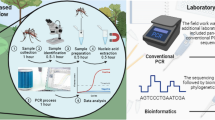
Abstract
Background
Aedes vigilax is one of the most significant arbovirus vector and pest species in Australia’s coastal regions. Occurring in multiple countries, this mosquito species occurs as a species complex which has been separated into three clades with two detected in Australia. Until recently, Ae. vigilax has largely been absent from Victoria, only occasionally caught over the years, with no reported detections from 2010 to 2016. Complicating the detection of Ae. vigilax is the shared sympatric distribution to the morphologically similar Ae. camptorhynchus, which can exceed 10,000 mosquitoes in a single trap night in Victoria. Currently, there are no molecular assays available for the detection of Ae. vigilax. We aim to develop a quantitative PCR (qPCR) for the detection of Ae. vigilax, with the specificity and sensitivity of this assay assessed as well as a method to process whole mosquito traps.
Methods
Trapping was performed during the 2017–2020 mosquito season in Victoria in two coastal areas across these 3 consecutive years. A qPCR assay was designed to allow rapid identification of Ae. vigilax as well as a whole mosquito trap homogenizing and processing methodology. Phylogenetic analysis was performed to determine which clade Ae. vigilax from Victoria was closest to.
Results
Aedes vigilax was successfully detected each year across two coastal areas of Victoria, confirming the presence of this species. The qPCR assay was proven to be sensitive and specific to Ae. vigilax, with trap sizes up to 1000 mosquitoes showing no inhibition in detection sensitivity. Phylogenetic analysis revealed that Ae. vigilax from Victoria is associated with clade III, showing high sequence similarity to those previously collected in New South Wales, Queensland and Western Australia.
Conclusions
Aedes vigilax is a significant vector species that shares an overlapping distribution to the morphologically similar Ae. camptorhynchus, making detection difficult. Here, we have outlined the implementation of a specific and sensitive molecular screening assay coupled with a method to process samples for detection of Ae. vigilax in collections with large numbers of non-target species.
Graphical abstract

Similar content being viewed by others
Background
Aedes vigilax is regarded as an aggressive mosquito species, with a host preference for humans, other mammals and occasionally avian species [1, 2]. Aedes vigilax is regarded as the principal vector of Ross River virus (RRV) in coastal and sub-coastal areas of Australia [3, 4] as well as vectoring a range of other arboviruses such as Barmah Forest virus (BFV) and has experimentally been proven competent to exotic arboviruses such as chikungunya virus (CHIKV), West Nile virus and Japanese encephalitis virus [3,4,5]. Active during crepuscular periods, Ae. vigilax can bite during the day, particularly around larval habitats where large populations can occur. This mosquito species has been classified as a diverse generalist feeder increasing this mosquitoe species’ ability to potentially act as a bridge vector of a range of pathogens [2].
Aedes vigilax (known as the northern saltmarsh mosquito) breeds episodically in coastal regions of Australia, with the female depositing her eggs in damp soil associated with floodplains, mudflats and brackish to hypersaline pools, with a high density of eggs occurring at vegetated sites amongst mangroves and artificial drainage areas [6, 7]. These coastal sites get flooded by above average or “king” tides causing large hatching events of this species [8]. Aedes vigilax populations can sporadically explode (with overnight collections in the 1000s) with the coincidence of high tides, warmer air temperatures and day length [6]. Along Australia’s coastline, Ae. vigilax occurs in New South Wales, Queensland, Northern Territory, Western Australia and South Australia and has generally been thought to be absent or not well established in Victoria and Tasmania [9, 10]. Although first reported in the northwest of Victoria in the original surveys by Lee et al. in 1984, there have only been sporadic detections of this species in the state-wide mosquito and arbovirus surveillance program (the Victorian Arbovirus Disease Control Program). More recently, two individuals were detected in 2005 in the Bass coast, a single individual in 2008 in Attwood, 17 in Wellington in 2009 and 8 individuals in Moira 2010 [11, 12], with this representing the last detection of this species until the recent 2017 trapping reported here.
Aedes camptorhynchus (known as the southern saltmarsh mosquito) has a similar habitat to Ae. vigilax and a sympatric distribution but prefers lower mean temperatures and is common in the coastal areas of Victoria and other parts of southern Australia [6]. Outbreaks of Ross River virus (RRV) [13,14,15] and Barmah Forest virus (BFV) [16] have been associated with a high abundance of this mosquito species. Previous records have indicated a potentially more extensive geographic distribution of Ae. camptorhynchus, with reported detections in non-coastal sites such as inland regions with high salinity, including Mildura in Northern Victoria and the Wheatbelt Valleys in Western Australia [17,18,19,20]. Morphologically, these two species are similar, apart from a few distinguishing characteristics such as pale scales on the wings and hook-shaped scales on the Ae. vigilax tergites [21], which can be easily missed if the specimens are damaged or in large collections. In addition to the morphological similarities to other species, Ae. vigilax occurs as a complex of three clades with morphological and molecular variations, with two of these occurring in Australia [22,23,24]. Variation in vector competence has been detected between Ae. vigilax populations' ability to transmit viruses and filarial parasites, with these variations possibly reflecting differences between clades [23, 25].
Molecular assays, such as conventional PCR [26] or PCR-restriction fragment length polymorphism (RFLP) [27], have widely been used to identify significant mosquito species. However, these techniques require a time-consuming visualization process after amplification and are not suited for rapid processing of traps containing large numbers of mosquitoes. The use of species-specific qPCR assays have been implemented to rapidly and effectively identify exotic species [28, 29] and inform public health risk assessments for arboviral diseases [30]. Sequencing-based detection of mosquitoes and the viruses they transmit in recent years has increased in popularity [31, 32]. However, in many cases, this can result in reduced sensitivity compared to qPCR due to the non-specific nature of these techniques and the need for higher quality DNA due to the larger fragments being targeted [29, 33, 34]. Methods that are based on the processing of whole mosquito traps can also have compromised detection sensitivity as a result of the presences of PCR inhibitors; however, rigorous method development for whole mosquito trap processing can overcome these inhibitors, as has been documented.
During the 2017/2018 mosquito season, Ae. vigilax was first detected in the saltmarsh areas of the Gippsland Lakes in Victoria, Australia, a region historically dominated by Ae. camptorhynchus. These detections led to expanded surveillance to understand the distribution of Ae. vigilax and determine whether this species had established in the area. Here, we present the development of a specific qPCR assay and a whole trap processing method that can be used to efficiently detect Ae. vigilax in whole trap collections. The phylogenetic relationship of the newly detected Ae. vigilax, with established Ae. vigilax clades was also investigated by sequencing three loci, one mitochondrial cytochrome c oxidase subunit 1 (COI) [35] and two nuclear genes, alpha amylase and the zinc finger gene [23]. This investigation provides further understanding of the occurrence of Ae. vigilax in Victoria as well as the development of a new molecular method for the detection of individual mosquitoes in whole traps.
Methods
Mosquito trapping
Mosquitoes were trapped using encephalitis virus surveillance (EVS) traps baited with dry ice pellets as a source of carbon dioxide [36]. The traps were set as part of the Victoria Arbovirus Disease Control Program, a Department of Health and Human Services funded program that supports local government to carry out larval and adult mosquito surveillance and vector control [37]. Traps were set once a week before dusk and collected after dawn, between September to April each year from 2017 to 2020 at a series of sites across Wellington Shire Council and East Gippsland Shire Council, Victoria, Australia (Fig. 1, Additional file 1: Table S1). After collection, mosquitoes were anesthetized by placing the catch bag into an esky with dry-ice for 30 min before transferring to a Petri dish. Petri dishes were sent to the laboratory by express post and on wet ice. Once received at the laboratory, samples were maintained at – 20 °C before processing. All mosquito samples tested in this study were field collected and stored under the same condition as are standard surveillance samples and hence are representative of the condition and quality of these samples. Insects were morphologically identified on a pre-chilled cold bench to preserve the quality of the specimens using a stereo-dissecting microscope and taxonomic keys [21, 38, 39].
Map of Australia and New Caledonia with dots representing Ae. vigilax collection sites, with sequences of individuals for each location included in the Ae. vigilax phylogenetic analysis. Yellow dots indicating Ae. vigilax collection sites for this study. Blue dots indicating sites from previous studies [23]. Trap locations include Victoria, East Gippsland (EAS) and Wellington (WEL); South Australia, St Kilda (SK), Port Adelaide (PA), Adelaide (AD) and Mypolonga (MY); New South Wales, Byron Bay (BY), Port Stephens (PS), Sydney (SY), Shellharbour (SH) and Batemans Bay (BA); Queensland, Cairns (CA); Northern Territory, Darwin (DA); Western Australia, Derby (DE), Broome (BR), Goegrup Lake (GL) and Mandurah (MA); New Caledonia, Noumea (NO). The map was created in QGIS v2.18.20, using ESRI Shapefiles for Australia [70] and New Caledonia [71]
Primers and probe design
Aedes vigilax primer and probes were designed using published sequences available in GenBank (JN228453-506, GQ143720-32 and MG242526), which included Ae. vigilax from each of the three clades and sequences generated in this study (GenBank: MW542561-71) from individuals collected in Victoria. Sequences were aligned using Mega v7.0.26 before trimming to a conserved region and exporting the alignment for primer selection [40]. PrimerHunter v1.0.2 was used to design the forward Vig_F_S1_9–5′–TTATCCCCTTTGTCATCTG–3′ and reverse Vig_R_S1_23–5′–AAGTAATTCCAGCAGATCGT–3′ primers targeting the COI with default parameters except for an optimal amplicon size of < 150 bp [41]. The probe was designed manually and in association with the PrimerHunter software, with a five prime FAM dye and a BHQ1 quencher, 5′- FAM-CATGCAGGAGCTTCAGTAG-BHQ1- 3′ [41]. The final amplicon size procedure by the assay was 149 bp. The designed primers and probe ability to detect all three clades of Ae. vigilax from multiple regions were also assessed by performing an in silico analysis. Primers and probe were aligned to the sequences outlined above in Mega v7.0.26 and analyzed for their ability to detect all three Ae. vigilax clades; in total, 5 sequences from clade I, 16 from clade II and 45 from clade III were compared.
Aedes vigilax qPCR assay parameters
qPCR primers and probe were optimized for concentration using Ae. vigilax extracted DNA over a ten-fold dilution series, with an optimal concentration of 400 nM for both primers (Sigma-Aldrich) and probe (Macrogen) determined. Aedes vigilax qPCR reactions were performed using TaqManTM Fast Universal PCR Master Mix (2X), no AmpEraseTM UNG (Applied Biosystems), with 10 µl of TaqMan 2X Universal PCR Master Mix, 1 µl of the primer-probe mix and 2.5 µl of DNA, made up to a total reaction volume of 20 µl with nuclease-free water. Inhibition in the qPCR reaction was assessed by adding 2 µl of VetMax Xeno Internal Positive Control DNA (Applied Biosystems) to each reaction. Detection of the Xeno positive control was performed as per the manufacturer’s instruction with the VetMax Xeno Internal Positive Control-VIC Assay (Applied Biosystems). No temple control was added to every qPCR run to assess for contamination and cross primer-dimer formation. The Ae. vigilax qPCR was performed on a QuantStudio 5 Real-Time PCR System (Applied Biosystems), with cycling conditions as follows: an initial denaturation at 95 °C for 20 s and 45 cycles at 95 °C for 10 s and 60 °C for 30 s. All qPCR reactions were performed in triplicate. Positives were classified as reactions that produced a cycle quantification (Cq) value < 38, equivocal results were classified between 38 and 40, and negative results of Cq 40 or greater. All data were analyzed using the QuantStudioTM Design and Analysis Software v1.4.3 with the Delta Rn threshold set at 0.05.
Mosquito pool preparation and processing
The mosquito pools used to assess the sensitivity of the Ae. vigilax assay were sourced from Wood Pile, Wellington, and consisted of Ae. camptorhynchus (98%) and Anopheles annulipes (s.l.) (2%). Mosquito pools were prepared by determining the weight of 50 mosquitoes, counted individually and then extrapolating the weight to obtain the required mosquito pools sizes of 199, 399, 599, 799 and 999. Pools were homogenized in 50 ml conical tubes containing a single 9.5-mm stainless steel grinding bead with 2 ml of MEM medium added (8% FBS, 0.1% amphotericin, 1% antibiotics [penicillin and streptomycin], 10% L-glutamine and 1% HEPES) per 100 mosquitoes. Homogenization was performed using a 2010 Geno/Grinder (Thomas Scientific) automated tissue homogenizer at two cycles of 1000 strokes/min for 1.5 min, with the samples kept on ice between cycles. An 80 µl aliquot was taken from each pool and tested in triplicate with the Ae. vigilax qPCR to confirm the absence of this species. Pools were subsequently spiked with a single Ae. vigilax and homogenized again as outlined above. A single Ae. vigilax in a volume of 20 ml of media was included as a positive control, representing the amount of media added to the maximum pool size (1000 mosquitoes) to assess the diluting factor of the media and any inhibitors present in the mosquito pools. Homogenized samples were clarified using a double centrifuge method: first, at 2000 g for 15 min, with the supernatant transferred into a clean tube, and then centrifuged for another 2000 g for 5 min.
DNA extraction
Eighty microliters of supernatant was extracted using a DNeasy Blood and Tissue kit (Qiagen) following the insect protocol. Homogenates were spiked with 2 µl of VetMax Xeno Internal Positive Control DNA (Applied Biosystems) to assess inhibition. Homogenates were incubated for 1 h with 180 µl of buffer ATL and 20 µl of Proteinase K at 56 °C before completing the extraction protocol as per the manufacturer’s instructions.
Individual Ae. vigilax for the phylogenetic analysis, qPCR efficiency and the 20 mosquito species used in the Ae. vigilax qPCR specificity study was extracted using the ISOLATE II Genomic DNA kit (Bioline). Individuals were incubated in 180 µl of Lysis Buffer GL and 25 µl of Proteinase K at 56 °C for 3 h before removing the individual and completing extraction protocol as per the manufacturer’s instructions, with the exception of the elution performed using 40 µl of preheated (70 °C) elution buffer G.
Aedes vigilax qPCR analytical specificity and analytical sensitivity
To assess the analytical specificity of the Ae. vigilax qPCR assay, a selection of the 20 mosquito species frequently detected in the region covering five genera were tested (Ae. alternans, Ae. bancroftianus, Ae. camptorhynchus, Ae. clelandi, Ae. flavifrons, Ae. imperfectus Dobrotworsky, Ae. notoscriptus, Ae. rubrithorax, Ae. sagax, Ae. theobaldi, Ae. vittiger, Anopheles annulipes, Coquillettidia linealis, Cx. annulirostris, Cx. australicus Dobrotworsky & Drummond, Cx. cylindricus, Cx. globocoxitus Dobrotworsky, Cx. molestus, Cx. quinquefasciatus Say and Tripteroides atripes. Analytical sensitivity of the Ae. vigilax assay was assessed by screening mosquito pools with increasing numbers of other mosquitoes (200, 400, 600, 800, 1000) which had a single Ae. vigilax added to them. Each mosquito pool was homogenized as outlined above, with three subsamples removed, extracted and tested with the qPCR in triplicate.
The efficiency of the qPCR assay was assessed by generating a standard curve with six ten-fold serial dilutions of an extracted Ae. vigilax individual. The Ae. vigilax individual was extracted as outlined above, with the DNA concentration determined by testing on a dsDNA HS Assay Kit on a QubitTM 2.0 (Invitrogen) fluorometer. A starting DNA concentration of 2.08 ng/µl was determined, with ten-fold serial dilutions performed in EB buffer (Qiagen), with the dynamic range of 2.08E-1 ng/µl to 2.08E-6 ng/µl being tested; 2.5 µl of each dilution was tested four times using the reaction setup outlined above, with a linear regression fit to the replicates. Data were analyzed in RStudio v4.0.2 [42] using the ggplot2 v3.21 and ggmisc [43] packages. The qPCR assay efficiency was derived from the slope of the standard curve, using the following equation E = − 1 + 10(-1/slope).
Phylogenetic analysis and haplotype networks
Phylogenetic analysis of Ae. vigilax was performed by amplifying the mitochondrial cytochrome c oxidase subunit 1 [35] and the two nuclear genes alpha amylase [23] and zinc finger [23]. Amplification was performed using MyTaq HS DNA Polymerase (Bioline), with 5 µl of 5 × MyTaq Reaction Buffer, 1 µl of the respective forward and reverse primer at 10 µM each, 0.5 µl of MyTaq HS DNA Polymerase and 5 µl of DNA with the reactions made up to 25 µl with nuclease-free water. Cycling conditions were as follows: 95 °C for 1 min, 40 cycles at 95 °C for 15 s, annealing at 49 °C, 56 °C or 56 °C for COI, alpha amylase or zinc finger, respectively, and extension at 72 °C for 10 s before a final extension at 72 °C for 2 min. PCR products were purified using the ISOLATE II PCR and Gel Kit (Bioline) before capillary sequencing using both the forward and reverse PCR primers for each gene.
Alignment of Ae. vigilax sequences from this and other studies [23] was performed using ClustalW in Mega v7.0.26 and trimmed to a consensus region of 591, 828 and 786 bp for COI, alpha amylase and zinc finger gene, respectively. Consensus regions for each gene were analyzed using jModelTest2 v2.1.10, topology taking the best of nearest neighbor interchange, subtree pruning and regrafting [44]. Akaike information criterion was used to select the most appropriate substitution model. Maximum-likelihood trees were constructed in PhyML v3.3.2 with 1000 bootstrap replicates; the proportion of gamma distribution and invariable sites were both estimated [45]. The general time-reversible (GTR) model was selected for all trees. Phylogenetic relationships of Ae. vigilax were further investigated through the construction of haplotype networks. Statistical parsimony networks were constructed in PopArt using the 591-bp region of COI with 95% connection limits in TCS 1.21 [46]. DnasSP v5 [47] was used to investigate the number of polymorphic sites, haplotype and nucleotide diversity. DnasSP was used to test for population expansion using the neutrality tests Tajima’s D [48] and Fu’s Fs [49]. Sequence data accession number can be found in Additional file 2: Table S2 and Additional file 3: Table S3.
Results
Repeat detection of Ae. vigilax between 2017 and 2020 season
Mosquito trapping over 2017 and 2020 identified the presence of Ae. vigilax in the East Gippsland and Wellington Shire councils (Fig. 1). The rediscovery of Ae. vigilax in these councils in recent years indicates the possible establishment of this mosquito species. Trapping records from 2017 to 2020 showed the maintained presences of this species in both regions (Fig. 2). Peak numbers of Ae. vigilax collected during a single catch night was 42 individuals from East Gippsland during the 2019–2020 season, with the largest catch night occurring around March of each trapping season. Aedes camptorhynchus numbers were higher than those of Ae. vigilax during the annual trapping period with peak numbers reaching 8452 in a single night with a mean trap collection of 204 (range: 0–1993) and 836 (range: 0–8452) individuals from East Gippsland and Wellington, respectively, collected (Fig. 2).
The occurrence of Ae. vigilax (stacked area chart) and Ae. camptorhynchus (bar chart) at one trapping site in East Gippsland (A) shire and one trapping site in Wellington (B) shire. Mosquito abundance for Ae. camptorhynchus is shown on the left axis and Ae. vigilax on the right axis. Trapping events consist of a single trap night with mosquitoes morphologically identified. Mosquito seasons are colored for 2017/2018 (orange), 2018/2019 (gray) and 2019/2020 (green)
Development and analytical specificity of the Ae. vigilax assay
Primers and probes were successfully designed and optimized to detect the presences of Ae. vigilax. The qPCR amplification efficiency of the Ae. vigilax assay was assessed and determined to have a good level of efficiency at 94.9% over six ten-fold dilution series, with the mean and standard deviation of each dilution displayed (Fig. 3). The assay was successful at detecting the sixth ten-fold dilution with a mean Cq value of 35.2 (log10 = 4.5 copies/reaction(rxn)); however, with a seventh dilution (log10 = 3.5 copies/rxn) (data not shown), not all replicates were detected indicating the limit of detection for the assay. The analytical specificity of the assay indicated that it is specific to Ae. vigilax with no cross-reaction detected between the 20 species tested covering five genera of mosquitoes. No amplification was detected in the no template controls. As no specimens from clade I or II were tested in this study, an in silico sequence analysis was performed with the designed primers and probe. Primers and probes were aligned to sequences covering individuals in all three clades and from multiple different regions. A maximum of two nucleotide mismatches were observed across the primers/probe regions when comparing individual sequences across the three clades and never more than one mismatch in the probe sequence.
Analytical sensitivity and assessment of the whole trap extraction technique using a Xeno internal control
The whole trap mosquito processing method and detection of Ae. vigilax were optimized and assessed for inhibition based on Xeno internal control spikes (Table 1). The homogenization of mosquitoes in 2 ml of media per 100 mosquitoes was determined to be optimal and resulted in no significant (ANOVA: F(5, 48) = 1.41, P = 0.23) detectable inhibition with increasing pool size (Table 1). The qPCR was shown to be effective in the detection of a single Ae. vigilax in the extrapolated mosquito pool sizes of 200, 400, 600, 800 and 1000 (Table 1). Three aliquots of each pool were processed and screened; each aliquot repeated in triplicates for qPCR testing with an average Cq value of 31.6 ± SD 2 observed across the five pool sizes. There was no significant difference in the Cq values of Ae. vigilax in the 200, 400 or 800 pool size (ANOVA: F(2, 24) = 0.37, P = 0.69) (Table 1). However, pool sizes of 600 and 1000 were three Cq values higher than the smaller pool sizes.
Phylogenetics of Victorian Ae. vigilax specimens
COI sequences were successfully obtained for 12 Ae. vigilax from the Wellington (n = 7) and East Gippsland (n = 5) region in Victoria from 2018 to 2019. Phylogenetic analysis based on COI of Ae. vigilax from Australia and New Caledonia identified that all Victorian specimens cluster in clade III (Fig. 4). Intraspecies divergence based on COI among the three clades varied from 0.5 to 4.1% (average, 2.4%) (Table 2). Within the Ae. vigilax group collected in Victoria, the COI divergence ranged from 0.2 to 1.4% (average, 0.8%), with no distinct clustering of Ae. vigilax from Wellington or East Gippsland observed (Figs. 4 and 5). All 12 Ae. vigilax from Victoria were also found to have between 99 and 100% identity based on COI to Ae. vigilax that had been previously collected from Sydney (NSW), Shellharbour (NSW), Byron Bay (NSW), Cairns (QLD), Broome (WA), Darwin (WA) and Derby (WA) (Fig. 1). This high level of sequence similarity indicates potential gene flow between these regions.
Aedes vigilax cytochrome c oxidase subunit 1, maximum-likelihood phylogenetic tree including sequences from the two capture locations within Victoria, denoted in boldface. All other sequences were obtained from Puslednik et al. [26]. Trap locations include Victoria, East Gippsland (EAS) and Wellington(WEL); South Australia, St Kilda (SK), Port Adelaide (PA), Adelaide (AD) and Mypolonga (MY); New South Wales, Byron Bay (BY), Port Stephens (PS), Sydney (SY), Shellharbour (SH) and Batemans Bay (BA); Queensland, Cairns (CA); Northern Territory, Darwin (DA); Western Australia, Derby (DE), Broome (BR), Goegrup Lake (GL) and Mandurah (MA); New Caledonia, Noumea (NO). Based on a 591-bp region of the COI gene. General time-reversible (GTR) substitution model was used with 1000 bootstrap replicates. Bootstrap proportions (BSP ≥ 70%) are indicated beside nodes. The number of nucleotide substitutions per site is represented by the scale bar. Aedes procax and Ae. theobaldi were used as an outgroup
Cytochrome oxidase subunit I haplotype network of Ae. vigilax sampled across Australia and New Caledonia. Dashes along lines represent the number of mutations; color of each circle represents the geographical location the insect was collected from. Trap locations include Victoria, East Gippsland (EAS) and Wellington VIC (WEL); South Australia, St Kilda (SK), Port Adelaide (PA), Adelaide (AD) and Mypolonga (MY); New South Wales, Byron Bay (BY), Port Stephens (PS), Sydney (SY), Shellharbour (SH) and Batemans Bay (BA); Queensland, Cairns (CA); Northern Territory, Darwin (DA); Western Australia, Derby (DE), Broome (BR), Goegrup Lake (GL) and Mandurah (MA); New Caledonia, Noumea (NO)
Across all sampling locations, both alpha amylase and the zinc finger (Additional file 4: Figure S1) region showed lower divergence on average compared to the COI region, with alpha amylase gene showing 0.1–3.9% (average, 1.3%) divergence (Table 2), and zinc finger 0–3.6% (average, 0.9%) (Table 2) between the clades. Upon comparison between the Ae. vigilax collected in Victoria, alpha amylase 0.4–1.3% (0.8%) and zinc finger 0–1.0% (average, 0.5%) showed a similar level of divergence to that of COI (Table 2). Phylogenetic analysis of alpha amylase and zinc finger failed to show any clear supported separation between the three clades (Additional file 4: Figure S1).
A haplotype network was used to further investigate the COI diversity, with three distinct groups observed. The haplotype network showed a star like pattern for clade III indicating population expansion. A high level of haplotype diversity was observed for COI [47] (Fig. 5, Table 3) as well as alpha amylase [63] and for the zinc finger [56] gene (Table 3). The highest level of nucleotide diversity was seen for the COI (Table 3) gene. Across all groups, the number of polymorphic sites was highest for clade III. For all clades, low nucleotide diversity but high haplotype diversity was observed, indicating only small differences occurring between the haplotypes. This is also highlighted in the COI haplotype networks with only a few nucleotide substitutions between haplotypes shown (Fig. 5). Neutrality test using Tajima’s D and Fu’s Fs were both negative and significant for COI clade III, with a similar result observed for the Ae. vigilax from Victoria, supporting the haplotype network and indicating past population expansion (Table 3).
Discussion
Recent years have seen an increase in the number of arboviral outbreaks occurring around the world as a result of the increased movement and establishment of significant vector species [50,51,52]. Complicating the detection of important vector species is that mosquito trapping can provide poor quality specimens and often many morphologically similar species that require highly specialized taxonomists to morphologically identify individual mosquitoes using a microscope. Molecular-based screening assays for the detection of mosquito species is an expanding area of research that has been successfully applied to detect a range of species [28, 31, 53]. Although Ae. vigilax occurs throughout most states of Australia [23] as well as in New Caledonia [23], Seychelles, Fiji [54], the Philippines, Indonesia, Malaysia, Taiwan and Timor-Leste [55], there is currently no specific molecular assay for the detection of this species. We have developed a specific and sensitive Ae. vigilax qPCR assay that can be used to screen whole mosquito traps and successfully detect a single Ae. vigilax among 1000 mosquitoes of other species. The specificity of the Ae. vigilax qPCR to detect all three clades of Ae. vigilax was assessed by in-silico analysis aligning multiple individuals from all three clades to the primer and probe sequences. A maximum of two mismatches across these three regions was seen for some individuals, but never more than one mismatch in the probe region indicating there would be successful binding, amplification and detection of all three clades. The Ae. vigilax assay was also assessed against 20 mosquito species covering five genera commonly collected in Victoria with no amplification detected, highlighting the specificity of the assay.
During mosquito season peak collections in excess of 20,000 mosquitoes can be collected in a single trap night in coastal areas, requiring subsampling of traps, increasing the likelihood that taxonomists may miss this important vector species [56]. The whole trap extraction methodology developed in this study is an accurate and efficient way to process mosquito traps of up to 1000 insects, which typically takes an experienced taxonomist up to 45 min to process. Previous studies have identified that whole trap extraction of arthropods can be negatively impacted by PCR inhibitors, affecting PCR amplification and detections [57, 58]. However, our results show that through the assessment of spiked exogenous internal positive controls, PCR inhibition can be negated for by proportionally increasing homogenization media, thereby ensuring detection sensitivity. The Ae. vigilax qPCR assay efficacy was tested by performing a six ten-fold serial dilution through a dynamic range of 2.08E-1 ng/µl to 2.08E-6 ng/µl (9.5 to 4.5 log10 copies/rxn) (Fig. 3) being tested. The assay was determined to have 94.9% efficiency. A seventh dilution was performed (3.5 log10 copies/rxn); however, not all replicates were detected, indicating the limit of detection for the assay.
Detection sensitivity of the assay was assessed by spiking pools of mosquitoes with a single Ae. vigilax. Aedes vigilax was successfully detected in all mosquito pool sizes of 200, 400, 600, 800 and 1000. There was no difference in the Cq vale for the detection of Ae. vigilax in pools sizes of 200, 400 and 800. However, there was a three Cq value increase for detections of Ae. vigilax in pools sizes of 600 and 1000. As majority traps collected during the state-wide surveillance program are below 1000 mosquitoes (Fig. 2), this highlights the suitability of this assay for routine screening of whole mosquito traps.
Aedes vigilax has been historically detected in Victoria on several occasions during the annual mosquito trapping program of the VADCP [11, 12]. However, in this study, we present the possible establishment of this mosquito species in two coastal regions of Victoria with this species being detected over 3 consecutive years. Aedes vigilax numbers over the 3 years typically peaked in mid-February to March (Fig. 2), with a similar trend observed in Ae. vigilax populations from South Australia [6] and New South Wales [59]. In general, the overall capture number of Ae. vigilax individuals was low compared to trapping in other states, which can capture ~ 20,000 individuals in a single trap night [56]. The lower number of Ae. vigilax collected in Victoria compared to the northern states may also be a result of factors such as tidal inundation events of breeding sites, rainfall [60] and temperature [61]. Additionally, many of the trapping sites are historical and were not moved in this study; hence, there may be more optimal trap locations closer to larval habitats. Additional methods could also be used to increase the chance of trapping Ae. vigilax in the area such as adding 1-octen-3-ol lures to the CO2-baited EVS traps, which has been shown to significantly increase collections of Ae. vigilax [63,64,64].
Phylogenetic analysis of Ae. vigilax identified that individuals collected from Victoria were positioned in clade III. COI sequences obtained from Victorian Ae. vigilax had up to 100% sequencing identity to those Ae. vigilax previously collected from a range of locations including Sydney (NSW), Shellharbour (NSW), Byron Bay (NSW), Cairns (QLD), Broome (WA), Darwin (WA) and Derby (WA) [23], indicating that one of these sites might have been the source of the original introduction to Victoria (Fig. 1). No phylogenetic separation was seen between Ae. vigilax collected from the two capture sites in Victoria, as has been observed for different locations across Australia [25]. This was not surprising because of the relative proximity between these two populations (closest trapping sites approximately 12 km apart). Ae. vigilax are capable of flying large distances, having been recorded to travel up to 9 km from their larval habitats [24], and previous reports of wind-borne dispersal of up to 50 km [55], highlight the potential of movement and mixing between these two Victorian populations. A high level of haplotype diversity was identified within the COI sequences of Victorian specimens with 12 haplotypes found (Table 3). The higher number of haplotypes and the shared sequence variation to the Ae. vigilax clade III have been seen in previous studies [23]. Neutrality test indicates that clade III has gone through a population expansion, which was also seen when investigating the Victorian Ae. vigilax individuals within this clade (Table 3). Puslednik et al. [26] identified that clade II and III subdivided many years ago and have developed separate lineages, followed by secondary contact and the current sympatric distribution of clade II and III [25]. This work is supported by a study performed by Shibani [25], which used microsatellite data to show there is no reproductive isolation between these Ae. vigilax clades [65].
Aedes camptorhynchus and Ae. vigilax are the primary vector species of RRV in coastal areas of Australia [10]. Currently, in Victoria Ae. camptorhynchus is the primary vector species of RRV along the coastline [14]. If Ae. vigilax successfully established in this area, it could result in changes to virus transmission dynamics. Previous studies have indicated that Ae. vigilax is a more competent vector of RRV compared to Ae. camptorhynchus [66]. Aedes vigilax has experimentally also been shown to be a more efficient vector compared with other mosquitoes in its ability to transmit BFV [67] and CHIKV [4]. If Ae. vigilax becomes more widespread and abundant across Victoria, this could increase the length of the RRV transmission season. Previous studies in South Australia have highlighted the occurrence of seasonal succession with Ae. camptorhynchus occurring from spring to early summer and Ae. vigilax occurring from mid-late summer and autumn [20]; with the establishment of Ae. vigilax in Victoria, a similar extended RRV transmission season could occur.
Conclusions
Here we present the development of a specific and sensitive molecular assay for the detection of Ae. vigilax from whole mosquito traps. An optimized processing method was developed to screen up to 1000 mosquitoes without compromising detection sensitivity, allowing for rapid and specific detection of this species in surveillance samples. Further development and validation of this assay could allow the identification of the different life stages of Ae. vigilax and potentially enable detection of this mosquito species larvae in water samples, as has been outlined by previous studies [68]. Aedes vigilax collected in Victoria were seen to have a high sequence identity to those of clade III, with no separation seen between individuals from the two capture locations in Victoria, indicating mixing between these populations. Globally, recent years have seen the increased movement and establishment of mosquito species in new regions. At times these new establishments can result in increased disease burden in these areas [69]. This highlights the need to implement accurate and rapid testing techniques that can be used alongside traditional mosquito surveillance programs to detect the presences of significant mosquito vector species such as Ae. vigilax.
Availability of data and materials
All sequences generated in this study are publicly available from NCBI under accession numbers MW 351796-819 and MW542560-71.
Abbreviations
- Ae:
-
Aedes
- BFV:
-
Barmah Forest virus
- CHIKV:
-
Chikungunya virus
- CO2 :
-
Carbon dioxide
- COI :
-
Cytochrome c oxidase subunit 1
- Cx:
-
Culex
- Cq:
-
Cycle quantification
- EVS:
-
Encephalitis virus surveillance
- GTR:
-
general time-reversible
- NSW:
-
New South Wales
- PCR:
-
Polymerase chain reaction
- QLD:
-
Queensland
- qPCR:
-
Quantitative polymerase chain reaction
- RFLP:
-
Restriction fragment length polymorphism
- RRV:
-
Ross River virus
- SD:
-
Standard deviation
- WA:
-
Western Australia
References
Jansen CC, Webb CE, Graham GC, Craig SB, Zborowski P, Ritchie SA, et al. Blood sources of mosquitoes collected from urban and peri-urban environments in eastern Australia with species–specific molecular analysis of avian blood meals. Am J Trop Med Hyg. 2009;81:849–57.
Stephenson EB, Murphy AK, Jansen CC, Peel AJ, McCallum H. Interpreting mosquito feeding patterns in Australia through an ecological lens: an analysis of blood meal studies. Parasit Vectors. 2019;12:156.
Jansen CC, Williams CR, van den Hurk AF. The usual suspects: comparison of the relative roles of potential urban chikungunya virus vectors in Australia. PLoS One. 2015;10:e0134975.
van den Hurk AF, Hall-Mendelin S, Pyke AT, Smith GA, Mackenzie JS. Vector competence of Australian mosquitoes for chikungunya virus. Vector Borne Zoonotic Dis. 2010;10:489–95.
van den Hurk AF, Nisbet DJ, Hall RA, Kay BH, MacKenzie JS, Ritchie SA. Vector competence of Australian mosquitoes (Diptera: Culicidae) for Japanese encephalitis virus. J Med Entomol. 2003;40:82–90.
Kokkinn MJ, Duval DJ, Williams CR. Modelling the ecology of the coastal mosquitoes Aedes vigilax and Aedes camptorhynchus at Port Pirie, South Australia. Med Vet Entomol. 2009;23:85–91.
Jacups SP, Kurucz N, Whelan PI, Carter JM. A comparison of Aedes vigilax larval population densities and associated vegetation categories in a coastal wetland, Northern Territory, Australia. J Vector Ecol. 2009;34:311–6.
Knight JM. A model of mosquito–mangrove basin ecosystems with implications for management. Ecosystems. 2011;14:1382–95.
Gislason GM, Russell RC. Oviposition sites of the saltmarsh mosquito, Aedes vigilax (Skuse) (Diptera: Culicidae), at Homebush Bay, Sydney, NSW—a preliminary investigation. Aust J Entomol. 1997;36:97–100.
Russell RC. Ross River virus: ecology and distribution. Annu Rev Entomol. 2002;47:1–31.
Andrews K, Brown K, Azuolas J. Victorian Arbovirus Disease Control Program Annual Report 2009/2010. Department of Primary Industries Victoria; 2010.
Azuolas J, Brown K, Wishart E. Victorian Arbovirus Disease Control Program mosquito monitoring report 2005/2006. Department of Primary Industries Victoria; 2006.
Barton PS, Weaver HJ. Mosquito (Diptera: Culicidae) and rainfall associations with arbovirus disease in eastern Victoria. Trans R Soc S Aust. 2009;133:257–64.
Dhileepan K, Peters C, Porter A. Prevalence of Aedes camptorhynchus (Thomson) (Diptera: Culicidae) and other mosquitoes in the eastern coast of Victoria. Aust J Entomol. 1997;36:183–90.
Cutcher Z, Williamson E, Lynch SE, Rowe S, Clothier HJ, Firestone SM. Predictive modelling of Ross River virus notifications in southeastern Australia. Epidemiol Infect. 2017;145:440–50.
Passmore J, O’Grady KA, Moran R, Wishart E. An outbreak of Barmah Forest virus disease in Victoria. Commun Dis Intell. 2002;26:600–4.
Carver S, Spafford H, Storey A, Weinstein P. Colonization of ephemeral water bodies in the wheatbelt of Western Australia by assemblages of mosquitoes (Diptera: Culicidae): role of environmental factors, habitat, and disturbance. Environ Entomol. 2009;38:1585–94.
Wishart E. Species composition and population studies of mosquitoes (Diptera: Culicidae) in the Mildura district in the Murray Valley of southern Australia. Aust J Entomol. 2002;41:45–8.
Lee DJ, Hicks MM, Griffiths RC, Russell RC. Entomology monograph no. 2 the Culicidae of the Australiasian region, vol. 7. Canberra: Australian Government Publishing Service; 1987.
Williams CR, Williams SR, Nicholson J, Little SM, Riordan J, Fricker SR, et al. Diversity and seasonal succession of coastal mosquitoes (Diptera: Culicidae) in the northern Adelaide region of South Australia. Aust J Entomol. 2009;48:107–12.
Russell RC. A colour photo atlas of mosquitoes of Southeastern Australia. The Department of Medical Entomology, Westmead Hospital and University of Sydney. University of Sydney Printing Service, Sydney. 1996.
Ballard JWO, Puslednik L, Wolff JN, Russell RC. Variation under nature: a sesquicentennial DNA barcoding perspective. Chiang Mai J Sci. 2009;36:188–99.
Puslednik L, Russell RC, Ballard JWO. Phylogeography of the medically important mosquito Aedes (Ochlerotatus) vigilax (Diptera: Culicidae) in Australasia. J Biogeogr. 2012;39:1333–46.
Chapman HF, Hughes JM, Jennings C, Kay BH, Ritchie SA. Population structure and dispersal of the saltmarsh mosquito Aedes vigilax in Queensland, Australia. Med Vet Entomol. 1999;13:423–30.
Ryan PA, Do KA, Kay BH. Definition of Ross River virus vectors at Maroochy Shire, Australia. J Med Entomol. 2000;37:146–52.
Smith JL, Fonseca DM. Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: Culicidae). Am J Trop Med Hyg. 2004;70:339–45.
Alam MT, Das MK, Dev V, Ansari MA, Sharma YD. PCR-RFLP method for the identification of four members of the Anopheles annularis group of mosquitoes (Diptera: Culicidae). Trans R Soc Trop Med Hyg. 2007;101:239–44.
Hill LA, Davis JB, Hapgood G, Whelan PI, Smith GA, Ritchie SA, et al. Rapid identification of Aedes albopictus, Aedes scutellaris, and Aedes aegypti life stages using real-time polymerase chain reaction assays. Am J Trop Med Hyg. 2008;79:866–75.
van de Vossenberg BT, Ibanez-Justicia A, Metz-Verschure E, van Veen EJ, Bruil-Dieters ML, Scholte EJ. Real-time PCR tests in Dutch exotic mosquito surveys; implementation of Aedes aegypti and Aedes albopictus identification tests, and the development of tests for the identification of Aedes atropalpus and Aedes japonicus japonicus (Diptera: Culicidae). J Med Entomol. 2015;52:336–50.
Shahhosseini N, Kayedi MH, Sedaghat MM, Racine TP, Kobinger G, Moosa-Kazemi SH. DNA barcodes corroborating identification of mosquito species and multiplex real-time PCR differentiating Culex pipiens complex and Culex torrentium in Iran. PLoS One. 2018;13:e0207308.
Batovska J, Lynch SE, Cogan NOI, Brown K, Darbro JM, Kho EA, et al. Effective mosquito and arbovirus surveillance using metabarcoding. Mol Ecol Resour. 2018;18:32–40.
Batovska J, Mee PT, Lynch SE, Sawbridge TI, Rodoni BC. Sensitivity and specificity of metatranscriptomics as an arbovirus surveillance tool. Sci Rep. 2019;9:19398.
Schneider J, Valentini A, Dejean T, Montarsi F, Taberlet P, Glaizot O, et al. Detection of invasive mosquito vectors using environmental DNA (eDNA) from water samples. PLoS One. 2016;11:e0162493.
Harper LR, Lawson Handley L, Hahn C, Boonham N, Rees HC, Gough KC, et al. Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol Evol. 2018;8:6330–41.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Rohe D, Fall RP. A miniature battery powered CO2 baited light trap for mosquito borne encephalitis surveillance. Bull Soc Vect Ecol. 1979;4:24–7.
Knope K, Doggett SL, Jansen CC, Johansen CA, Kurucz N, Feldman R, et al. Arboviral diseases and malaria in Australia, 2014–15: Annual report of the National Arbovirus and Malaria Advisory Committee. Commun Dis Intell. 2019;43:1–69.
Dobrotworsky NV. The mosquitoes of Victoria (Diptera, Culicidae). London: Melbourne University Press; 1965.
Liehne PFS. An atlas of the mosquitoes of Western Australia. Perth: Health Department of Western Australia; 1991.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Duitama J, Kumar DM, Hemphill E, Khan M, Mandoiu II, Nelson CE. PrimerHunter: a primer design tool for PCR-based virus subtype identification. Nucleic Acids Res. 2009;37:2483–92.
RStudio Team. RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. 2020. http://www.rstudio.com/. Accessed 18 Jan 2021.
Wickham H. Elegant graphics for data analysis. In: Wickham H, editor. ggplot2. New York: Springer; 2016.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704.
Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–9.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585.
Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–25.
Pagès F, Peyrefitte CN, Mve MT, Jarjaval F, Brisse S, Iteman I, et al. Aedes albopictus mosquito the main vector of the 2007 Chikungunya outbreak in Gabon. PLOS One. 2009;4:e4691-e.
Grandadam M, Caro V, Plumet S, Thiberge JM, Souarès Y, Failloux A-B, et al. Chikungunya virus, southeastern France. Emerg Infect Dis. 2011;17:910–3.
Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–6.
Batovska J, Blacket MJ, Brown K, Lynch SE. Molecular identification of mosquitoes (Diptera: Culicidae) in southeastern Australia. Ecol Evol. 2016;6:3001–11.
Burnett GF. The arrival of Aedes (Ochlerotatus) vigilax (Skuse) in Fiji. Pac Sci. 1960;14:389–94.
Lee DJ, Hicks MM, Griffiths M, Russell RC, Marks EN. The Culicidae of the Australasian region, vol. 3. Canberra: Australian Government Publishing Service; 1984.
Webb CE, Russell RC. Dispersal of the mosquito Aedes vigilax (Diptera: Culicidae) from urban estuarine wetlands in Sydney, Australia. J Med Entomol. 2019;56:1290–5.
Murray DC, Coghlan ML, Bunce M. From benchtop to desktop: important considerations when designing amplicon sequencing workflows. PLoS One. 2015;10:e0124671.
Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors—occurrence, properties and removal. J Appl Microbiol. 2012;113:1014–26.
Webb CE, Russell RC. Towards management of mosquitoes at Homebush Bay, Sydney, Australia. I. Seasonal activity and relative abundance of adults of Aedes vigilax, Culex sitiens, and other salt-marsh species, 1993–94 through 1997–98. J Am Mosq Control Assoc. 1999;15:242–9.
Yang G-J, Brook BW, Whelan PI, Cleland S, Bradshaw CJA. Endogenous and exogenous factors controlling temporal abundance patterns of tropical mosquitoes. Ecol Appl. 2008;18:2028–40.
Kay B, Jennings C. Enhancement or modulation of the vector competence of Ochlerotatus vigilax (Diptera: Culicidae) for Ross River virus by temperature. J Med Entomol. 2002;39:99.
Kemme JA, Van Essen PHA, Ritchie SA, Kay BH. Response of mosquitoes to carbon dioxide and 1-octen-3-ol in southeast Queensland Australia. J Am Mosq Control Assoc. 1993;9:431–5.
Van Essen PHA, Kemme JA, Ritchie SA, Kay BH. Differential responses of Aedes and Culex mosquitoes to octenol or light in combination with carbon dioxide in Queensland, Australia. Med Vet Entomol. 1994;8:63–7.
Ritchie SA, Kline DL. Comparison of CDC and EVS light traps baited with carbon dioxide and octenol for trapping mosquitoes in Brisbane, Queensland (Diptera: Culicidae). Aust J Entomol. 1995;34:215–8.
Shibani NS. An investigation of patterns of connectivity among populations of the Australian mosquito (Aedes vigilax) using mitochondrial sequences and microsatellite loci [PhD Doctorate]. Brisbane: Griffith University; 2009.
Ballard JW, Marshall ID. An investigation of the potential of Aedes camptorhynchus (Thom.) as a vector of Ross River virus. Aust J Exp Biol Med Sci. 1986;64:197–200.
Boyd AM, Kay BH. Vector Competence of Aedes aegypti, Culex sitiens, Culex annulirostris, and Culex quinquefasciatus (Diptera: Culicidae) for Barmah Forest virus. J Med Entomol. 2000;37:660–3.
Odero J, Gomes B, Fillinger U, Weetman D. Detection and quantification of Anopheles gambiae sensu lato mosquito larvae in experimental aquatic habitats using environmental DNA (eDNA). Wellcome Open Res. 2018;3:26.
Schaffner F, Medlock JM, Bortel WV. Public health significance of invasive mosquitoes in Europe. Clin Micobiol Infect. 2013;19:685–92.
Australian Bureau of Statistics. Australian Standard Geographical Classification (ASGC) Urban Centres and Localities (UC/L) Digital Boundaries, Australia, 2006. 2007. https://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/1259.0.30.0032006?OpenDocument#Publications. Accessed 3 May 2021.
GADM. New Caledonia Shapefile v2.8. 2015. https://gadm.org/. Accessed 3 May 2021.
Acknowledgements
We acknowledge the Victorian Arbovirus Disease Control Program at the Department of Health and Human Services, East Gippsland Shire Council and Wellington Shire Council for funding the mosquito field surveillance. We thank Rebecca Feldman from the Department of Health and Human Services and Dr. Fiona Constable from Agriculture Victoria Research for their support of this project. We also thank Barry Curtain from Wellington Shire Council and Kate Allen from East Gippsland Shire Council and their teams for performing the mosquito trapping.
Funding
Agriculture Victoria Research funded this work. The mosquito trapping was performed as part of the Victorian Arbovirus Disease Control Program, which is funded by the Department of Health and Human Services.
Author information
Authors and Affiliations
Contributions
Conceptualization: PM, SW, SL. Writing and preparation of original drafts: PM, SW, KB, SL. Visualization, validation, formal analysis and data curation: PM, SW, SL. Methodology: PM, SW, KB, SL. Investigation: PM, SL. Writing, reviewing and editing: PM, SW, KB, SL. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Aedes vigilax species information used in the multiple locus typing.
Additional file 2:
Table S2. Aedes vigilax NCBI reference numbers.
Additional file 3:
Table S3. Aedes procax and Aedes theobaldi NCBI reference numbers.
Additional file 4:
Figure S1. Phylogenetic analysis of Ae. vigilax based on an 828-bp region of the alpha amalyse (a) and 786 bp region of the zinc finger (b) gene. Maximum-likelihood phylogenetic tree including sequences from the Wellington (WEL) and East Gippsland (EAS) capture locations within Victoria, denoted in boldface. All other sequences were obtained from Puslednik et al. [26]. General time-reversible (GTR) substitution model was used for both trees with 1000 bootstrap replicates. Bootstrap proportions (BSP ≥ 70%) are indicated beside nodes. The number of nucleotide substitutions per site is represented by the scale bar. Aedes procax and Ae. theobaldi were used as an outgroup.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mee, P.T., Wong, S., Brown, K. et al. Quantitative PCR assay for the detection of Aedes vigilax in mosquito trap collections containing large numbers of morphologically similar species and phylogenetic analysis of specimens collected in Victoria, Australia. Parasites Vectors 14, 434 (2021). https://doi.org/10.1186/s13071-021-04923-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-04923-y