Abstract
Background
Culicoides kingi and Culicoides oxystoma belong to the Schultzei group of biting midges. These two species are vectors of disease in livestock of economic importance. As described in the literature, morphological identification for discrimination between them is still unclear. However, species-specific identification is necessary to solve taxonomic challenges between species and to understand their roles in disease transmission and epidemiology. This study aims to develop accurate tools to discriminate C. oxystoma from C. kingi using traditional morphometry and polymerase chain reaction-restriction fragment length polymorphism (PCR RFLP) assays for use in developing countries.
Methods
Specimens were collected from the region of Kairouan in central Tunisia. A total of 446 C. oxystoma/C. kingi individuals were identified using traditional morphometric analyses combined with PCR–RFLP of the cytochrome c oxidase subunit I gene. Thirteen morphometric measurements were performed from the head, wings, and abdomen of slide-mounted specimens, and six ratios were calculated between these measurements. Multivariate analyses of the morphometric measurements were explored to identify which variables could lead to accurate species identification.
Results
Four variables, namely antennae, wings, spermathecae, and palpus length, were suitable morphometric characteristics to differentiate between the species. Digestion with the SspI restriction enzyme of the PCR product led to good discriminative ability. Molecular procedures and phylogenetic analysis confirmed the efficiency of this simple and rapid PCR–RFLP method.
Conclusions
This study highlights for the first time in Tunisia the presence of C. oxystoma and its discrimination from C. kingi using abdominal measurements and the PCR–RFLP method. This approach could be applied in future epidemiological studies at the national and international levels.
Graphical Abstract

Similar content being viewed by others
Background
The biting midges in the genus Culicoides (Diptera: Ceratopogonidae) are small hematophagous flies (from 1 to 5 mm) that are widely distributed worldwide [1]. This genus includes 1347 valid species. Of these, 875 species are placed in subgenera, 336 species are placed in separate species groups, and 136 species are still unclassified [2].
They are biological vectors of a wide variety of important arbovirus diseases, including bluetongue virus (BTV), African horse sickness (AHS), epizootic hemorrhagic disease virus (EHDV), and Schmallenberg virus (SBV) [3, 4]. These diseases are considered a great threat to livestock, with important economic implications. In both the Mediterranean Basin and sub-Saharan Africa, the main vector of BTV and SBV is Culicoides imicola [1]. Other Culicoides species, such as C. obsoletus and C. scoticus, are reportedly able to transmit BTV in regions where C. imicola is absent. Moreover, several species of the subgenus Culicoides, including C. pulicaris, C. lupicaris, C. punctatus, C. newsteadi, and C. paolae, are considered potential vectors of BTV based on their ecological habitats and on virus isolation or viral genome detection from field-collected individuals [5].
Culicoides groupings such as the Schultzei species group (C. oxystoma, C. schultzei, C. subschultzei, C. kingi, C. rhizophorensis, C. enderleini, C. nevilli, and C. neoschultzei) are implicated in the transmission of some of these viruses [6]. In particular, in Australian and Asian regions, C. oxystoma is a vector of bovine arboviruses such as Akabane virus in Japan [7] and EHDV in Israel [8], and is a potential vector of BTV in India [9]. The emergence of these different diseases highlights the need to understand the epidemiology and transmission of Culicoides-borne viruses in Tunisia. This understanding requires detailed knowledge of the vector species, including their life characteristics, abundance, and vector competence [10].
In the investigation of vector-borne diseases, the proper identification of arthropod species is very important, and it is critical to differentiate them from other Culicoides species. Morphological identification includes many important morphological characteristics, such as the pigmentation pattern of the wings, length and shape of the antennal segments, characteristics of the genitalia in males, distribution of the sensilla on the antennae, and number and size of the spermathecae in females [11, 12]. Specific diagnosis between closely related species is often difficult, requiring intense work from expert taxonomists according to morphological parameters [12], without error [13]. Moreover, damage to midges during the collection step can result in identification difficulties or misidentification.
Consequently, other techniques such as morphometric discrimination [14, 15] and landmark-based geometric morphometrics [16,17,18] have been developed to separate intractable species. Many studies have used these methods to classify species and to examine variation among medically important mosquitoes that are morphologically similar, such as Anopheles spp. [19]. The landmark-based approach is considered the main method for wing venation patterns, helping to improve morphological species identification such as species of the Maculatus group [19].
Various biting midge species are found in the same habitat, and they share certain morphological features. Culicoides oxystoma shares many morphological features with C. kingi, leading to a great deal of confusion. Only expert entomologists can resolve such differentiation, as it requires consistent observation under a stereomicroscope using various identification keys. The problem is the presence of similarities in the wings for females. However, it has been shown that the presence of a clear spot under the radial cells is a typical characteristic of C. kingi [20]. On the other hand, according to Morag et al. [8], the presence of this spot has not been taken into account, and these species are then called atypical C. kingi. In fact, two morphological forms of C. kingi have been described [8, 21]: the Kenyan and the Senegalese forms. The two forms are separated by the presence of lightening behind the radial cells and by the spot of the second medial cell (m2) largely merged with its counterpart of the first medial cell (m1) for the Kenyan form. The Senegalese form is distinguished by the absence of lightening in the outer space of the radial cells and by a smaller spot of m2, which is sometimes separated from m1 or even absent. However, a literature review attributed atypical C. kingi as the corresponding C. oxystoma in the Japanese form described by Arnaud [21] (two spots in the cubital cell) and in Arabia by Boorman [22] (a stain in the cubital cell between rib median veins 3 and 4 [M3 + 4] and the edge of the wing). Cornet and Brunhes [23] reported that C. oxystoma is a species complex requiring taxonomic revision and that the published descriptions were not in concordance. Three different morphological features of C. oxystoma were found, one of which corresponded to C. oxystoma sensu Arnaud from Japan. Morag et al. [8] demonstrated that the other two features were present in Israel and that one of them is synonymous with the Japanese form. These authors suggest that molecular methods for analysis of cytochrome oxidase subunit I (COI) are useful to solve taxonomic problems associated with this group. Morphological and molecular identification techniques were used to establish the status of C. kingi and C. oxystoma present in Senegal. There are a number of PCR-based methods available to differentiate between closely related species and to supply unambiguous results. For these molecular methods, several genes have been used in large-scale Culicoides studies: COI, internal transcribed spacer 1 and 2 (ITS1, ITS2) of ribosomal DNA (rDNA), and the nuclear CAD gene [24]. The use of molecular data has renewed interest and activity in systematics [25]. The COI gene is the most commonly sequenced marker for Culicoides barcoding. However, PCR-based methods remain costly and cannot be used in routine or in large-scale epidemiological surveys. Augot et al. [26] combined a single step of PCR and restriction fragment length polymorphism (RFLP) as a tool for the identification of Culicoides species. This process is less time-consuming, while also enabling the processing of multiple samples simultaneously.
To the best of our knowledge, in Tunisia, no studies have been carried out combining morphological, morphometric, and molecular methods to discriminate between two closely related species in the subgenus Remmia: C. oxystoma and C. kingi. These species share many features, making their specific morphological identification difficult using microscopy on mounted specimens and impossible based on wing patterns using stereomicroscopy.
The aims of this study were (i) to identify Culicoides species trapped in different geographical regions in the district of Kairouan; (ii) to discriminate C. oxystoma from C. kingi by morphological and morphometric techniques; (iii) to develop a molecular tool combining a single step of PCR and the RFLP method to differentiate C. oxystoma from C. kingi; and (iv) to assess the phylogenetic utility to provide species identification in agreement with morphological and morphometric identifications.
Methods
Description of the study area
Kairouan is located in central Tunisia and occupied an area 6712 km2, with a population of 570,559 (Fig. 1). This population is predominantly rural, and all collections were performed in human-inhabited biotopes with the presence of domestic animals (i.e., cattle, horses, dogs, goats, and chickens) and muddy environments that formed in proximity to livestock troughs. The region has annual rainfall of 250–400 mm. The weather of Kairouan is semiarid, with hot and dry summers and cold and wet winters.
Location of study sites in Kairouan (modified according to [27]). N north, E eastern
Field sampling
Adult midges were captured once a week from August 2014 to September 2017 in nine geographical locations within Kairouan province (Fig. 1). Culicoides from all sites were trapped using a Centers for Disease Control and Prevention (CDC) light trap with a collection bucket. The bucket was filled with 50 ml water to kill trapped insects. Traps were placed at sunset and collected the next day before sunrise. All specimens of Culicoides were stored in 70% ethanol prior to morphological and molecular analysis.
Specimen identification and mounting
Culicoides midges were separated from other insects and identified according to wing characteristics using a stereomicroscope. The two species C. kingi and C. oxystoma were differentiated from each other on the basis of wing spot patterns (Additional file 1: Figure S1). The individual females were dissected on a slide using sterile dissecting needles. The head (dorsal side up), wings, and posterior abdominal segment (ventral side up) of each of these specimens were subsequently mounted on the slide under three separate coverslips using Canada balsam. The remaining thorax, legs, and anterior abdomen were stored in 75% ethanol for further molecular analysis. All identifications were performed using the Interactive Identification Key for Culicoides (IIKC) [28].
Morphometric analysis
Morphometric measurements
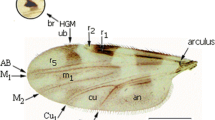
Slide-mounted specimens were examined and measured using binocular microscopy. Morphometric measurements were taken from the head, wings, and genitalia of individual females. Thirteen variables (Additional file 2: Figure S2) were recorded in Culicoides adult females: length of the wing from arculus to tip and width of wing from the location of the second radial cell to the base of the cubital vein (Cu1); length of the space of the two sensilla up the eyes; length of the five flagellar segments (11 to 15) and eight basal flagellar segments (3 to 10); length of the third segment of palpus and width of the third segment of palpus; length of flagellomeres 10 and 11; length and width of the first spermatheca; length and width of the second spermatheca. Six additional variables were calculated to determine the following ratios: wing ratio (wing length/wing width); spermathecal 1 ratio (spermatheca 1 length/spermatheca 1 width); spermathecal 2 ratio (spermatheca 2 length/spermatheca 2 width); palpal ratio (length of the third palpus/length of the first and second palpi); flagella ratio (length of flagellomere 11/length of flagellomere 10 (R: 11/10); antenna segment ratio (total length of five apical segments 11–15/total length of eight basal segments 3–10).
Statistical analysis
Here, we employed principal component analysis (PCA) to explore the correlation structure between variables and to determine which variables represented the greatest variance. PCA was carried out using Statistical Package for the Social Sciences (SPSS) version 22 (IBM Corp., Armonk, NY, USA) software. PCA calculates the correlation matrix, the principal component loading matrix, and respective eigenvalues to explain the structure of the parameters.
Molecular analysis
Extraction of genomic DNA
Genomic DNA was extracted from 30 individual Culicoides specimens (10 C. kingi and 20 C. oxystoma) using tissue extraction kits (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. DNA samples were eluted in 100 μl of Tris EDTA (TE) buffer and stored at −20 °C. Primer sequences used for the detection of mitochondrial DNA (mtDNA) have been described previously [29]. For the cytochrome oxidase I (COI) gene, PCR was performed in a 50 µl volume using 8 μl of the sample extracted or control DNA (water), two units of GoTaq DNA polymerase (Promega, Madison, WI, USA), 10 μl of associated 2× buffer containing MgCl2, 200 μM dNTPs, and 1 μM of each primer. PCR was carried out in a GeneAmp PCR System 9700 thermal cycler under the following program [30]. Five microliters of each amplified product was visualized after electrophoresis at 100 V for 50 min with 100-bp DNA ladder molecular weight markers (Boiron, Germany) in a 1.5% agarose gel stained with 5 µl ethidium bromide. The target DNA was visualized on an ultraviolet transilluminator. The PCR product of the COI gene was approximately 689 bp.
PCR–RFLP analysis
In silico screening for a rapid PCR–RFLP discriminative method between C. oxystoma and C. kingi was first carried out using an in silico assay. Sites for restriction enzymes were predicted for COI sequences of C. oxystoma and C. kingi (Table 1) using Restriction Analyzer (http://www.molbiotools.com/restrictionanalyzer.html) and CLC Sequence Viewer 8.0 (www.qiagenbioinformatics.com) software. A panel of restriction enzymes was tested. One restriction enzyme, SspI, provided an original digestion pattern per species for the mtDNA marker and was selected (Additional file 3: Figure S3).
In vitro PCR–RFLP assays were performed in a 20 μl total volume reaction mixture containing 7 μl of PCR product (from PCR vials), 0.4 μl of SspI, and 0.8 μl of supplied buffer. PCR products were digested for 1 h at 37 °C. The digested samples were separated by electrophoresis on a 2% agarose gel to produce DNA fragments and were sized by comparison with a 100-bp marker ladder (Boiron, Germany).
DNA sequencing and phylogenetic analysis
To confirm the results of the PCR–RFLP analysis, direct sequencing of the COI gene amplicons was performed using the same set of primers that were used in the PCR assay (Eurofins MWG Operon, Munich, Germany). The obtained sequences were edited using Chromas software version 2.33 (http://ww.technelysium.com. au/chromas.html) and identified by comparison with sequences available in GenBank using the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nlm.nih.gov/blast/). Species assignment was considered complete when a match of 98% or greater was found between our sequences and those in GenBank. DNA sequence-based analyses were performed using the maximum likelihood (ML) method (Tamura-Nei) with MEGA 5 software [31]. Genetic distances were computed using the Kimura 2-parameter (K2P) method. The tree topology was supported by 1000 bootstrap replicates to determine node reliability. COI sequences in this study were compared with sequences from the same species in GenBank (MF399736, MF399686, MF399705, MF399698, KJ729983, MF399780, MF399688, MF399776, MF399699, MF399697).
Results
Morphological identification
During the study period, 1190 biting midges were collected in the district of Kairouan (Fig. 2). Of these specimens, 82% and 18% females and males were identified, respectively. Morphological identification revealed the presence of 11 species: C. imicola, C. kingi, C. paolae, C. oxystoma, C. sahariensis, C. circumscriptus, C. sergenti, C. jumineri, C. puncticollis, C. langeroni, and C. newsteadi.
Culicoides imicola and C. kingi were the most abundant species, constituting 35.21% (n = 419) and 30% (n = 357), respectively (Fig. 2). Two species belonging to the Schultzei group were identified that were ascribable to C. kingi (190 ♀, 167♂) and C. oxystoma (88 ♀, 01 ♂). It should be noted that C. oxystoma is reported here for the first time in Tunisia. This species was collected from the district of Sbikha (Kairouan) (Fig. 1). However, C. kingi was more abundant and was sampled at the four collection sites. The wing structure was generally unique for each species, although slight variations in wing patterns were observed between the two species (Additional file 1: Figure S1). Culicoides kingi from Tunisia presented two pale round spots after the second radial cell (r2), one pale spot in the first medial cell (m1 of the wing crossing the median vein (M2), two separate pale spots in m4, two pale spots in anal cells (An), and one pale spot in m1 and m2 (Additional file 1: Figure S1). Culicoides oxystoma had pale spots under radial cells, pale spots in m1 that did not cross M2, another pale spot between Cu1) and the edge of the wing, and two pale spots merged in m4 (Additional file 1: Figure S1).
Morphometric analysis
Morphometric differences in the measurement data were studied via PCA. Kaiser’s [32] stopping rule states that only the number of axes with eigenvalues over 1.00 should be considered in the analysis. From the 13 morphometric measurements, five principal components (axes) had an eigenvalue greater than 1.00 (Table 2), and when combined, these factors accounted for 72.24% of the total variance. A scree plot (Fig. 3) suggested inclusion of only the two first axes PC1 and PC2.
The first axis (PC1) was positively correlated with the length of eight basal segments (loadings ≥ 0.8) and with the lengths and widths of the wing (loadings ≥ 0.65) and correlated with the third palpal segment and the length of flagellomere 11 (loadings ≥ 0.65). The second axis (PC2) was positively correlated with the length of spermathecae 1 and 2, as well as the width of the third segment palpus (loadings ≥ 0.4). Additionally, it correlated with the length of five apical segments and with the length of flagellomere 10 (loadings ≥ 0.5) (Table 3).
PCA was also applied to the six morphometric ratios to augur differences in shape between C. kingi and C. oxystoma. Kaiser’s stopping rule suggested the inclusion of the first three axes (Table 4), while the screen test (Fig. 3) suggested inclusion of first two axes. Indeed, four of the six axes accounted for similar amounts of variance (6–18%) (Table 5). PC1 was correlated with the palpal and flagella ratios, while PC2 was negatively correlated with the segment ratio and positively correlated with the spermatheca ratio (Table 4).
Molecular analyses
PCR amplification of the COI region of the mtDNA from the midges identified morphologically as C. kingi and C. oxystoma (n = 30) exhibited uniformity in band size (689 bp).
Species-diagnostic restriction enzyme sites
In silico PCR–RFLP simulation using different restriction enzymes demonstrated that SspI was suitable for discrimination between the closely related species C. oxystoma and C. kingi. The restriction fragments obtained from each DNA reference sample (Table 1) agreed with the predicted profiles for the COI sequence by in silico analysis (Additional file 3: Figure S3).
Digestion of the COI PCR products with SspI demonstrated that the patterns of the two species were different. For C. kingi (KJ729983.1), two restriction fragments were predicted (615 and 74 bp), but in agarose gel only one band (615 bp) could be seen, probably because of co-migration of the 74-bp band (Additional file 3: Figure S3). For C. oxystoma, three very close bands were visible in agarose gel (385, 210, 994 bp) (Additional file 3: Figure S3).
DNA sequencing and phylogenetic analysis
The obtained DNA sequences were compared with those deposited in the GenBank database. Phylogenetic analysis was performed to confirm the genetic relationship between species. The topology of the phylogenetic tree showed a clear subdivision in seven distinct and well-supported phylogenetic lineages corresponding to six species of the Schultzei group (C. oxystoma, C. kingi, C. nevilli, C. subschultzei, C. enderleini, and C. schultzei), and Forcipomyia sp. Specimens of C. oxystoma formed two separate clades, one including specimens from India (KT307834) and China (MK917536; MK917537) and the other including specimens from Senegal (MF399728 and MF399738). Tunisian specimens (E2, E5, and E7) identified as C. oxystoma and representing morphological variations (Additional file 1: Figure S1) were grouped in the same clade. One clade clustered the specimens identified as C. kingi, including those collected in Tunisia (KJ729983) and Cameroon (MF399780) (Fig. 4).
Maximum likelihood tree based on COI nucleotide sequences obtained in this study. The tree show phylogenetic analyses of three individual midges morphologically identified as C. oxystoma together with the most closely related published sequences and their GenBank accession numbers. Bootstrap values are shown on the branches
Discussion
To the best of our knowledge, this is the first study on the presence of C. oxystoma in Tunisia. However, this is also the first morphological and molecular investigation aiming to discriminate C. oxystoma from C. kingi.
Culicoides imicola was the dominant species of biting midges recorded during this study, representing 35.21% of the total Culicoides captured by CDC light traps. This result is consistent with previous findings in Tunisia [33,34,35,36], where C. imicola was typically the most dominant species, exceeding 80% of the total species composition and abundance at sites where BTV transmission was intense. The second most dominant species collected in this study comprised midges of the Schultzei group, C. kingi. This species represented 30% of the total Culicoides captured. Other species were collected but in minor numbers. It is important to note that the main weakness with this low proportion (e.g., C. sahariensis and others) is the inability for subsampling [37]. Furthermore, as traps were only operated from 1 h before dusk until 1 h after sunrise, Culicoides spp. that are active during daylight hours were likely underrepresented. However, Sanders et al. [38] found that estimation based on subsampling provided a good estimation of the total Culicoides captured, and it has been previously demonstrated that most Culicoides spp. are crepuscular [3].
The identification of C. oxystoma in Tunisia (Kairouan) is of great importance, as this species is known to transmit many important bovine arboviruses, including Akabane virus [7, 39]. Additionally, it is a suspected vector of EHDV in Israel [8]. It would therefore be interesting to determine whether this important potential vector species plays a role in the transmission of Culicoides-borne diseases in Tunisia and North Africa. This species shares many morphological characteristics with C. kingi, making morphological identification very difficult. Many previous studies have tried to separate closely related Culicoides species such as C. obsoletus and C. scoticus, and a number of studies [1, 39, 40] have been conducted to assess the reliability of traditional morphological or morphometric identification techniques to differentiate between the two species. Augot et al. [1] suggested that C. obsoletus and C. scoticus can be identified using multivariate analyses based on the length and width of spermatheca 1, the length of spermatheca 2, and the width between the chitinous plates. Foxi et al. [41] identified females in the Obsoletus group under a stereomicroscope by combining two characteristics: the shape of the third segment of the maxillary palp, and the number and location of hairs on the first abdominal tergite. It should be noted that previous studies have indicated that measurements of the total length of the five apical segments and eight basal segments of the antenna, which produce an antenna ratio, can significantly differentiate C. obsoletus and C. scoticus. Furthermore, a study by Kluiters et al. [42] highlighted that abdominal measurements, such as larger and smaller spermatheca lengths and widths, can be used to differentiate C. obsoletus from C. scoticus.
In the current study, the diagnostic value of 13 morphometric variables in females of C. kingi and C. oxystoma was used to verify statistical identification. The results suggest that abdominal measurements (length of eight basal segments and wing lengths and widths) could be used to reliably separate these two species, despite the small numbers of samples and individuals used, as C. kingi exhibits smaller measurements than those from C. oxystoma. Our research demonstrates that abdominal size combined with the form of the wing spot is a suitable characteristic for separating the two species.
We developed a PCR–RFLP-based tool to discriminate between the species C. kingi and C. oxystoma. Interestingly, it was found that this approach was able to produce different profiles using the SspI restriction enzyme and could therefore eliminate the difficulties present in systematic identification. Each of the two species can be discriminated by visualizing agarose gel in an ultraviolet transilluminator, as each species produces a unique pattern after restriction digestion. Furthermore, the same results were obtained with in silico digestion, demonstrating that C. oxystoma was distinguishable from C. kingi. Compared to classical molecular techniques, this technique is rarely used for the identification of Culicoides species [37, 43]; our PCR–RFLP method produced satisfying results and could be a convenient tool to separate the two closely related species.
There has been much discussion of the taxonomic status of C. oxystoma as a member of the Schultzei group, but it remains unclear [21, 44]. Here, we found that the morphologically and molecularly identified C. oxystoma from the present study was closely related to C. oxystoma from India, China, Israel, and Lebanon, and was separated from those of Senegal. Our results were consistent with those previously reported [8, 45, 46]. The maximum likelihood tree also revealed that C. oxystoma is discriminated from C. kingi. This result is consistent with morphometric analyses showing a high degree of divergence between the two species.
Furthermore, it was suggested that C. oxystoma is a complex of sibling species [21, 47], owing to the high level of intraspecific divergence observed in C. oxystoma based on the COI sequences [46, 48]. Further studies on a large number of specimens are needed to study the taxonomic status of C. oxystoma species in Tunisia.
Conclusions
In this study, we highlighted that C. oxystoma could be differentiated based on wing form and abdominal measurements. Accordingly, a PCR–RFLP assay was developed to discriminate between these two closely related species. This technique is cost-effective and rapid and does not require any sophisticated equipment. Characterization of cryptic species could be conducted using PCR–RFLP, and its utilization could be more common in the future. Finally, combining morphological and molecular identification of Culicoides specimens is very important for a better understanding of the systematics of C. oxystoma, which is a vector of disease.
Availability of data and materials
Not applicable.
Abbreviations
- PCR:
-
Polymerase chain reaction
- RFLP:
-
Restriction fragment length polymorphism
- BTV:
-
Bluetongue virus
- AHS:
-
African Horse Sickness
- EHDV:
-
Epizootic hemorrhagic disease virus
- SBV:
-
Schmallenberg virus
- COI:
-
Cytochrome oxidase subunit I
- ITS1:
-
Internal transcribed spacer 1
- ITS2:
-
Internal transcribed spacer 2
- rDNA:
-
Ribosomal DNA
- CDC:
-
Centers for Disease Control and Prevention
- IIKC:
-
Interactive Identification Key for Culicoides
- Cu1:
-
Cubital 1
- PCA:
-
Principal component analysis
- SPSS:
-
Statistical Package for the Social Sciences
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediamine tetraacetic acid
- TE:
-
Tris ethylenediamine tetraacetic acid
- MgCl2 :
-
Magnesium chloride 2
- dNTP:
-
Deoxyribonucleotide triphosphate
- BLAST:
-
Basic Local Alignment Search Tool
- ML:
-
Maximum Likelihood
- K2P:
-
Kimura 2-parameter
- r2 :
-
Second radial cell
- m1 :
-
First medial cell
- M2 :
-
Median vein
- An:
-
Anal cell
- PC1:
-
Principal component 1
- PC2:
-
Principal component 2
References
Augot D, Sauvage F, Jouet D, Simphal E, Veuille M, Couloux A, et al. Discrimination of Culicoides obsoletus and Culicoides scoticus, potential bluetongue vectors, by morphometrical and mitochondrial cytochrome oxidase subunit I analysis. Infect Genet Evol. 2010;10:629–37.
Borkent A, Dominiak P. Catalog of the biting midges of the world (Diptera: Ceratopogonidae). Zootaxa. 2020;4787:1–377.
Mellor PS, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol. 2000;45:307–40.
Balenghien T, Pagès N, Goffredo M, Carpenter S, Augot D, Jacquier E, et al. The emergence of Schmallenberg virus across Culicoides communities and ecosystems in Europe. Prev Vet Med. 2014;116:360–9.
Caracappa S, Torina A, Guercio A, Vitale F, Calabrò A, Purpari G, et al. Identification of a novel bluetongue virus vector species of Culicoides in Sicily. Vet Rec. 2003;153:71–4.
Mellor P, Osborne R, Jennings D. Isolation of bluetongue and related viruses from Culicoides spp. in the Sudan. J Hyg. 1984;93:621–8.
Yanase T, Kato T, Kubo T, Yoshida K, Ohashi S, Yamakawa M, et al. Isolation of bovine arboviruses from Culicoides biting midges (Diptera : Ceratopogonidae) in Southern Japan : 1985–2002. J Med Entomol. 2005;42:63–7.
Morag N, Saroya Y, Braverman Y, Klement E, Gottlieb Y. Molecular identification, phylogenetic status, and geographic distribution of Culicoides oxystoma (Diptera: Ceratopogonidae) in Israel. PLoS ONE. 2012;7:e33610.
Dadawala AI, Biswas SK, Rehman W, Chand K, Mathapati BS, Kumar P. Isolation of bluetongue virus serotype 1 from Culicoides vector captured in livestock farms and sequence analysis of the viral genome segment-2. Transboud Emerg Dis. 2012;59:361–8.
Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse BV. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Res. 2013;100:102–13.
Campbell JA, Clinton E. Taxonomic review of the British species of Culicoides Latreille (Diptera, Ceratopogonidae). Proc Roy Entomol Soc. 1960;67:181–302.
Wirth W, Hubert A. The Culicoides of Southeast Asia (Diptera: Ceratopogonidae). Amer Ent Inst 1989.
Pagés N, Sarto I. Differentiation of Culicoides obsoletus and Culicoides scoticus (Diptera : Ceratopogonidae) based on mitochondrial cytochrome oxidase subunit I. J Med Entomol. 2005;42:1026–34.
Borkent A. World species of biting midges (Diptera: Ceratopogonidae). 2012.
Borkent A. The pupae of Culicomorpha—morphology and a new phylogenetic tree. Zootaxa. 2012;3396:1–98.
Henni L, Sauvage F, Ninio C, Depaquit J, Augot D. Wing geometry as a tool for discrimination of Obsoletus group (Diptera : Ceratopogonidae : Culicoides) in France. Infect Genet Evol. 2014;21:110–7.
Muñoz-muñoz A, Talavera S, Pagès N. Geometric morphometrics of the wing in the subgenus Culicoides (Diptera : Ceratopogonidae): from practical implications to evolutionary interpretations. J Med Entomol. 2011;48:129–39.
Muñoz-muñoz A, Talavera S, Carpenter S, Nielsen S, Werner D, Pagès N. Phenotypic differentiation and phylogenetic signal of wing shape in western European biting midges, Culicoides spp., of the subgenus Avaritia. Entomol Vet. 2014;28:319–29.
Chaiphongpachara T, Sriwichai P, Samung Y, Ruangsittichai J, Morales Vargas RE, Cui L, et al. Geometric morphometrics approach towards discrimination of three member species of Maculatus group in Thailand. Acta Trop Elsevier. 2019;192:66–74.
Fall M. Ecologie et lutte contre les Culicoides vecteurs de la peste équine et de la fièvre catarrhale ovine au Sénégal. Université Cheikh Anta Diop. Thése de Doctorat; 2015.
Arnaud PHJ. The heleid genus Culicoides in Japan. Korea and Ryukyu Islands (Insecta: Diptera). Microentomology. 1956;21:84–207.
Boorman J. Culicoides (Diptera: Ceratopogonidae) of the Arabian peninsula with notes on their medical and veterinary importance. Fauna Saudi Arab. 1989;10:160–224.
Cornet M, Brunhes J. Révision des espèces de Culicoides apparentées à C. schultzei (Enderlein, 1908) dans la région afrotropicale (Diptera, Ceratopogonidae). Bull Soc Entomol Fr. 1908;1994(99):149–64.
Harrup L, Bellis G, Balenghien T, Garros C. Culicoides Latreille (Diptera: Ceratopogonidae) taxonomy: current challenges and future directions. Infect Genet Evol. 2015;30:249–66.
Pires A, Marinoni L. DNA barcoding and traditional taxonomy unified through integrative taxonomy : a view that challenges the debate questioning both methodologies. Biota Neotrop. 2010;10:339–46.
Augot D, Ninio C, Akhoundi M, Lehrter V, Couloux A, Jouet D, et al. Characterization of two cryptic species, Culicoides stigma and C. parroti (Diptera: Ceratopogonidae), based on barcode regions and morphology. J Vector Ecol. 2013. https://doi.org/10.1111/j.1948-7134.2013.12039.x.
Selmi M, Ben Hariz M, Krichen N, Ben Ahmed Selmi M, Khayatia K, Issaoui M, et al. Atlas du Gouvernorat de Kairouan. 2011; 1-86.
Mathieu B, Cêtre-Sossah C, Garros C, Chavernac D, Balenghien T, Carpenter S, et al. Development and validation of IIKC: An interactive identification key for Culicoides (Diptera: Ceratopogonidae) females from the Western Palaearctic region. Parasit Vectors. 2012;5:137.
Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci U S A. 2006;103:968–71.
Costa FO, DeWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, et al. Biological identifications through DNA barcodes: the case of the Crustacea. Can J Fish Aquat Sci. 2007;64:272–95.
Kumar S, Stecher G, Peterson D, Tamura K. MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28:2685–6.
Kaiser HF. An index of factorial simplicity. Psychometrika. 1974;39:31–6.
Chaker E, Sfari M, Rabhi M, Babba HAR. Note faunistique sur les Culicoides (Diptera: Ceratopogonidae) du gouvernoratt de Monastir (Tunisie). Parasite. 2005;12:359–61.
Slama D, Chaker E, Mathieu B, Babba H, Depaquit J, Augot D. Biting midges monitoring (Diptera: Ceratopogonidae: Culicoides Latreille) in the governate of Monastir (Tunisia): species composition and molecular investigations. Parasitol Res. 2014. https://doi.org/10.1007/s00436-014-3873-1.
Slama D, Haouas N, Mezhoud H, Babba H, Chaker E. Blood meal analysis of Culicoides (Diptera: Ceratopogonidae) in Central Tunisia. PLoS ONE. 2015. https://doi.org/10.1371/journal.pone.0120528.
Sghaier S, Hammami S, Goffredo M, Hammami M, Portanti O, Lorusso A, et al. New species of the genus Culicoides (Diptera Ceratopogonidae) for Tunisia, with detection of bluetongue viruses in vectors. Vet Ital. 2017;53:357–66.
Dallas JF, Cruickshank RH, Linton YM, Nolan DV, Patakakis MBY, Capela M, Capela R, Pena I, Meiswinkel R, Ortega MD, Baylis M, Mellor PMA. Phylogenetic status and matrilineal structure of the biting midge, Culicoides imicola, in Portugal Rhodes and Israel. Med Vet Entomol. 2003;17:379–87.
Sanders CJ, Shortall CR, Gubbins S, Burgin L, Gloster J, Harrington R, Reynolds DR, Mellor PS, Carpenter S. Influence of season and meteorological parameters on flight activity of Culicoides biting midges. J Appl Ecol. 2011;48:1355–64.
Jae-Ku O, Joon-Yee C, Mee-Soon K, Toh-Kyung K, Tae-Uk L, Bae YC. Abundance of biting midge species (Diptera : Ceratopogonidae, Culicoides spp.) on cattle farms in Korea. J Vet Sci. 2013;14:91–4.
Sanders CJ, Shortall CR, Gubbins S, Burgin L, Gloster J, Harrington R, et al. Influence of season and meteorological parameters on flight activity of Culicoides biting midges. J Appl Ecol. 2011;48:1355–64.
Foxi C, Meloni G, Puggioni G, Manunta D, Rocchigiani A, Vento L, et al. Bluetongue virus detection in new Culicoides species in Sardinia. Italy Vet Rec. 2019;184:621.
Kluiters G, Pagès N, Carpenter S, Gardès L, Guis H, Baylis M, et al. Morphometric discrimination of two sympatric sibling species in the Palaearctic region, Culicoides obsoletus Meigen and C. scoticus Downes & Kettle (Diptera: Ceratopogonidae), vectors of bluetongue and Schmallenberg viruses. Parasites Vectors. 2016;9:262.
Linton YM, Mordue AJ, Cruickshank RH, Meiswinkel R, Mellor PSDJ. Phylogenetic analysis of the mitochondrial cytochrome oxidase subunit I gene of five species of the Culicoides imicola species complex. Med Vet Entomol. 2002;16:139–46.
Wirth WW, Dyce AL. The current taxonomic status of the Culicoides vectors of bluetongue viruses. Prog Clin Biol Res. 1985;178:151–64.
Liu Y, Tao H, Yu Y, Yue L, Xia W, Zheng W, et al. Molecular differentiation and species composition of genus Culicoides biting midges (Diptera: Ceratopogonidae) in different habitats in southern China. Vet Parasitol. 2018;254:49–57.
Harrup LE, Miranda MA, Carpenter S. Advances in control techniques for Culicoides and future prospects. Vet Ital. 2016;52:247–64.
Bakhoum MT, Labuschagne K, Huber K, Fall M, Mathieu B, Venter G, et al. Phylogenetic relationships and molecular delimitation of Culicoides Latreille (Diptera: Ceratopogonidae) species in the Afrotropical region: interest for the subgenus Avaritia. Syst Entomol. 2017;43:355–71.
Bakhoum MT, Fall M, Fall AG, Bellis GA, Gottlieb Y, Labuschagne K, et al. First record of Culicoides oxystoma Kieffer and diversity of species within the Schultzei group of Culicoides Latreille (Diptera: Ceratopogonidae) biting midges in Senegal. PLoS ONE. 2013;8:e84316.
Acknowledgements
We gratefully acknowledge the valuable input of the anonymous reviewers on this paper.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: DS, HB, EC, and RB. Performed the experiments: DS and RB. Drafted the manuscript: DS, and EC. Participated in field missions: LR, DS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Additional file 1: Figure S1.
Wing structure of Culicoides species. a. Wings of C. kingi (b1) and C. oxystoma (b2), demonstrating the difficulty in differentiating between species of the Schultzei group. Arc arculus, Trans transverse, r1, r2 first and second radial cells, r5 radial cell, M1, M2 first and second median veins, Cu1, Cu2 first and second cubital veins, An anal cell; m1, m2 first and second medial cell.

Additional file 2: Figure S2.
Morphometric measurements of Culicoides species. a Wing (length wing [a1], width wing [a2]). b Head (length of the space of the two sensilla up the eyes [b1]; length of the five flagellar segments [b2] and eight basal flagellar segments [b3]; length of the third palpus [b4] and width of the third segment of the palp [b5]; length of flagellomeres 10 [b6] and 11 [b7]). c Spermathecae (length [c1] and width of the first spermatheca [c2]; length [c3] and width of the second spermatheca [c4]). Different lowercase letters indicate the measurements taken on different parts of the Culicoides body.

Additional file 3: Figure S3.
PCR products (lanes 1–4) of the Culicoides species (a). Lane C: negative control (no DNA); lane MW: molecular marker 1 Kb plus DNA ladder (Invitrogen™). b Restriction results revealing complete coincidence with the in silico analysis. c In silico analysis of restriction profiles using SspI for the same species (S1–S4, C. oxystoma; S5, C. kingi, accession number: KJ729983). M molecular marker 1 Kb.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Slama, D., Baraket, R., Remadi, L. et al. Morphological and molecular differentiation between Culicoides oxystoma and Culicoides kingi (Diptera: Ceratopogonidae) in Tunisia. Parasites Vectors 14, 607 (2021). https://doi.org/10.1186/s13071-021-05084-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-05084-8