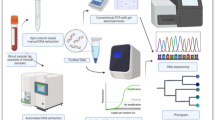
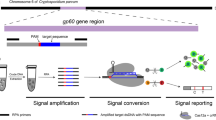
Abstract
Background
Toxoplasma gondii is an opportunistic protozoan that is ubiquitous in humans and animals. It can invade any human organ and cause severe diseases, including toxoplasma ophthalmopathy, meningoencephalitis, and liver necrosis. Porcine toxoplasmosis is prevalent in China. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and Cas (CRISPR-Associated Protein) systems are widely used for gene editing and pathogen detection. CRISPR-based diagnostics are molecular assays that have been developed to detect parasites with high sensitivity and specificity.
Methods
This study aimed to establish a combined CRISPR/Cas12a and RPA rapid detection method for T. gondii by targeting the B1 gene and 529 bp repeat element (529 RE). The detection results could be visualized by the fluorescence or lateral flow strips (LFS). The sensitivity and specificity of the method were evaluated, and T. gondii-infected mouse blood was used for detection.
Results
The results indicated that the established method for T. gondii detection was satisfactory, with a detection limit of 1.5 cp/μl for the two loci. Moreover, the B1 gene could detect 1 tachyzoite per reaction, and the 529 RE could detect 0.1 tachyzoite per reaction, consistently with the highly sensitive nested polymerase chain reaction (PCR) results. The method was suitable for strains, including RH, and did not cross-react with other protozoa DNA with similar habits. The T. gondii-infected mouse blood samples were all positive for T. gondii at 1, 3, and 5 days post infection (dpi).
Conclusions
This study established a rapid, sensitive, and time-saving DNA detection method for T. gondii that has the potential to be an alternative tool for T. gondii detection in the field.
Graphical abstract

Similar content being viewed by others
Introduction
Toxoplasmosis is a human-animal disease caused by a specialized intracellular parasite, Toxoplasma gondii, with a worldwide distribution [1]. Toxoplasma gondii gondii can infect all warm-blooded animals, including humans [2]. In immunocompetent individuals, infection with T. gondii can cause flu-like symptoms that persist for weeks to months before resolving. However, immunocompromised individuals, such as those with acquired immune deficiency syndrome (AIDS), the elderly, and those taking immunosuppressive drugs, are at a higher risk of severe illness and even death following T. gondii infection [3]. Pregnant women infected with T. gondii are also at risk of congenital toxoplasmosis in the fetus, which can lead to a range of adverse outcomes such as abortion, malformation, or stillbirth [4]. The serological positive rate of T. gondii in humans in some regions of China has been reported to be as high as 23.41%, indicating that the infection is widespread in the country [5]. In addition to its impact on human health, T. gondii has a significant impact on animal husbandry, with studies showing that the global seroprevalence of T. gondii in pigs is 19% [6]. Infection with T. gondii in pigs can exhibit a range of symptoms, including missed fever, diarrhea, pulmonary edema, and abortion in sows. Given that pork is a primary source of meat for humans, porcine toxoplasmosis increases the possibility of human T. gondii infection. Prevention and screening of T. gondii are becoming increasingly important. However, the lack of specific clinical symptoms of toxoplasmosis poses a challenge for diagnosis, highlighting the need for the development of more effective T. gondii detection methods.
Toxoplasma gondii gondii detection can be achieved through a variety of methods, including etiological, immunological, and molecular techniques. Etiological methods, such as peritoneal fluid smears and histological section staining, are time-consuming and have a high false-negative rate. Immunological tests, such as enzyme-linked immunosorbent assay (ELISA), are the most common methods for detecting toxoplasmosis. While ELISA is simple and rapid, patients in the window period of infection may be missedm and there are various interfering factors, such as rheumatoid factor [7]. Furthermore, patients with chronic lymphocytic leukemia (CLL) and secondary hypogammaglobulinemia may produce false-negative results due to their inability to produce IgG antibodies [8]. In molecular biology, PCR technology is the most widely used. Burg et al. successfully used PCR technology to detect T. gondii DNA from blood using B1 gene as a target gene in the twentieth century [9]. The use of conserved genes, such as the P30 gene and the SAG1 gene, has been successful in detecting T. gondii [10, 11]. Quantitative PCR (Q-PCR) is a useful derivative of PCR that can be used for both qualitative and quantitative analysis. However, Q-PCR requires expensive and precise instruments, limiting its use to the laboratory, making it unsuitable for field applications.
In addition to the high specificity, high sensitivity, and short detection window, the ideal pathogen detection should also ensure the portability and low cost of detection equipment. In recent years, the emergence of isothermal amplification techniques such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) [12, 13], as well as the discovery of Cas12a, Cas13a, and Cas14 in the CRISPR/Cas family [14,15,16], have provided viable approaches for point-of-care testing (POCT). RPA can achieve exponential amplification of template DNA in a short time and conventional temperature environment and eliminate the dependence on PCR instrument. The CRISPR/Cas system has shown potential in pathogen detection, with the Cas12a system able to non-specifically cleave other single-stranded DNA in the system after specific cleavage of double-stranded DNA by the crRNA targeting the target sequence [17]. In 2018, Doudna et al. established the Cas12a-based DNA endonuclease-targeted CRISPR trans-reporter system (DETECTR), which has been used to rapidly detect pathogens such as SARS-CoV-2 and African swine fever virus (ASFV) [14, 18, 19]. Moreover, CRISPR-based detection methods have been applied to the detection of Cryptosporidium parvum and Plasmodium [20,21,22], indicating the broad potential of CRISPR in parasite detection.
The B1 gene of T. gondii, with 35 copies in the parasite's genomic DNA, is now considered the most used gene in the field of T. gondii nucleic acid detection [7, 23, 24]. Similarly, the 529 RE gene fragment, with a repetition rate ranging from 200 to 300 copies in the T. gondii genome, is thought to contribute to the improved sensitivity and specificity of the detection [25]. In this study, we have developed a molecular detection method for T. gondii B1 gene and 529 RE using the newly developed DETECTR technology and compared its efficacy with that of the nested PCR approach. The established method was used to detect the blood samples of host animals. We demonstrate that our method is characterized by a high level of simplicity, speed, and accuracy and is for point-of-care testing (POCT), using of simple visualization techniques like placing the test tube in a dark box under UV light in an outdoor environment or application of a lateral flow strip (LFS) for direct naked-eye readout of results.
Materials and methods
Mice and parasites
The strains RH, Pru, wh3, and wh6, utilized in the present investigation, were generously provided by the Anhui Key Laboratory of Zoonoses, Anhui Medical University. The DNA of C. parvum and Babesia were procured from Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Furthermore, Plasmodium was obtained from Chao Zhang of the Department of Pathogen Biology, Anhui Medical University. Six- to eight-week-old female BALB/c mice were purchased from the Animal Experiment Center of Anhui Medical University.
Protein expression and purification
Cas12a plasmid (pMBP-LbCas12a) was purchased from Addgene (Plasmid #113431). Expression vectors containing N-terminal 10 × His-tag, MBP, and TEV protease cleavage sites were transformed into receptor cells Escherichia coli BL21 (DE3). Cells were cultured in LB medium at 37 ℃ until logarithmic growth phase; 8 ml cells was added to 400 ml LB medium at 16 ℃, 200 rpm, and incubated overnight. Then cells were collected and centrifuged. The supernatant was discarded and resuspended in lysis buffer (50 mM Tris–HCl, 500 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, 0.5 mM PMSF, and 0.25 mg/ml lysozyme, pH 7.5) and lysed by ultrasonication. The precipitate was removed by centrifugation with 10,000 rcf for 1 h at 4 °C. The protein supernatant was filtered after centrifugation using a 0.22-μm bacterial filter. The filtrate was added to a Ni–NTA purification resin pre-loaded column (Sangon Biotech, Shanghai, China), linear elution with buffer. After holding overnight, TEV cleavage at 4 °C was carried out to remove the 10 × His-MBP tag, and the sample was loaded into a heparin column (GE, Hi-Trap) for further purification to obtain Cas12a protein. The buffer was replaced with the final storage solution for Cas12a (20 mM Tris–HCl, 2.0 M NaCl, pH 8.5) for protein concentration by Amicon® Ultra-4 centrifugal filtration device (Millipore, China). The BCA method was used for concentration determination, identification was made using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Additional file 1: Fig. S1), with storage at − 20 °C.
Design and preparation of crRNA
Conserved and specific T. gondii genome sequences were identified using the National Center of Biotechnology Information (NCBI), and the final selection was the T. gondii B1 gene (GenBank: AF179871) and 529 RE (GenBank: AF146527) as the target genes. After 66 nt ssDNA oligonucleotide was synthesized by the company (Additional file 1: Table S1), ssDNA oligonucleotide was transcribed into ssRNA in vitro using HiScribeT7 Quick High Yield RNA Synthesis Kit (NEB, USA). The transcription system was 10 μl NTP buffer mix, 2 μl T7 RNA polymerase mix, and 1 ug ssDNA oligonucleotide, and 20 μl was reached with RNase-free water. Incubation was carried out at 37 ℃ for 7 h. After incubation, 2 μl DNaseI was added, and water was added to reach a volume of 50 μl. The DNA template was removed by incubation at 37 ℃ for 15 min. The resulting crRNA was obtained by RNA purification using Monarch® RNA Cleanup Kit (NEB, USA). Attention should be paid to prevent RNase contamination during all operations. The purified product was used for concentration determination and dispensed into a − 80 ℃ cryogenic refrigerator for storage before use.
DNA extraction, recombinant plasmid constructs, and tachyzoite DNA dilution
The suspension containing the tachyzoites of T. gondii RH was centrifuged at 2000 rpm for 5 min, and the supernatant was discarded. Toxoplasma gondii DNA was extracted using the TIANamp Genomic DNA Kit (Tiangen, China). Target genes were amplified by PCR primers (Additional file 1: Table S2) and purified with a DNA purification kit (Axygen, China). The purification product was inserted into T-Vector pMD19 vector by T-A cloning. After transformation to the DH5α-receptive state, extraction was performed with the Axygen Plasmid Small Volume DNA Extraction Kit (Axygen, China). Concentration was measured by NanoDrop 2000 (Thermo Fisher, USA), recorded, and then stored at − 20 °C.
DNA was extracted after counting 106 T. gondii tachyzoites with a cell counting plate, and genomic DNA was diluted doubling with ddH2O to contain 105 tachyzoite DNA per tube to 10–1 tachyzoite DNA for subsequent testing.
DETECTR assays
The RPA Nucleic Acid Amplification Kit (Hangzhou ZC, China) was used for the isothermal amplification of nucleic acids. The total reaction system for amplification was 50 μl; 41.5 μl Buffer A solution, 2 μl (10 μM) each forward and reverse primers (Additional file 1: Table S3), and 2 μl template DNA were added to an RPA unit tube containing enzyme lyophilized powder; 2.5 μl Buffer B solution was added to its cap. The cap was covered, inverted, and mixed 5–6 times. The reaction tube was bathed in 37 ℃ water for 30 min to obtain an amplification product that could be used for subsequent CRISPR assays.
The metal PCR tube rack required for CRISPR reactions was precooled in advance in a − 80 °C refrigerator, after which the following system was prepared on the cooled PCR tube rack: 1 μM LbCas12a, 2.5 μM crRNA, and 2 μl 10 × NEB buffer 2.1, and finally 20 μl was made up with RNase-free water. Ten minutes incubation at 37 °C was followed by the addition of 0.5 μM DNA fluorescent probe (5'-FAM-TTATTATT-Bio-3'). Taking 18 μl of the above reaction system, 2 μl of RPA amplification product was added to it, and the fluorescence was monitored at the FAM channel using BioRad CFX96 to read the fluorescence value.
Lateral flow strips
Aspirated 2 μl of the completed pre-amplified sample was added to the CRISPR/Cas12a cleavage reaction system; 20 μl of the reaction system was incubated for 1 h at 37 °C. Then, 80 μl of 1 × PBS was added and mixed well; 100 μl was aspirated to the sample pad and the results observed after 5–10 min at room temperature.
Nested PCR
The nested PCR primers (Additional file 1: Table S4) were used as reported in the article by Shirzad Fallahi et al. [26]. The length of the nested PCR product was 194 bp for the B1 gene and 164 bp for the 529 RE. The first round of PCR reaction system was 50 µl, containing 10 µl 5 × PrimeSTAR buffer, 4 µl dNTP Mix, 1 µl (10 µM) each of upstream and downstream primers, 1 µl DNA template, 1 µl Taq DNA polymerase, and 50 µl made up with ddH2O.
The amplification procedure was as follows: initial denaturation was done at 95 °C for 5 min, followed by 34 cycles of denaturation (10 s at 98 °C), annealing (15 s at 56 °C), and extension (60 s/kb at 72 °C), at the end of the cycle extended at 72 °C for 5 min. The products of the first round were added to the system of the second round as the template of the second round, and the procedure was the same as in the first round. The products were visualized by agarose gel electrophoresis.
Establishment of mouse model of T. gondii infection
Counting was performed using a cell counting plate. Each mouse was injected with 1000 tachyzoites, and the control group was injected with the same volume of saline. Five mice from each group were selected for eye blood sampling on 1, 3, and 5 dpi, and control mice were dissected and killed on the last day. The blood needed to be anticoagulated, after which 200 µl of anticoagulated blood was taken for testing, using samples from the saline group as a negative control. The use of animals was approved by the Experimental Animal Ethics Committee of Anhui Medical University.
Results
Design and optimization of DETECTR detection system for T. gondii
In this study, we developed a rapid detection method for T. gondii infection by combining RPA and CRISPR/Cas12a (Fig. 1a). The method involves three simple steps: nucleic acid extraction, RPA amplification, and CRISPR/Cas12a specific detection. To increase the sensitivity of the detection method for T. gondii, we conducted a comparison of targets, screening of primers, and system optimization. For the B1 and 529 RE targets, we designed three crRNAs for each and evaluated their performance in the DETECTR system by assessing fluorescence intensity. Ultimately, we selected B1-L3 and 529 RE-L3 for detecting T. gondii (Fig. 1b). To identify the most efficient primers for RPA amplification, we designed three forward and reverse primers for each locus. We fixed the forward primers to screen out the optimal reverse primers and similarly screened out the forward primers (Fig. 1c, d). We optimized the system by adjusting the concentration of protein and crRNA and determined that the best detection effect was achieved when the protein final concentration was 5 μM and the crRNA final concentration was 10 μM. To reduce assay costs, we chose to use a Cas12a final concentration of 1 μM and a crRNA final concentration of 2.5 μM, which were employed in subsequent experiments (Fig. 1e).
Establishment of CRISPR/Cas12a combined with RPA for detection of Toxoplasma gondii. a DETECTR workflow of T. gondii. b Comparison of different crRNAs targeting B1and 529 RE by fluorescence assay. c Screening of RPA primers for B1-L3. d Screening of RPA primers for 529 RE-L3. e Optimization of the concentration of Cas12a and crRNA
DETECTR has robust sensitivity for detecting T. gondii
We evaluated the limit of detection of the DETECTR assay for T. gondii using recombinant plasmids and found that the detection sensitivity for the B1 gene and 529 RE loci was as low as 1.5 cp/μl (Fig. 2a, b). Notably, positive samples emitted strong yellow-green fluorescence under UV, which was observable by the naked eye, making this method particularly suitable for use in resource-limited settings (Fig. 2a, b).
Sensitivity of the DETECTR system: (a) B1-DETECTR and (b) 529 RE-DETECTR reaction with plasmid standards. The tube shows the fluorescence signal of the sample under UV light. 1–8 represent 3000 cp/μl, 300 cp/μl, 30 cp/μl, 3 cp/μl, 1.5 cp/μl, 0.3 cp/μl, 0.03 cp/μl, negative control. c B1-DETECTR and d529 RE-DETECTR reaction with T. gondii tachyzoite DNA. e B1-nested PCR and f 529 RE-nested PCR reaction with T. gondii tachyzoite DNA. 1–7 represent 10,000, 1000, 100, 10, 1, 0.1, 0.01 tachyzoite DNA per reaction. ****P < 0.0001; ns, not significant
We further tested the DETECTR assay using serial dilutions of tachyzoite DNA and found that the detection limit was 1 tachyzoite per reaction for B1 (Fig. 2c) and 0.1 tachyzoite per reaction for 529 RE (Fig. 2d); 529 RE was more sensitive than B1, perhaps because 529 RE has 200–300 copies in the T. gondii genome, while the B1 gene has only 35 copies. Furthermore, the DETECTR assay was found to be comparable to nested PCR as both methods were able to detect 1 tachyzoite per reaction for B1 (Fig. 2e), while 529 RE could detect 0.1 tachyzoite per reaction (Fig. 2f). These results suggested that the DETECTR assay was a sensitive and reliable method for the detection of T. gondii tachyzoite DNA, particularly when targeting the 529 RE locus.
DETECTR has robust specificity for detecting T. gondii
To investigate the universality of the DETECTR assay for T. gondii, we detected the DNA extracted from four different strains, including RH in type I, Pru in type II, and wh3 and wh6 strains of Chinese 1 genotype. The results revealed that this method was effective for all four strains, exhibiting strong fluorescence values, and there was no significant difference in the detection effect among the four strains (Fig. 3a, b).
Specificity of the DETECTR system: (a) B1-DETECTR and (b)529 RE-DETECTR reaction of DNA of four different strains of T. gondii; all four strains can be detected. NC, negative control. c B1-DETECTR and d529 RE-DETECTR reaction of four types of parasite DNA, except DNA of T. gondii, could not be detected. NC, negative control; ***P < 0.001; ****P < 0.0001; ns, not significant
In addition, we selected Plasmodium, C. parvum, and Babesia, which have similar habits or infection symptoms to T. gondii, for specific detection. The DETECTR system for T. gondii showed positivity only for T. gondii DNA, and the fluorescence values of the other three protozoa were consistent with those of the negative control group (Fig. 3c, d). This result was also evident under UV light (Additional file 1: Fig. S2). Hence, the DETECTR system demonstrated good specificity.
DETECTR and nested PCR detection of mouse blood samples
We have developed a murine model of toxoplasmosis by administering 1000 RH tachyzoites intraperitoneally to BALB/c mice. We employed the DETECTR system to perform rapid detection of T. gondii in whole blood samples collected from infected mice. Our findings demonstrated that T. gondii DNA could be successfully detected in all blood samples obtained on 1, 3, and 5 dpi, while the negative control group injected with saline produced negative results (Fig. 4a, c). Nested PCR was also performed on blood samples from mice infected with 1000 RH tachyzoites, and all samples were positive at 1, 3, and 5 dpi, while all samples in the control group were negative (Fig. 4b, d). Our study indicates that the DETECTR system is capable of detecting T. gondii infection in its early stages.
Analysis results of blood samples from Toxoplasma gondii-infected mice. a B1-DETECTR and c 529 RE-DETECTR reaction with mouse blood DNA. 1 dpi, 1 day post-infection mice #1; NS-1, normal saline #1; PC, positive control; NC, negative control; b B1-nested PCR and d 529 RE-nested PCR reaction with mouse blood DNA. 1–15 represent sample mouse blood samples collected 1, 3, 5 days post injection; 16–20 represent samples after saline injection; 21 represents positive control; 22 represents negative control
Portability of DETECTR
In this study, we combined the CRISPR/Cas12a-based DETECTR detection system with LFS to detect target DNA. The LFS technology [27] utilizes the principle of sandwich immunochromatography to detect the presence of target DNA in a sample (Fig. 5a). Specifically, the LFS incorporates a sample binding area, a test line, and a control line. Gold nanoparticle-coupled FITC antibodies are immobilized in the sample binding region, the test line is labeled with streptavidin, and the control line is labeled with anti-FITC antibodies. After adding the reaction system, the ssDNA reporter molecule (FITC-TTATTATT-biotin) can bind to the gold nanoparticle-coupled FITC antibody as a complex. If it is a negative sample, the complex is captured at the test line and both lines will show up; if it is a positive sample, Cas12a is activated and cleaves the reporter molecule, and the complex is not captured by the test line; only the control line will be shown. If neither or only the test line is shown, this indicates that the operation is incorrect or the test strip has failed, so the results are not informative. Overall, the developed method provides a promising tool for portable detection of target DNA in various settings.
DETECTR-based LFS detection of T. gondii. a Principle of LFS. b B1-LFS and c 529 RE-LFS reaction with Specificity detection. 1–5 represent Toxoplasma gondii, Cryptosporidium parvum, Babesia, Plasmodium, negative control. d B1-LFS and e 529 RE-LFS reaction with mouse blood DNA. 1–5 represent 5 days post injection; 6–8 represent samples after saline injection; 9 represents positive control, 10 represents negative control
In this study, we evaluated the sensitivity and specificity of LFS detection method for the diagnosis of T. gondii infection using the CRISPR-based detection system. Specifically, we mixed the amplified samples with a CRISPR detection system and incubated them for 1 h before performing the LFS. We then performed a specificity assay with the LFS and found that there was no cross-reactivity with other parasites, indicating the high specificity of the method (Fig. 5b, c). Moreover, we used the LFS to detect T. gondii in whole blood samples collected from five mice on the 5 dpi. Remarkably, all samples were positive for T. gondii, and the results were consistent with fluorescence detection (Fig. 5d, e). These findings collectively demonstrated that the LFS detection method exhibited good sensitivity and specificity for T. gondii detection and can be used as a promising tool for accurate diagnosis of T. gondii infection.
Discussion
To date, there is no effective therapeutic drug available for T. gondii infection. Therefore, the development of a rapid and accurate detection method for this pathogen is crucial for disease control and prevention of its spread. In this study, we developed a novel T. gondii detection approach that combined RPA and CRISPR/Cas12a technology, utilizing fluorescence or LFS as the reporting method. We conducted a comparative analysis of the effectiveness of nested PCR and DETECTR assays targeting B1 and 529 RE, respectively. Results indicated that DETECTR offered comparable sensitivity and specificity to nested PCR, but was more user-friendly because of the lack of complex technical requirements. The DETECTR reaction was performed at a consistent temperature of 37 °C, and the entire process did not necessitate complex technical expertise, facilitating prompt on-site detection of T. gondii DNA within a brief span of time.
At present, nucleic acid detection methods for pathogen identification primarily involve three main stages: sample processing, nucleic acid amplification, and signal transduction and reporting. Traditional amplification methods such as PCR and Q-PCR are time-consuming, taking up to 2 h or even longer, and require thermal cycling. Although LAMP is a constant temperature amplification method, its primer design is complex, and the amplification reaction requires incubation at 65 °C for 1 h [28]. In contrast, RPA primer design is simple, and the amplification reaction can be performed at 37 °C for only 20–30 min. Additionally, the CRISPR/Cas12a trans-cleavage system can be activated at 37 °C, and the entire reaction can be performed in a water bath, constant temperature incubator, or even in the palm of one's hand. The results can be read under UV light or through the use of lateral flow strips, making detection more feasible in non-laboratory settings.
In this investigation, B1 and 529 RE were selected as targets due to their frequent use in nucleic acid detection of T. gondii. Upon recognition of the target double-stranded DNA, the CRISPR/Cas12a system was activated, leading to massive cleavage of the single-stranded DNA reporter, which served as a signal transducer for converting the target dsDNA recognition to the fluorescence or LFS bands [29, 30]. The combination of RPA amplification effect and DETECTR technology results in highly sensitive detection of T. gondii DNA.
The sensitivity of DETECTR assays for the detection of serially diluted recombinant plasmids can reach 1.5 cp/μl. For T. gondii tachyzoite DNA, DETECTR can detect a minimum of 1 tachyzoite by B1 gene (35 copies) and 0.1 tachyzoite by 529 RE (200–300 copies), with 529 RE being a better choice than B1, as corroborated by Shirzad Fallahi and colleagues [26]. Notably, the high sensitivity of nested PCR [31] for T. gondii DNA detection is comparable to that of DETECTR, indicating that the DETECTR technology can identify low concentrations of T. gondii in the environment. Furthermore, when evaluating whole blood samples from mice, DETECTR could detect positive samples on 1 dpi and distinguish infected from uninfected samples. The method we have developed is mainly for the detection of host blood samples; the collection of blood samples is a minimally invasive or even non-invasive process, unlike tissue samples, which can cause harm to the host. This method has the potential to be an early and powerful diagnostic tool for T. gondii DNA.
The validation of the DETECTR assay's specificity for T. gondii detection was performed by analyzing the DNA of Plasmodium, C. parvum, and Babesia, which are parasites that are either similar in development to T. gondii (C. parvum) or can be present in blood samples (Plasmodium, Babesia). Notably, all results were negative for these parasites, with the exception of T. gondii, confirming the high specificity of the DETECTR assay for T. gondii detection.
In summary, DETECTR is a straightforward and speedy method for detecting T. gondii that can be applied to various settings with limited resources. In addition to the detection of infected animals, the detection of T. gondii DNA in the environment can also be performed, which has great potential as an alternative to current detection methods. In addition to the above advantages, DETECTR has some limitations in the DNA extraction stage. CRISPR-based nucleic acid detection systems have been demonstrated to be compatible with methods that rapidly expose nucleic acids, such as heating or chemical lysis [32]. Therefore, the subsequent research has focused on developing techniques to quickly release T. gondii DNA from samples without affecting detection sensitivity, thus further shortening the detection time. Because differential genes exist between T. gondii strains [33], leading to variations in virulence and pathogenicity, current typing methods rely on sequencing and some PCR-derived techniques [34, 35]. Strain-specific sgRNAs can be designed for CRISPR targeting, which can be particularly beneficial for identifying and typing T. gondii genes. In addition to its usefulness in detecting T. gondii, DETECTR technology holds great promise for broader applications in T. gondii research.
Availability of data and materials
The original contributions presented in the study are included in the article/Supplementary material; further inquiries can be directed to the author, Xiaofeng Wang: wangxiaofeng9516@163.com.
Abbreviations
- CRISPR:
-
Clustered Regularly Interspaced Short Palindromic Repeats
- Cas:
-
CRISPR-Associated Protein
- 529 RE:
-
529 bp repeat element
- PCR:
-
Polymerase chain reaction
- AIDS:
-
Acquired immune deficiency syndrome
- ELISA:
-
Enzyme-linked immunosorbent assay
- CLL:
-
Chronic lymphocytic leukemia
- Q-PCR:
-
Quantitative PCR
- LAMP:
-
Loop-mediated isothermal amplification
- RPA:
-
Recombinase polymerase amplification
- DETECTR:
-
DNA endonuclease-targeted CRISPR trans reporter
- POCT:
-
Point-of-care testing
- NCBI:
-
National Center of Biotechnology Information
- LFS:
-
Lateral flow strip
- dpi:
-
Day post infection
References
Flegr J, et al. Toxoplasmosis—a global threat. Correlation of latent toxoplasmosis with specific disease burden in a set of 88 countries. PLoS ONE. 2014;9:e90203.
Su C, et al. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137:1–11.
Robert-Gangneux F, et al. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25:264–96.
Montoya JG, et al. Toxoplasmosis. Lancet. 2004;363:1965–76.
Su YJ, et al. Geospatial epidemiology of T. gondii infection in livestock, pets, and humans in China, 1984–2020. Parasitol Res. 2022;121:743–50.
Foroutan M, et al. The global seroprevalence of T. gondii in pigs: a systematic review and meta-analysis. Vet Parasitol. 2019;269:42–52.
Naot Y, et al. IgM enzyme-linked immunosorbent assay test for the diagnosis of congenital Toxoplasma infection. J Pediatr. 1981;98:32–6.
Rajput R, et al. False negative toxoplasma serology in an immunocompromised patient with PCR positive ocular toxoplasmosis. Ocul Immunol Inflamm. 2018;26:1200–2.
Burg JL, et al. Direct and sensitive detection of a pathogenic protozoan, Toxoplasma gondii, by polymerase chain reaction. J Clin Microbiol. 1989;27:1787–92.
Buchbinder S, et al. Comparison of real-time PCR detection methods for B1 and P30 genes of Toxoplasma gondii. Diagn Microbiol Infect Dis. 2003;45:269–71.
Yu H, et al. Evaluation of a real-time PCR assay based on the single-copy SAG1 gene for the detection of Toxoplasma gondii. Vet Parasitol. 2013;197:670–3.
Zhao Y, et al. Isothermal amplification of nucleic acids. Chem Rev. 2015;115:12491–545.
Mayboroda O, et al. Multiplexed isothermal nucleic acid amplification. Anal Biochem. 2018;545:20–30.
Chen JS, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–9.
Abudayyeh OO, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–4.
Harrington LB, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–42.
Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–71.
Broughton JP, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–4.
He Q, et al. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens Bioelectron. 2020;154:112068.
Yu F, et al. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum IId-subtype-family from human and cattle fecal samples. Parasites Vectors. 2021;14:208.
Lee RA, et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc Natl Acad Sci USA. 2020;117:25722–31.
Cunningham CH, et al. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. EBioMedicine. 2021;68:103415.
da Silva, et al. The detection of T. gondii by comparing cytology, histopathology, bioassay in mice, and the polymerase chain reaction (PCR). Veterinary Parasitol. 2001;97:191–8.
Dai F, et al. Early detection of T. gondii infection in Mongolian Gerbil by quantitative real-time PCR. J Parasitol. 2019;105:52–7.
Homan WL, et al. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in T. gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol. 2000;30:69–75.
Fallahi S, et al. Challenging loop-mediated isothermal amplification (LAMP) technique for molecular detection of Toxoplasma gondii. Asian Pac J Trop Med. 2015;8:366–72.
He C, et al. Rapid and accurate detection of SARS-CoV-2 mutations using a Cas12a-based sensing platform. Biosens Bioelectron. 2022;198:113857.
Lalle M, et al. Loop-mediated isothermal amplification-lateral-flow dipstick (LAMP-LFD) to detect Toxoplasma gondii oocyst in ready-to-eat salad. Food Microbiol. 2018;70:137–42.
Li Y, et al. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–43.
Zhang M, et al. Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application. Talanta. 2020;214:120818.
Pujol-Riqué M, et al. Design of a one-tube hemi-nested PCR for detection of Toxoplasma gondii and comparison of three DNA purification methods. J Med Microbiol. 1999;48:857–62.
Myhrvold C, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science (New York NY). 2018;360:444–8.
Kong JT, et al. Serotyping of Toxoplasma gondii infections in humans using synthetic peptides. J Infect Dis. 2003;187:1484–95.
Frazão-Teixeira E, et al. Multi-locus DNA sequencing of Toxoplasma gondii isolated from Brazilian pigs identifies genetically divergent strains. Vet Parasitol. 2011;175:33–9.
Howe DK, et al. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–6.
Acknowledgements
We acknowledge Li Yu at Anhui Key Laboratory of Zoonoses, Anhui Medical University, for providing the Toxoplasma gondii. We acknowledge Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, for providing DNA of Cryptosporidium parvum and Babesia. We acknowledge Chao Zhang at the Department of Pathogen Biology, Anhui Medical University, for providing the Plasmodium.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2301100).
Author information
Authors and Affiliations
Contributions
Design of the work: XW, CX, YJ, YD. Acquisition and analysis of the data: XW, MC, SY, QL, YZ. Writing of the original manuscript: XW. Supervision of the project: YJ, YD. Funding acquisition: YD. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments in this study have been approved by the Laboratory Animal Ethics Committee of Anhui Medical University (approval no. LLSC20221092).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Protein expression and purification. Coomassie blue-stained acrylamide gel. Figure S2. Specificity of the DETECTR reaction. The left tube represents B1-L3; the right tube represents 529 RE-L3. Table S1. ssDNA oligonucleotide sequences. Table S2. PCR primers for the construction of standards. Table S3. RPA primers. Table S4. Nested PCR primers
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Cheng, M., Yang, S. et al. CRISPR/Cas12a combined with RPA for detection of T. gondii in mouse whole blood. Parasites Vectors 16, 256 (2023). https://doi.org/10.1186/s13071-023-05868-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05868-0