Abstract
Background
Birds play an important role in short- and long-distance transportation of ticks and tick-borne pathogens. The aim of the present study was to provide comprehensive information on the species and genetic diversity of ixodid ticks transported by migratory and non-migratory bird species in Central Europe, and to evaluate relevant data in a geographical, as well as in an ecological context.
Methods
During a three year period (2012-2014), altogether 3339 ixodid ticks were collected from 1167 passerine birds (representatives of 47 species) at ringing stations in Hungary. These ticks were identified, and the tick-infestations of bird species were compared according to various traits. In addition, PCR and sequencing of part of the cytochrome oxidase subunit-I (COI) and 16S rDNA genes were performed from representatives of five tick species.
Results
The most abundant tick species found were Ixodes ricinus and Haemaphysalis concinna (with 2296 and 989 immature stages, respectively). In addition, 48 I. frontalis (all stages), three Hyalomma rufipes nymphs, one I. lividus and two I. festai females were collected. The majority of I. ricinus and I. frontalis specimens occurred on ground-feeding bird species, as contrasted to Ha. concinna. Hy. rufipes showed the highest degree of sequence identity to an Ethiopian hybrid of the same tick species. Based on both COI and 16S rDNA gene analyses, two genetic lineages of I. frontalis were recognized (with only 91.4 % identity in their partial COI gene). These were highly similar to South-Western European isolates of the same tick species. Phylogenetic analysis of Ha. concinna specimens collected from birds in Hungary also revealed two genetic lineages, one of which showed high (≥99 %) degree of 16S rDNA sequence identity to conspecific East Asian isolates.
Conclusions
Two genetic lineages of I. frontalis and Ha. concinna are transported by birds in Central Europe, which reflect a high degree of sequence identity to South-Western European and East Asian isolates of the same tick species, respectively. In addition, I. festai was collected for the first time in Hungary. These findings highlight the importance of western and eastern migratory connections by birds (in addition to the southern direction), which are also relevant to the epidemiology of tick-borne diseases.
Similar content being viewed by others
Background
The epidemiological role of birds has been increasingly recognized. They are carriers of important viruses, bacteria and parasites, some of which may pose a risk to humans and domestic or game/wild animals [1]. In Europe, similarly to other parts of the globe, migratory birds play an important role in short- and long-distance transportation of ixodid ticks (Acari: Ixodidae) and tick-borne pathogens [1]. Non-migrating bird species contribute significantly to the local tick populations, as they are preferred hosts of immature stages (larvae and nymphs) of several tick species of medical and veterinary importance, such as Ixodes ricinus [2] and Haemaphysalis concinna [3]. Since numerous exotic and local tick species are transported by birds and may infest humans, avian hosts may contribute to zoonotic pathogen transmission, particularly in urban habitats [4]. Furthermore, birds may harbour ornithophilic tick species, such as I. frontalis and I. arboricola [5], which may be relevant to the transmission of pathogens within or between bird populations [6].
Accordingly, most molecular studies focus on the detection of pathogens that are associated with bird ticks [1], or on avian hosts as potential pathogen reservoirs [7]. Reports on the molecular taxonomic comparison of birds ticks are rare (e.g. [5]), particularly in a geographical context [8].
In the eastern part of Central Europe (including Poland, Slovakia and Hungary) birds were reported to harbour exotic tick species [4, 9, 10]. It has also been shown that tick-carrier birds are important as hosts of local/indigenous tick species in the region [11, 12]. The large scale survey of this study aimed to extend the scope of these previous works by providing comprehensive information on the species and genetic diversity of ixodid ticks transported by migratory and non-migratory bird species in Central Europe, while evaluating relevant data in a geographical, as well as in an ecological context.
Methods
Sample collection
During a three year period (from January 2012 until December 2014) ixodid ticks were collected from passerine birds at three ringing stations in Hungary (Ócsa: 47.2967°, 19.2101°; Fenékpuszta: 46.7088°, 17.2427°; Bódva-völgy: 48.2934°, 20.7385°). Birds were captured by standard Ecotone mist-nets (Gdynia, Poland), 12 m in length, 2.5 m in height and with 16 mm mesh as described [7]. All captured birds were examined for the presence of ticks, which were removed with fine tweezers and put into 70 % ethanol (for storage) in separate vials according to their hosts. Morphological identification was done with a stereo microscope (SMZ-2 T, Nikon Instruments, Japan, illuminated with model 5000-1, Intralux, Switzerland) according to standard keys [13, 14]. Characteristics (feeding site preference, migration distance and weight) were assigned to bird species based on ornithological observations [15].
Molecular analyses
The DNA was extracted from 46 specimens of I. frontalis, 12 larvae/nymphs of Ha. concinna, and one Hyalomma nymph, as well as from one hind leg of two I. festai and one I. lividus females as described [7].
The cytochrome oxidase subunit I (COI) gene was chosen as the first target for molecular analysis, on account of its suitability as a DNA-barcode sequence for tick species identification. The PCR was modified from Folmer et al. [16] and amplifies a 710 bp fragment of the gene. The primers HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) and LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) were used in a reaction volume of 25 μl, containing 1 U (0.2 μl) HotStarTaq Plus DNA polymerase, 2.5 μl 10× CoralLoad Reaction buffer (including 15 mM MgCl2), 0.5 μl PCR nucleotide Mix (0.2 mM each), 0.5 μl (1 μM final concentration) of each primer, 15.8 μl ddH2O and 5 μl template DNA. For amplification, an initial denaturation step at 95 °C for 5 min was followed by 40 cycles of denaturation at 94 °C for 40 s, annealing at 48 °C for 1 min and extension at 72 °C for 1 min. Final extension was performed at 72 °C for 10 min.
Another PCR [17] was used to amplify a 460 bp fragment of the 16S rDNA gene from one sample among those that yielded the same COI genotype, with the primers 16S + 1 (5′-CTG CTC AAT GAT TTT TTA AAT TGC TGT GG-3′) and 16S-1 (5′-CCG GTC TGA ACT CAG ATC AAG T-3′). Other reaction components, as well as cycling conditions were the same as above, except for annealing at 51 °C.
PCR products were electrophoresed in a 1.5 % agarose gel (100 V, 60 min), stained with ethidium-bromide and visualized under ultra-violet light. Purification and sequencing was done by Biomi Inc. (Gödöllő, Hungary). Sequences were submitted to GenBank (Table 1). The phylogenetic analyses were conducted with the Tamura-Nei model and Maximum Composite Likelihood method by using MEGA version 5.2 as reported [18].
Ethical approval
The study was carried out according to the national animal welfare regulations (28/1998). Licence for bird ringing was provided by the National Inspectorate for Environment and Nature (No 14/3858-9/2012.).
Statistical analysis
Exact confidence intervals (CI) for the prevalence rates were calculated at a 95 % level. Sample prevalence data were analyzed by Fisher’s exact test. Mean values for the intensity of tick infestation (number of all ticks collected from a bird species, divided by the number of all tick-infested individuals of the same bird species) were compared between bird categories by Mann-Whitney U-Test. Differences were considered significant when P < 0.05.
Results
Tick-infestation of birds according to tick and bird species
In the period between 2012-2014, altogether 3339 ixodid ticks were collected from 1167 passerine birds (representatives of 47 species). The most abundant tick species found were I. ricinus and Ha. concinna, with 2296 (68.8 %, CI: 67.2–70.3 %) and 989 (29.6 %, CI: 28.1–31.2 %) specimens (only larvae and nymphs), respectively. The presence of I. ricinus on birds was noted between March and November, and that of Ha. concinna (both larvae and nymphs) from March to October.
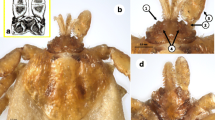
Forty-eight I. frontalis specimens (including three adults) were also collected (Fig. 1a), with a majority (79.2 %, CI: 65–89.5 %) from Robins (Erithacus rubecula) (Table 2). This tick species occurred during all seasons (August to November and January to April), however, most specimens were collected in springtime (Table 3).
Morphology of tick species identified in the relevant stage for the first time in Hungary. a: I. frontalis nymph showing parallel sides of palps and “frons” (arrows); b: Hy. rufipes nymph with broadly rounded posterior margin of the scutum (arrow) and elongated spiracular plate (insert); c: I. festai female, dorsal view – the scutum with deep punctuations and few long hairs, distinct cornuae on the basis capituli (arrows); d: I. festai female, ventral view – broad auriculae curved backwards, long internal spur on coxa I (arrows)
Regarding exotic tick species, three Hyalomma nymphs, which resembled Hy. rufipes based on the spiracular plates and the scutum (Fig. 1b), were found on a Common Whitethroat (Sylvia communis) in May (2014).
In addition, two I. festai females, identified on morphological bases (Fig. 1c, d), were removed from a Greenfinch (Carduelis chloris) and a Dunnock (Prunella modularis) in March (2014). One I. lividus female was collected from a Sand Martin (Riparia riparia) in July (2014).
Among the most important tick-infested bird species in this survey (Table 2), the majority of I. ricinus and I. frontalis larvae/nymphs (1756 of 2239: 78.4 %, CI: 76.7–80.1 % and 44 of 48: 91.7 %, CI: 80–97.7 %, respectively) occurred on ground-feeding bird species, whereas 73.1 % of Ha. concinna immatures (705 of 964, CI: 70.2–75.9 %) were found on arboreal birds, reflecting a highly significant difference (Fisher’s exact test: P < 0.0001). On the other hand, the intensity of tick infestation (Table 2) had no significant association with bird species of smaller (6–30 g) or larger (31–140 g) body weight, or with bird species that have long vs. short distance (or no) migration (Mann-Whitney U-Test: P > 0.05).
Molecular taxonomic analysis of bird ticks in a geographical context
Ixodes frontalis
Among the 46 I. frontalis specimens for which part of the COI gene was sequenced, two genetic lineages (each containing nine genotypes) were clearly recognizable (“A”: KU170492-500, and “B”: KU170501-9) and the separation of these lineages had high bootstrap support on the phylogenetic tree (Fig. 2). The relevant genotypes had 1-2 nucleotide differences within lineage “A” and 1-4 within lineage “B”, but up to 56 nucleotide difference (598/654 bp, i.e. only 91.4 % identity) between the two lineages. The subsequent 16S rDNA gene analysis included DNA samples of each different COI genotype, but revealed only two distinct genetic variants (KU170518: genotype A-Hu16S, KU170519: genotype B-Hu16S), which showed a 4 bp difference, i.e. 99 % (402/406 bp) identity. These two 16S rDNA genotypes had 100 % sequence identity to South-Western European isolates (KP769863 and KP769862, respectively, from Azores). The phylogenetic analyses of 16S rDNA sequences confirmed the separation of two I. frontalis lineages (Fig. 3). The isolation sources of I. frontalis genotypes are shown in Table 3.
Ixodes festai
In the case of I. festai the sequencing of the amplified part of the COI gene was not successful. The 16S rDNA sequences of the two specimens (KU170521-2) differed in three nucleotides (373/376 bp, i.e. 99.2 % identity), but clustered together on the phylogenetic tree (Fig. 3).
Ixodes lividus
The COI sequence of I. lividus obtained in this study (KU170510) had 100 % identity with an isolate of the same tick species from the UK (GU124743). The partial 16S rDNA sequence of the Hungarian specimen (KU170520) had 99.7 % (398/399 bp) identity with another isolate from Western Europe, Belgium.
Hyalomma rufipes
The partial COI sequence of one Hyalomma nymph (KU170491) showed the highest (645/649 bp, i.e. 99.4 %) degree of identity to a Hy. rufipes × Hy. dromedarii hybrid from Ethiopia (AJ437079), whereas a 99.2 % (644/649 bp) identity to Hy. marginatum (AJ437091). Based on the partial sequence of its 16S rDNA gene (KU170517), this specimen showed the highest degree of identity to Hy. rufipes (405/406 bp, i.e. 99.8 % identity to L34307, and only 403/407 bp: 99 % identity to KP776645, Hy. marginatum).
Haemaphysalis concinna
Among the molecularly analysed 12 Ha. concinna specimens six different COI genotypes were found (KU170511-6), with a difference in up to eight nucleotides (meaning 622/630, i.e. 98.7 % identity). The six COI genotypes clustered in two lineages (supported by high bootstrap value) on the phylogenetic tree (Fig. 2). These COI genotypes represented three 16S rDNA genotypes (KU170523-5), with 1–2 bp differences. The 16S rDNA phylogenetic analysis confirmed the separation of genotype Hu167 (encompassing COI genotypes Hu1-3) from the others (Hu168-9) (Fig. 3). The latter (e.g. KU170524) showed high (99.7 %, i.e. 387/388 bp) degree of identity to Far Eastern isolates of Ha. concinna (from Japan: e.g. AB819171) and another from East Siberia (KP866207: 384/387 bp, 99.2 % identity), with which they clustered together on the phylogenetic tree (Fig. 3). The isolation sources (bird species) and seasonality of COI and 16S rDNA genotypes of Ha. concinna are shown in Table 3.
Discussion
In the present study molecular evidence is provided for the first time on the transportation of immature stages of Hy. rufipes by birds in Central Europe. In addition, I. festai is reported for the first time in Hungary. While two females of I. frontalis (infesting birds) had been reported in Hungary more than half a century ago [19], the present results attest that this tick species is transported by birds regularly in the region. Interestingly, another ornithophilic tick species, I. arboricola was not found in this survey, although its preferred hosts (i.e. cavity-nesting bird species) were included in the study, and it has formerly been reported in Hungary [13] as well as in neighbouring countries (Slovakia: [6]; Romania: [20]).
The seasonal presence of I. ricinus on birds coincided with the reported questing activity of this tick species in Hungary [21]. In contrast to this, Ha. concinna larvae or nymphs were found two or one months earlier (respectively) on avian hosts (i.e. from March), compared to the initiation of their known seasonal activity in the region [3].
The presence of I. ricinus and I. frontalis larvae/nymphs showed a significant association with ground-feeding bird species (demonstrated here for the first time in case of I. frontalis), whereas Ha. concinna larvae and nymphs occurred significantly more frequently on arboreal birds. The latter finding can be explained by the relatively high questing height of Ha. concinna larvae and nymphs on the vegetation, as an adaptation to the size of their preferred host species (roe deer) in the region [22].
In the present study the great majority of I. frontalis specimens were collected from Robins. This bird species is known to have predominantly south-west to north-east spring migration from the Mediterranean region to Hungary [23]. Therefore, the present data suggest that I. frontalis is mainly transported from South-Western Europe to Central Europe. In support of this migratory connection, the 16S rDNA genotypes reported here were 100 % identical to corresponding sequences from the Azores, and the spring predominance of I. frontalis larvae and nymphs on birds in Hungary (as observed in the present study) followed the late winter seasonal peak reported in Portugal [24].
Based on the analysis of two genetic markers, the present data clearly indicate that two distinct genetic lineages of I. frontalis are transported by birds in Central Europe. The separation of these lineages is supported by high bootstrap values on the COI and 16S rDNA phylogenetic trees (Figs. 2, 3). Interestingly, the degree of COI sequence divergence between the two lineages (9 %) exceeds the proposed approximated sequence difference as species boundary for ticks (6.1 % of COI gene: [25]).
Previously, I. festai was not reported from Hungary. In the present study this tick species was found on a Greenfinch and a Dunnock. Both bird species migrate for wintering to the Mediterranean Basin, in the direction of Italy [15], where I. festai is known to occur [26].
The tick I. lividus is host specific for the Sand Martin (Riparia riparia). The sequence identity between the isolates of this tick species from Hungary (reported here) and the UK [27] indicates that the same genotype is present in Western and Central European populations of I. lividus, despite the fact that no direct migration of Sand Martins is known between Hungary and the UK [15].
In the present study Hyalomma nymphs were removed from a Common Whitethroat. Populations of this bird species, which breed in Central Europe, are known to migrate to sub-Saharan Africa for wintering [15]. Morphologically all three ticks resembled Hy. rufipes, and one of them showed the closest identity in its partial COI gene to an Ethiopian Hy. rufipes hybrid. The GenBank reference strains used for comparison in this context were reliably identified according to taxonomic keys [28]. In general, larvae and nymphs of the Hy. marginatum complex are known to be transported by birds to both Western and Central Europe from the south [10, 29]. Hy. rufipes in particular, was reported on birds in Northern Europe [30]. In Europe, birds carrying Hy. rufipes most likely arrive from the Middle East (which is a stopover region along the Black Sea – Mediterranean flyway), where larvae and nymphs of this tick species predominate on birds [31, 32]. Importantly, even under the continental climate in Central Europe nymphs of Hy. rufipes are able to moult to adults, previously reported to infest cattle in Hungary [33].
Based on the present results Ha. concinna larvae/nymphs carried by song birds in Central Europe have a high degree of 16S rDNA gene identity with conspecific ticks from East Siberia [34] and the Far East, Japan [35]. Although the reasons for this close identity can be multiple, tick exchange between Central Europe and East Asia via migratory birds may significantly contribute to it. The East Atlantic and the Black Sea – Mediterranean flyways connect Europe to Asia (the latter being more relevant to the study area). Migratory birds wintering in Western Europe and those having spring migration towards Russian Far East may have overlapping breeding grounds [36]. In this survey Ha. concinna genotypes, highly similar to East Asian isolates, were only collected during spring and autumn bird migration, i.e. from Robins, a Blackbird (Turdus merula) and a Dunnock (Table 2). These bird species have eastern migratory connections, i.e. towards Eastern Europe and Asia [15, 37].
Conclusions
Two genetic lineages of I. frontalis and Ha. concinna are transported by birds in Central Europe, which reflect a high degree of sequence identity to South-Western European and East Asian isolates of the same tick species, respectively. In addition, I. festai was collected for the first time in Hungary. These findings highlight the importance of western and eastern migratory connections by birds (in addition to the southern direction), which are also relevant to the epidemiology of tick-borne diseases.
References
Hasle G. Transport of ixodid ticks and tick-borne pathogens by migratory birds. Front Cell Infect Microbiol. 2013;3:48.
Gray JS. The ecology of ticks transmitting Lyme borreliosis. Exp Appl Acarol. 1998;22:249–58.
Nosek J. The ecology, bionomics and behaviour of H aemaphysalis (Haemaphysalis) concinna tick. Zentralbl Parasitenknd. 1971;36:233–41.
Hornok S, Csörgő T, de la Fuente J, Gyuranecz M, Privigyei C, Meli ML, et al. Synanthropic birdsassociated with high prevalence of tick-borne rickettsiae and with the first detection ofRickettsia aeschlimannii in Hungary. Vector Borne Zoonotic Dis. 2013;13:77–83.
Heylen D, De Coninck E, Jansen F, Madder M. Differential diagnosis of three common Ixodes spp. ticks infesting songbirds of Western Europe: Ixodes arboricola, I. frontalis and I. ricinus. Ticks Tick Borne Dis. 2014;5:693–700.
Novakova M, Bulkova A, Costa FB, Kristin A, Krist M, Krause F, et al. Molecular characterization of ‘Candidatus Rickettsia vini’ in Ixodes arboricola from the Czech Republic and Slovakia. Ticks Tick Borne Dis. 2015;6:330–3.
Hornok S, Kováts D, Csörgő T, Meli ML, Gönczi E, Hadnagy Z, et al. Birds as potential reservoirs of tick-borne pathogens: first evidence of bacteraemia with Rickettsia helvetica. Parasit Vectors. 2014;7:128.
Gylfe A, Yabuki M, Drotz M, Bergström S, Fukunaga M, Olsen B. Phylogeographic relationships of Ixodes uriae (Acari: Ixodidae) and their significance to transequatorial dispersal of Borrelia garinii. Hereditas. 2001;134:195–9.
Nowak-Chmura M. Ixodes eldaricus Djaparidze, 1950 (Ixodidae) on migrating birds – Reported first time in Poland. Vet Parasitol. 2012;186:399–402.
Capek M, Literak I, Kocianova E, Sychra O, Najer T, Trnka A, et al. Ticks of the Hyalomma marginatum complex transported by migratory birds into Central Europe. Ticks Tick Borne Dis. 2014;5:489–93.
Spitalská E, Literák I, Sparagano OA, Golovchenko M, Kocianová E. Ticks (Ixodidae) from passerine birds in the Carpathian region. Wien Klin Wochenschr. 2006;118:759–64.
Nowak-Chmura M, Siuda K, Wegner Z, Piksa K. Species diversity of ticks (Acari:Ixodida) on migrating birds on the Baltic Sea coast of Poland. Zool Stud. 2012;51:1411–7.
Babos S. Kullancsok − Ixodidea. Fauna Hungariae. 1965;18:1–38 [In Hungarian].
Apanaskevich DA, Horak IG. The genus Hyalomma Koch, 1844.V. Re-evaluation of the taxonomic rank of taxa comprising the H. (Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int J Acarol. 2008;34:13–42.
Csörgő T, Karcza Z, Halmos G, Gyurácz J, Magyar G, Szép T, et al. Hungarian Bird Migration Atlas. Budapest: Kossuth Publishing Zrt; 2009 [In Hungarian with English summary].
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mel Marine Biol Biot. 1994;3:294–9.
Black WC, Piesman J. Phylogeny of hard and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16 s rDNA sequences. Proc Natl Acad Sci U S A. 1994;91:10034–8.
Hornok S, Estók P, Kováts D, Flaisz B, Takács N, Szőke K, et al. Screening of bat faeces for arthropod-borne apicomplexan protozoa: Babesia canis and Besnoitia besnoiti-like sequences from Chiroptera. Parasit Vectors. 2015;8:441.
Janisch M. Geographical distribution of tick species in Hungary. Állatt Közl. 1959;47:103–10 [In Hungarian].
Mihalca AD, Dumitrache MO, Magdaş C, Gherman CM, Domşa C, Mircean V, et al. Synopsis of the hard ticks (Acari: Ixodidae) of Romania with update on host associations and geographical distribution. Exp Appl Acarol. 2012;58:183–206.
Hornok S. Allochronic seasonal peak activities of Dermacentor and Haemaphysalis spp. under continental climate in Hungary. Vet Parasitol. 2009;163:366–9.
Hornok S, Horváth G, Jongejan F, Farkas R. Ixodid ticks on ruminants, with on-host initiated moulting (apolysis) of Ixodes, Haemaphysalis and Dermacentor larvae. Vet Parasitol. 2012;187:350–3.
Hornok S, Karcza Z, Csörgő T. Birds as disseminators of ixodid ticks and tick-borne pathogens: note on the relevance to migratory routes. Ornis Hung. 2012;20:86–9.
Norte AC, da Silva LP, Tenreiro PJ, Felgueiras MS, Araújo PM, Lopes PB, et al. Patterns of tick infestation and their Borrelia burgdorferi s.l. infection in wild birds in Portugal. Ticks Tick Borne Dis. 2015;6:743–50.
Lv J, Wu S, Zhang Y, Chen Y, Feng C, Yuan X, et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasit Vectors. 2014;7:93.
Contini C, Palmas C, Seu V, Stancampiano L, Usai F. Redescription of the male of Ixodes festai Rondelli, 1926 (Ixodida: Ixodidae) on specimens from Sardinia (Italy). Parasite. 2011;18:235–40.
Graham RI, Mainwaring MC, Du Feu R. Detection of spotted fever group Rickettsia spp. from bird ticks in the U.K. Med Vet Entomol. 2010;24:340–3.
Rees DJ, Dioli M, Kirkendall LR. Molecules and morphology: evidence for cryptic hybridization in African Hyalomma (Acari: Ixodidae). Mol Phylogenet Evol. 2003;27:131–42.
Jameson LJ, Morgan PJ, Medlock JM, Watola G, Vaux AG. Importation of Hyalomma marginatum, vector of Crimean-Congo haemorrhagic fever virus, into the United Kingdom by migratory birds. Ticks Tick Borne Dis. 2012;3:95–9.
Hasle G, Bjune G, Edvardsen E, Jakobsen C, Linnehol B, Røer JE, et al. Transport of ticks by migratory passerine birds to Norway. J Parasitol. 2009;95:1342–51.
Hoogstraal H, Kaiser MN. Observation on Egyptian Hyalomma ticks (Ixodoidea, Ixodidae). 2.Parasitism of migrating birds by immature H. rufipes Koch. Ann Entomol Soc Am. 1958;51:12–6.
Fain A, Roggeman W, Symens P, Al Suhaibani A. Notes on a small collection of immature Hyalomma ticks from migrating birds during their Spring migration in Karan Island, Persian Gulf, Saudi Arabia. Bull Inst Roy Sci Natur Belg Entomol. 1995;65:101–3.
Hornok S, Horváth G. First report of adult Hyalomma marginatum rufipes (vector of Crimean-Congo haemorrhagic fever virus) on cattle under a continental climate in Hungary. Parasit Vectors. 2012;5:170.
Khasnatinov MA, Liapunov AV, Manzarova EL, Kulakova NV, Petrova IV, Danchinova GA. The diversity and prevalence of hard ticks attacking human hosts in Eastern Siberia (Russian Federation) with first description of invasion of non-endemic tick species. Parasitol Res. 2016;115:501–10.
Takano A, Fujita H, Kadosaka T, Takahashi M, Yamauchi T, Ishiguro F, et al. Construction of a DNA database for ticks collected in Japan: application of molecular identification based on the mitochondrial 16S rDNA gene. Eisei Dobutsu. 2014;65:13–21.
Dobrynina DV, Kharitonov SP. The Russian waterbird migration atlas: temporal variation in migration routes. In: Boere GC, Galbraith CA, Stroud DA, editors. Waterbirds around the world. Edinbrugh: The Stationary Office; 2006. p. 582–9.
Collar N. Common Blackbird (Turdus merula). In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E, editors. Handbook of the Birds of the WorldAlive. Barcelona: Lynx Edicions; 2014. Retrieved from http://www.hbw.com/node/58261 on1 December 2015.
Acknowledgements
Part of the study was funded by OTKA 115854. The authors would like to thank the contributions of Maria Mulvihill (Ireland) to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
SH initiated the study, participated in tick identification, supervised parasitological work, wrote the manuscript. BF identified ticks, participated in tick collection, extracted the DNA. NT and JK performed molecular and phylogenetic analyses, respectively. CT and DK collected ticks, supervised ornithological work and contributed relevant information to the manuscript. ÁC, BRJ participated in tick collection. All authors read and approved the final version of the manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hornok, S., Flaisz, B., Takács, N. et al. Bird ticks in Hungary reflect western, southern, eastern flyway connections and two genetic lineages of Ixodes frontalis and Haemaphysalis concinna . Parasites Vectors 9, 101 (2016). https://doi.org/10.1186/s13071-016-1365-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1365-0