The temporal distribution of louse fly species found on birds during the 8-year-long study period is visualized in Fig. 10. It should be noted that in this study no sample collection was conducted during the winter. Ornithomya avicularia specimens were recovered from the second half of May up to the end of October, reaching their peak abundance in the first half of July. Ornithomya biloba specimens were only collected from the first half of August until the first half of October and were most common in the first half of September, even though its most common hosts (Hirundo rustica) were occasionally caught and checked for the presence of louse flies during the springtime. The occurrence of O. fringillina was at its highest in the first half of September; this species was present from the second half of August until the first half of October. Ornithoica turdi showed activity from the first half of July until the second half of October, with peak abundance in the second half of July. Only three specimens of O. chloropus were collected: one on 5 September 2022 and one on 31 October 2020; for the third specimen, the precise date of collection was not recorded, but is estimated to be October 2019 (Additional file 1: Table S1). Ornithoctona laticornis was collected only once, on 9 October 2016.
Discussion
Six bird-associated louse fly species were identified and analyzed. For several species, we report here for the first time molecular and host-related data from either Central European or European countries. Therefore, our results supplement missing information on the ecology and phenology of relevant louse fly species in this geographical region. We also addressed host-related factors of louse-fly infestation and the taxonomic uniformity of this group.
Identified louse fly species
To our best knowledge, this is the first time that O. laticornis has been found in Europe. In 1984, Hutson [15] suggested that the appearance of this louse fly species in Europe could be expected, since it had been found on multiple Palearctic migrants in Africa. However, up to the present study, it had only been reported in Central and South Africa [34] and Madagascar [24]. Interestingly enough, our specimen was found feeding on a Blue Tit on 9 October 2016, which is long after the spring migration period. Not much is known about the life-cycle of O. laticornis, but other hippoboscid adults can survive for around 4 months, and the duration of the pupal stage varies from 19 to 23 days in the summer and from 20 to 36 days in the winter [35]. Also, bird-specific louse flies tend to overwinter in pupal form in Europe, which may indicate even longer pupal stages [21]. The pupae of hippoboscids (in general) usually can be found in bird nests, on the hair of mammalian hosts or on the ground [3]. Based on this information, one hypothesis is that a bird carried an imago from Africa during the spring migration and that the adult fly survived until October in Hungary. However, due to the relatively large temporal distance (approx. 5–7 months) between the spring migration of birds (March–May) and the finding of the O. laticornis specimen, it is also conceivable that an already fertilized female O. laticornis had arrived in Hungary on a migrating bird in the spring and had been able to lay a larva ready to pupate that later hatched. In the latter scenario, the imago was probably second-generation. A second hypothesis is suggested from the work of Hutson [15]: Ornithoctona laticornis can occasionally be found in Europe, but due to its close morphological resemblance to Ornithomya avicularia (Figs. 4, 6) some specimens may have been misidentified in the past. According to European keys, the morphological similarity between O. laticornis and Ornithomya rupes may be even more deceiving [3]. Therefore, population(s) of O. laticornis may be currently present in Europe. At this time we do not have enough information to draw accurate conclusions. However, regardless of how this specimen came to Hungary, it was found on a resident bird, which shows that O. laticornis can survive Central European conditions, not just in the summer, but in the autumn. This finding highlights the importance of comprehensive research on wild bird parasites as due to the migratory nature of their hosts, these animals can be indicators of the direct effects of climate change.
The other louse fly species identified in this study as Ornithomya avicularia, O. fringillina, O. biloba, O. chloropus and Ornithoica turdi have all been previously reported in Hungary [22]. The euryxenous natures of O. avicularia, O. turdi and O. fringillina have long been known, as has the host specificity of O. biloba towards the Hirundinidae family (especially the Barn Swallow) [10, 13, 16, 22, 36, 37].
Ornithomya avicularia was the most abundant louse fly in the present study, the second most abundant was O. turdi, followed by O. biloba, O. fringillina and O. chloropus. A similar study recently performed in another Central European country, the Czech Republic, reported different relative abundancies [13]; for example, O. biloba and other stenoxenous species were represented in much larger numbers. The main reason for this difference is that in the study from the Czech Republic, nestlings of Barn Swallows and Swifts (Apus apus) were also checked for potential louse flies. Therefore, not only were an enormous number of O. biloba specimens collected but other stenoxenous louse flies were also found in the nests of the birds, namely Crataerina hirundinis and Crataerina pallida. In our study, no bird nests were examined and Barn Swallows were only occasionally caught, which explains the relatively low number of O. biloba, as due to the random nature of the sample collection, the relative presence of O. biloba (and other host-specific species) is also highly affected by the relative number of swallows among the examined hosts.
Molecular identification and phylogenetic analysis
We report here the nucleotide sequence of O. laticornis (PP111350) for the first time. Although our sequence showed 99.83% identity to an Ornithoctona sp. (EF531223) [38], the host and site of origin of EF531223 are unknown (from the collection of the North Carolina State University). Base on the cox1 gene, the closest relatives (that are available in the GenBank database) of our O. laticornis (PP111350) specimen are this Ornithoctona sp. from the North Carolina State University collection (EF531223) and Ornithoctona erythrocephala from Brazil (JQ246707) [39] (Fig. 9).
Based on the cox1 gene, the genus Ornithoica seems to show a more distant genetic relationship to genera belonging to the subfamily Ornithomyinae (Ornithomya, Ornithoctona, Icosta, Pseudolynchia, Crataerina) than it does to certain representatives of the subfamilies Hippoboscinae (Hippobosca equina, H. longipennis, H. variegata) and Lipopteninae (Lipoptena mazamae, L. cervi, L. fortisetosa and Melophagus ovinus) (Fig. 9). This is in line with a recent report from Russia [40]. As shown in a previous study, [39] this genetic relationship also holds when the phylogenetic analysis is based on 18S ribosomal DNA. These results suggest that the genus Ornithoica might belong to a different subfamily, and that the taxonomy of Ornithomyinae should be revised.
Statistical analyses
According to our statistical analyses, the migration habit, the habitat type and the feeding habit of birds affect their potential as louse fly hosts in Hungary. In our study, three euryxenous louse fly species were examined in this regard and significant differences were found between the migration habits of the hosts of O. avicularia and O. fringillina. In a previous study from the Czech Republic, significant differences were found between the migration habits of the hosts of O. avicularia and O. fringillina as well; however, in that study O. avicularia was far less common on long-distance migrant birds than on short-distance migrants [13]. In contrast, a study from Finland found no significant differences between the migration habits of the hosts [14]. These results might suggest that host preference patterns may differ under different climates, or that the migratory habit alone cannot explain host specificity patterns, but other factors (e.g. ornithological and geographic) may influence the results as well. Nevertheless, both species were the most common on long-distance migrants, and only differed by their ratios. The difference between the hosts of O. avicularia and O. turdi in the same context was not significant; however, the difference between the hosts O. turdi and O. fringillina was significant. The latter significance may strengthen the results of the previous two tests.
The habitat-association of hosts also seemed to be different in the case of O. avicularia and O. turdi (P < 0.0001), as O. avicularia was the most common on reed-associated birds, and O. turdi was the most abundant on forest- and meadow-associated birds. The difference was less pronounced and non-significant between O. avicularia and O. fringillina, and also non-significant between O. fringillina and O. turdi. In contrast to this, both O. avicularia and O. fringillina preferred birds with a forest habitat, but were uncommon on birds from “wetlands” in Finland [14].
Birds were categorized according to their feeding place as well. Interestingly, each comparison demonstrated significant differences (O. turdi vs O. avicularia, O. avicularia vs O. fringillina and O. fringillina vs O. turdi). These results show that despite all three of the hippoboscid species that were statistically examined being able to develop wings and to fly [15, 40], their host selection was influenced by the hosts feeding height. Specifically, O. turdi predominated on birds feeding at ground level, whereas O. fringillina was absent on birds exclusively feeding at ground level. Ornithomya avicularia was approximately twice as abundant on birds feeding above ground compared to those feeding on the ground (Table 4).
Temporal distribution of louse fly species
In the present study, most of the louse fly species (O. turdi, O. fringillina, O. chloropus) were the most active at the end of the summer and in the autumn. Ornithomya biloba flies were only collected during the autumn migration of their hosts (Barn Swallow, and other species of Hirundinidae). During the spring migration and the roosting season, none of the previously mentioned louse fly species were found, despite the fact that in other countries O. biloba has been found on early swallows [41]. In terms of temporal distribution, the appearance of O. avicularia preceded all other louse flies, as this species was active from the second half of May and remained active during the sample collection period. This means that this species was the only one that was found to be active on foraging birds during the main nesting season (Fig. 10). It is important to note that our sample collection started in March and ended in November each year. Although this interval includes the spring (March–May) and autumn migration (September–November) periods [42], only randomly caught birds were examined for the presence of louse flies and no bird nests were checked during the study. Therefore, these results are only relevant to the activity of flies on flying and/or foraging birds, except the wintering period.
Conclusions
This is the first report of O. laticornis in Europe, as well as the first molecular-phylogenetic analysis of this louse fly species. In accordance with previous studies, the migration habit, the habitat type and the feeding habits of birds were found to affect their potential role as the hosts of O. avicularia, O. fringillina and O. turdi, but these patterns may vary in different geographical regions. According to our analyses and data from available literature, members of the genus Ornithoica show distant phylogenetic clustering to genera belonging to the subfamily Ornithomyinae (where it was hitherto assigned), necessitating taxonomic revision of this group in the near future.
Availability of data and materials
The sequences obtained in the current study were deposited in the GenBank database and are available under accession numbers PP111350–PP111356 (https://www.ncbi.nlm.nih.gov/genbank/). All other relevant data are included in the manuscript and its appendices.
References
Janisch M. Kullancsgazda madarak különféle betegségek közvetítői [Tick carrier birds spreading disease agents]. Aquila. 1960;67–68:191–4.
Keve G, Csörgő T, Benke A, Huber A, Mórocz A, Németh Á, et al. Ornithological and molecular evidence of a reproducing Hyalomma rufipes population under continental climate in Europe. Front Vet Sci. 2023;10:272.
Oboňa J, Sychra O, Greš S, Heřman P, Manko P, Roháček J, et al. A revised annotated checklist of louse flies (Diptera, Hippoboscidae) from Slovakia. Zookeys. 2019;862:129–52.
Nartshuk EP, Matyukhin AV, Shokhrin VP, Markovets MY. New records of ornithophilous louse-flies (Diptera: Hippoboscidae: Ornithomyinae) from the Russian Far East. Far Eastern Entomol. 2019;384:15–20.
Maa TC. A revised checklist and concise host index of Hippoboscidae (Diptera). Pacific Insects Monogr. 1969;20:261–99.
Lee L, Tan DJX, Oboňa J, Gustafsson DR, Ang Y, Meier R. Hitchhiking into the future on a fly: Toward a better understanding of phoresy and avian louse evolution (Phthiraptera) by screening bird carcasses for phoretic lice on hippoboscid flies (Diptera). Syst Entomol. 2022;47:420–9.
Farajollahi A, Crans WJ, Nickerson D, Bryant P, Wolf B, Glaser A, et al. Detection of West Nile virus RNA from the louse fly Icosta americana (Diptera: Hippoboscidae). J Am Mosq Control Assoc. 2005;21:474–6.
Čisovská Bazsalovicsová E, Víchová B, Oboňa J, Radačovská A, Blažeková V, Králová-Hromadová I. Bird louse flies Ornithomya spp. (Diptera: Hippoboscidae) as potential vectors of mammalian Babesia and other pathogens. Vector-Borne Zoonotic Dis. 2023;23:275–83.
Peña-Espinoza M, Em D, Shahi-Barogh B, Berer D, Duscher GG, Van Der Vloedt L, et al. Molecular pathogen screening of louse flies (Diptera: Hippoboscidae) from domestic and wild ruminants in Austria. Parasit Vectors. 2023;16:179.
Rekecki T, Rajkovic D. Diversity and prevalence of ornithophilic louse flies (Diptera: Hippoboscidae: Ornithomyinae) in Serbia. Turk J Zool. 2023;47:261–7.
Andreani A, Belcari A, Sacchetti P, Romani R. Antennal morphology and fine structure of flagellar sensilla in Hippoboscid flies with special reference to Lipoptena fortisetosa (Diptera: Hippoboscidae). Insects. 2022;13:236.
Svobodová M, Volf P, Votýpka J. Trypanosomatids in ornithophilic bloodsucking Diptera. Med Vet Entomol. 2015;29:444–7.
Santolíková A, Brzoňová J, Čepička I, Svobodová M. Avian louse flies and their trypanosomes: new vectors, new lineages and host–parasite associations. Microorganisms. 2022;10:584.
Lehikoinen A, Pohjola P, Valkama J, Mutanen M, Pohjoismäki JLO. Promiscuous specialists: host specificity patterns among generalist louse flies. PLoS ONE. 2021;16:e0247698.
Hutson AM. Keds, flat-flies and bat-flies. Diptera, Hippoboscidae and Nycteribiidae. Handbooks for the identification of British insects, vol 10, part 7. London: Royal Entomological Society of London; 1984.
Oboňa J, Bazsalovicsová EČ, Pintilioaie A-M, Dumitru V, Gavril OCV, Topală L-E, et al. Checklist of Hippoboscidae (Diptera) from Romania. Historia Natural Bulgar. 2023;45:229–38.
Hornok S, Kováts D, Csörgő T, Meli ML, Gönczi E, Hadnagy Z, et al. Birds as potential reservoirs of tick-borne pathogens: first evidence of bacteraemia with Rickettsia helvetica. Parasit Vectors. 2014;7:128.
Flaisz B, Sulyok KM, Kováts D, Kontschán J, Csörgő T, Csipak Á, et al. Babesia genotypes in Haemaphysalis concinna collected from birds in Hungary reflect phylogeographic connections with Siberia and the Far East. Ticks Tick Borne Dis. 2017;8:666–70.
Hornok S, Flaisz B, Takács N, Kontschán J, Csörgő T, Csipak Á, et al. Bird ticks in Hungary reflect western, southern, eastern flyway connections and two genetic lineages of Ixodes frontalis and Haemaphysalis concinna. Parasit Vectors. 2016;9:101.
Kemenesi G, Dallos B, Oldal M, Kutas A, Földes F, Németh V, et al. Putative novel lineage of West Nile virus in Uranotaenia unguiculata mosquito, Hungary. Virusdisease. 2014;25:500–3.
Kaufman G. Magyarország kullancslégy fajai és a fecske-kullancslégy biológiája [The louse fly species of Hungary, and the biology of the swallow-louse fly]. Budapest: Állatorvostudományi Egyetem; 1988.
Papp L. Checklist of the Diptera of Hungary. Budapest: Hungarian Natural History Museum; 2001.
Csörgő T, Karcza Z, Halmos G, Magyar G, Gyurácz J, Szép T, et al. Magyar madárvonulási atlasz [Atlas of bird migration in Hungary]. Budapest: Kossuth kiadó; 2009.
Rahola N, Goodman SM, Robert V. The Hippoboscidae (Insecta: Diptera) from Madagascar, with new records from the “Parc National de Midongy Befotaka.” Parasite. 2011;18:127.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Reiczigel J, Marozzi M, Fábián I, Rózsa L. Biostatistics for parasitologists—a primer to quantitative parasitology. Trends Parasitol. 2019;35:277–81.
R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2023. Accessed 01 Dec 2023.
Dormann CF, Gruber B, Fruend J. Introducing the bipartite Package: analysing ecological networks. R News. 2008;8:8–11.
Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. 2005;Complex Systems:1695. https://igraph.org/. Accessed 01 Dec 2023.
Petersen T. ggraph: an implementation of grammar of graphics for graphs and networks. 2022. https://CRAN.R-project.org/package=ggraph. Accessed 01 Dec 2023.
Neuwirth E. RColorBrewer: ColorBrewer Palettes. 2022. https://CRAN.R-project.org/package=RColorBrewer. Accessed 01 Dec 2023.
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7.
Nei M, Kumar S, Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford, New York: Oxford University Press; 2000.
Nartshuk EP, Matyukhin AV, Redkin YA. Association of the louse-flies of the genus Ornithoctona Speiser, 1902 (Diptera: Hippoboscidae) with birds and first record of O. australasiae (Fabricius, 1805) from the Russian Far East. Far Eastern Entomol. 2018;355:23–8.
Reeves WK, Lloyd JE. Louse flies, keds, and bat flies (Hippoboscoidea). In: Medical and veterinary entomology, 3rd edition. Amsterdam: Elsevier; 2019. p. 421–38. https://www.sciencedirect.com/science/article/pii/B9780128140437000200.
Trilar T, Krčmar S. Contribution to the knowledge of louse flies of Croatia (Diptera: Hippoboscidae). Nat Croat. 2005;14:131–40.
González MA, Hidalgo JC, Talabante C, Bernal I. Nuevos datos faunísticos e interacciones de moscas piojo (Diptera: Hippoboscidae) parasitando aves capturadas con redes japonesas en el Sistema Central (España). Graellsia. 2023;79:e200–e200.
Petersen FT, Meier R, Kutty SN, Wiegmann BM. The phylogeny and evolution of host choice in the Hippoboscoidea (Diptera) as reconstructed using four molecular markers. Mol Phylogenet Evol. 2007;45:111–22.
Nirmala X, Hypša V, Žurovec M. Molecular phylogeny of Calyptratae (Diptera: Brachycera): the evolution of 18S and 16S ribosomal rDNAs in higher dipterans and their use in phylogenetic inference. Insect Mol Biol. 2001;10:475–85.
Yatsuk AA, Triseleva TA, Narchuk EP, Matyukhin AV, Safonkin AF. Morphology of the wings and attachment apparatus in the evolution of the family Hippoboscidae (Diptera). Integr Zool. 2023;1749–4877.12786.
Wawman DC. Ornithomya biloba, Pseudolynchia garzettae and Pseudolynchia canariensis (Diptera: Hippoboscidae): three new United Kingdom colonists and potential disease vectors. Med Vet Entomol. 2024;38:160-71.
Szép T. Madarak monitorozása: Ökológiai és evolúciós folyamatok feltárásának lehetőségei [Monitoring birds: Possibilities for exploring ecological and evolutionary processes]. Nyíregyháza. 2007. http://real-d.mtak.hu/91/1/Szep_Tibor_MTA_doktori_2007.pdf. Accessed 29 Dec 2023.
Acknowledgements
The authors are grateful to Ms. Nóra Takács for performing PCRs and to Ms. Adrienn Gréta Tóth for her support.
Funding
Open access funding provided by University of Veterinary Medicine. Molecular work was funded by the Office for Supported Research Groups, Hungarian Research Network (HUN-REN), Hungary (Project No. 1500107).
Author information
Authors and Affiliations
Contributions
GK: Conceptualization, study design, morphological identification, data curation, statistical analyses, phylogenetic analyses, manuscript writing, and preparation of figures and tables. TC, AB, ATB, HA, EAT, AN, AH, JG: Sample collection and data curation. DK: Ornithological categorization, sample collection and data curation. JK: Photography and phylogenetics. GK: Conceptualization and supervision. AN: Conceptualization, collection of literature data and preparation of figures. SH: Conceptualization, study design, DNA extraction, molecular analyses, manuscript writing and photography.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All songbirds were handled and released by experienced ringers of BirdLife Hungary. The license for bird ringing was issued by the Pest County Government Authority (https://www.mme.hu/sites/default/files/pe_ktf_97_13_2017_vvt.pdf). Hooded Crows (Corvus cornix) were culled by experienced hunters. The licence for hunting was issued by the National Hungarian Hunting Chamber, in accordance with Act No. 55 of 1996 and Government Decree No. 79 of 2004 on the protection and management of wildlife and on hunting. The aim of capturing or killing birds was never to collect parasites, but to either ring the captured bird, or pest control. Therefore, the study presented here is in accordance with Act No. 28 of 1998 and Government Decree No. 40 of 2013 on animal experiments. Notification or permission from the Animal Protection Authority was not required. Bird ringing registration numbers with the Hungarian Bird Ringing Centre are: DK: 517; EAT: 614; GJ: 109; AB: 586; AH: 488; AM: 618.
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
All individual findings of the study.
Additional file 2 Table S2.
Categorization of the bird species found to be louse fly infested during the study, according to their migration habit, habitat and feeding place.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Keve, G., Csörgő, T., Kováts, D. et al. Contributions to our knowledge on avian louse flies (Hippoboscidae: Ornithomyinae) with the first European record of the African species Ornithoctona laticornis. Parasites Vectors 17, 237 (2024). https://doi.org/10.1186/s13071-024-06303-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-024-06303-8