Abstract
Background
Previously, twelve Rickettsia species were identified in ticks, fleas, sheep keds (Melophagus ovinus), bats (Pipistrellus pipistrellus) and a tick-bitten patient in the Xinjiang Uygur Autonomous Region (XUAR) in northwestern China. Here we aimed to molecularly detect rickettsial agents in red fox (Vulpes vulpes), marbled polecat (Vormela peregusna) and their ticks.
Methods
During 2018–2019, 12 red foxes, one marbled polecat and their ticks were sampled in two counties and a city of the XUAR. The heart, liver, spleen, lung and kidney of these 13 carnivores were dissected, followed by DNA extraction. Hard ticks were identified both morphologically and molecularly. All samples were examined for the presence of rickettsiae by amplifying four genetic markers (17-kDa, gltA, ompA, sca1).
Results
A total of 26 adult ticks and 28 nymphs (38 Ixodes canisuga, nine Ixodes kaiseri, six Haemaphysalis erinacei and one Dermacentor marginatus) were collected from red foxes, and four Ha. erinacei ticks were removed from the marbled polecat. Analysis of cytochrome c oxidase subunit I (COI) gene sequences indicated that 2–32 nucleotides differed between I. canisuga, I. kaiseri and Ha. erinacei from northwestern China and Europe. Rickettsia raoultii was detected in three red foxes, Candidatus Rickettsia barbariae in a red fox, Rickettsia sibirica in a red fox and a marbled polecat, and R. raoultii in two tick species (I. canisuga and D. marginatus).
Conclusions
To the best of our knowledge, I. canisuga and I. kaiseri have not been previously reported from red foxes in China. The DNA of R. sibirica and R. raoultii was detected for the first time in the organs of red foxes, and R. sibirica in the organs of a marbled polecat. This is also the first molecular evidence for the presence of R. raoultii in I. canisuga. Our findings expand the range of tick-borne pathogens in wildlife species and associated ticks in China.
Graphical Abstract

Similar content being viewed by others
Background
The red fox (Vulpes vulpes) is widely distributed throughout Europe, Asia, North Africa and North America [1]. Its habitats strongly overlap with those of other wildlife species, domestic animals and even humans [2]. Previously, red foxes were reported to harbor several vector-borne pathogens of public health concern, including tick-borne encephalitis virus [3], Borrelia burgdorferi [4], Ehrlichia canis [5], Leishmania infantum [6], Hepatozoon canis [7] and Babesia vulpes [8, 9]. A serological investigation of red foxes in Spain indicated that 50.3% had antigens of spotted fever group rickettsiae, including Rickettsia massiliae and R. conorii [10]. In addition, immunofluorescence assay showed that 1.9% of red foxes had antibodies to R. typhi, and 6.7% to R. slovaca in Spain [4].
The geographic range of the marbled polecat (Vormela peregusna) covers Central Asia, northwestern China and Europe [2]. In studies on its epidemiological role, seroconversion to plague F1 antigen was detected in a marbled polecat in the Xinjiang Uygur Autonomous Region (XUAR) [11] in northwestern China. Borrelia burgdorferi sensu lato and Babesia sp. were molecularly identified in a marbled polecat in Romania and China, respectively [12, 13]. Furthermore, R. raoultii and Candidatus Rickettsia barbariae were molecularly identified in marbled polecats in the XUAR [13].
In the temperate climate zone, hard ticks (Acari: Ixodidae) are regarded as the most important vectors of pathogens [14]. Among these, Ixodes persulcatus, I. ricinus, I. hexagonus, I. kaiseri, I. canisuga, Dermacentor reticulatus, D. marginatus, Haemaphysalis punctata and Rhipicephalus sanguineus have been reported from red foxes [10, 15,16,17]. In Spain, R. massiliae, R. aeschlimannii and R. slovaca were detected in red fox ticks [10]. In addition, Ha. erinacei from marbled polecats contained the DNA of R. raoultii in China [18].
In China, at least 19 spotted fever group (SFG) rickettsial species have been detected in ticks, including R. heilongjiangensis, R. hulinii, R. mongolotimonae, R. sibirica [19], R. raoultii, R. slovaca [20], R. felis [21], R. aeschlimannii, R. massiliae [22], R. monacensis [23], R. japonica [24], Candidatus R. barbariae, R. conorii [25], R. parkeri, R. lusitaniae, R. rickettsii [26], Candidatus R. jingxinensis, Candidatus R. tarasevichiae [27] and Candidatus R. leptotrombidium [28]. The aim of the present study was to investigate rickettsial agents in 12 red foxes, a marbled polecat and their ticks in China.
Methods
Sample collection and species identification
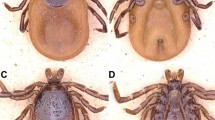
A total of 12 red foxes killed by illegal hunting or found as roadkill and one marbled polecat that died of natural causes were sampled in two counties and a city of the XUAR during 2018–2019 (data shown in Additional file 1). The red foxes and the marbled polecat were morphologically identified by an experienced zoologist. The heart, liver, spleen, lung and kidney of all 13 carcasses were removed. Simultaneously, the entire body surface of each individual was checked for ticks, all of which were removed. The ticks were morphologically identified to the species level according to the standard taxonomic keys as described previously [29]. This was also confirmed by molecular and phylogenetic analyses based on two mitochondrial markers, the 16S rRNA and the cytochrome c oxidase subunit I (COI) genes [17].
Detection, sequencing and phylogenetic analysis of rickettsiae
Genomic DNA was extracted from the organs (heart, liver, spleen, lung and kidney) of wild carnivores as well as from their ticks using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). To investigate the presence of rickettsiae in ticks, four genetic markers were targeted, including the 17 kDa antigen (17-kDa), citrate synthase (gltA), outer membrane protein A (ompA) and surface cell antigen 1 (sca1) genes. Two genes (gltA and ompA) were used to detect rickettsiae in the organs of wild carnivores [30]. The primers and polymerase chain reaction (PCR) cycling conditions used in this study are shown in Additional file 2. Each PCR assay included a negative control (distilled water instead of DNA template) and a positive control (containing sequence-verified DNA of R. massiliae from Rh. turanicus ticks collected in the XUAR) [22]. Purification and sequencing of the PCR products were performed as described previously [31, 32]. Sequences were manually edited, aligned and compared to reference GenBank sequences by the nucleotide BLASTn program (https://blast.ncbi.nlm.nih.gov). A phylogenetic tree was constructed using the maximum likelihood method in MEGA 7.0 software [18].
Results
Tick identification
A total of 26 adult ticks and 28 nymphs (i.e., 38 I. canisuga, 9 I. kaiseri, 6 Ha. erinacei and 1 D. marginatus) were collected from 12 red foxes, and four Ha. erinacei ticks were found on the marbled polecat. Morphological features are shown in Additional file 3: Fig. S1.
Molecular and phylogenetic analyses
Analysis of COI sequences revealed 2–32 nucleotide differences in the case of I. canisuga (3–6 bp), I. kaiseri (2–7 bp) and Ha. erinacei (30–32 bp) between Europe and China. Phylogenetic analysis showed that (i) I. canisuga in the XUAR was in a basal position to 11 European haplotypes (“A to K”) [17] (Fig. 1a); (ii) I. kaiseri from red foxes in the XUAR was also in basal position to nine European haplotypes (“L to T”), and had an identical sequence with conspecific ticks from long-tailed ground squirrels and Asian badgers [17, 30, 33] (Fig. 1b); (iii) Ha. erinacei from red foxes and marbled polecat had identical sequences, and formed a distinct clade from those reported in Turkey, Italy and Romania (Fig. 1c); and (iv) D. marginatus from red fox #2 had an identical sequence with the off-host tick collected formerly in Altaw City, XUAR.
Phylogenetic tree based on COI sequences of ticks collected from 12 red foxes and a marbled polecat in northwestern China. The evolutionary history was inferred using the maximum likelihood method (bootstrap replicates: 1000) with MEGA 7.0. New sequences obtained in this study are indicated by black triangles. a Ixodes canisuga, b Ixodes kaiseri, c Haemaphysalis erinacei and d Dermacentor marginatus
Red foxes #3, #5 and #11 were positive for R. raoultii, and red fox #8 was positive for Candidatus R. barbariae. At the same time, red fox #12 and the marbled polecat were positive for R. sibirica. In addition, R. raoultii was detected in I. canisuga from red fox #11 (Manas County) and D. marginatus from red fox #2 (Nilka County). Nucleotide sequences of rickettsial agents were deposited in the GenBank database (MT890502-MT890525). Phylogenetic analyses are shown in Fig. 2 and Additional file 4: Fig. S2.
Discussion
Molecular studies on I. canisuga and I. kaiseri have been reported mostly from Europe, where these tick species typically infest Eurasian badgers (Meles meles), red foxes (Vulpes vulpes), steppe polecats (Mustela eversmanni), raccoon dogs (Nyctereutes procyonoides), hedgehogs (Erinaceus europaeus) and domestic dogs. Among these, dogs and red foxes can also be co-infested with I. canisuga and I. kaiseri [17, 34]. In this study, I. canisuga and I. kaiseri were found on red foxes, as also confirmed by 16S rRNA gene sequences (GenBank: MT889694-MT889698 and MT889701-MT889705). To the best of our knowledge, I. canisuga and I. kaiseri have been discovered here for the first time on red foxes in China. Phylogenetic analysis of the COI gene showed that (i) I. canisuga specimens collected from the same host species (red fox) were genetically different between China (MT890495-MT890498) and Germany (KY962044-KY962045), Croatia (KY962037-KY962040), Bosnia-Herzegovina (KY962016-KY962017), Serbia (KY962030-KY962031) and Romania (KY962025, KY962021-KY962023); (ii) I. kaiseri from red foxes and long-tailed ground squirrels in the XUAR had 100% identity, but clustered in a separate phylogenetic position compared to European ticks; and (iii) Ha. erinacei infesting the red fox and marbled polecat in the XUAR shared identical COI gene sequences (MT890493 and MT890494), but differed from those collected in Italy, Turkey and Romania (Fig. 1c). These findings support the contention that the genetic diversity of I. kaiseri, I. canisuga and Ha. erinacei might reflect geographic distribution rather than host associations, consistent with Klompen et al. [35].
In previous reports, R. helvetica was detected in the blood sample of a red fox in Switzerland [36]. In addition, R. raoultii and Candidatus R. barbariae were reported in a marbled polecat in Altaw City, the XUAR [13]. R. raoultii was also found in the tissues of bats (Pipistrellus pipistrellus) [26]. Here, R. raoultii was detected for the first time in the organs of red foxes, and R. sibirica in the lung and kidney of a marbled polecat and in the liver of a red fox.
Moreover, the DNA of R. raoultii was shown to be present in I. canisuga. Previously, R. raoultii has been detected in at least 15 different tick species, including D. nuttalli, D. silvarum, D. marginatus, D. reticulatus, I. kaiseri, I. persulcatus, I. boliviensis, I. ricinus, Ha. concinna, Ha. japonica, Ha. erinacei, Rh. bursa, Rh. sanguineus, Hyalomma asiaticum and Amblyomma maculatum [13, 18, 20, 27, 30, 37,38,39,40,41,42,43,44]. On the other hand, R. sibirica has been detected in at least 12 different tick species, including Rh. turanicus, Rh. pusillus, Hy. asiaticum, Hy. truncatum, Hy. wellingtoni, Hy. yeni, D. marginatus, D. nuttalli, D. silvarum, D. sinicus, D. auratus and Ha. concinna [22, 45,46,47,48,49]. Based on the above findings, the epidemiological role of red foxes, marbled polecats and their ticks in maintaining and transmitting rickettsiae can be suggested. However, further studies (including those on experimental transmission) are likely required to verify this hypothesis. In addition, the scope of this work should be extended to more wildlife species from China and Central Asia to better understand the epidemiological implications among rickettsial species and wildlife.
Conclusions
To our knowledge, I. canisuga and I. kaiseri have not been previously identified from red foxes in China. The genetic diversity of I. kaiseri, I. canisuga and Ha. erinacei might be related to their geographic distribution rather than parasitized hosts. The results include the first detection of R. raoultii in I. canisuga and in red fox. We also provide the first molecular evidence of R. sibirica in red fox and marbled polecat. Our findings expand the range of tick-borne pathogens in wildlife species and associated ticks.
Availability of data and materials
The sequences obtained and analyzed during the present study are deposited in the GenBank database under the accession numbers MT890502-MT890525 (Rickettsial), MT889693-MT889705 and MT890492-MT890501 (Tick 16S rRNA and COI gene).
Abbreviations
- COI:
-
Cytochrome c oxidase subunit I
- ompA:
-
Outer membrane protein A
- gltA:
-
Citrate synthase
- 17-kDa:
-
17-KDa antigen
- sca1:
-
Cell surface antigen 1
- XUAR:
-
Xinjiang Uygur Autonomous Region
References
Wilson DE, Reeder DM. Mammal species of the world. A taxonomic and geographic reference. 3rd ed. Baltimore: Johns Hopkins University Press; 2005.
Hunter L, Barrett P. A field guide to the carnivores of the world. London: Bloomsbury Publishing; 2018.
Wurm R, Dobler G, Peters M, Kiessig ST. Serological investigations of red foxes (Vulpes vulpes L.) for determination of the spread of tick-borne encephalitis in Northrhine-Westphalia. J Vet Med B Infect Dis Vet Public Health. 2000;47:503–9.
Lledó L, Serrano JL, Isabel Gegúndez M, Giménez-Pardo C, Saz JV. Antibodies to Rickettsia spp. and Borrelia burgdorferi in Spanish Wild Red Foxes (Vulpes vulpes). J Wildl Dis. 2016;52:122–5.
Fishman Z, Gonen L, Harrus S, Strauss-Ayali D, King R, Baneth G. A serosurvey of Hepatozoon canis and Ehrlichia canis antibodies in wild red foxes (Vulpes vulpes) from Israel. Vet Parasitol. 2004;119:21–6.
Davoust B, Mary C, Marié JL. Detection of Leishmania in red foxes (Vulpes vulpes) from southeastern France using real-time quantitative PCR. J Wildl Dis. 2014;50:130–2.
Gabrielli S, Kumlien S, Calderini P, Brozzi A, Iori A, Cancrini G. The first report of Hepatozoon canis identified in Vulpes vulpes and ticks from Italy. Vector Borne Zoonotic Dis. 2010;10:855–9.
Checa R, López-Beceiro AM, Montoya A, Barrera JP, Ortega N, Gálvez R, et al. Babesia microti-like piroplasm (syn. Babesia vulpes) infection in red foxes (Vulpes vulpes) in NW Spain (Galicia) and its relationship with Ixodes hexagonus. Vet Parasitol. 2018;252:22–8.
Dezdek D, Vojta L, Curković S, Lipej Z, Mihaljević Z, Cvetnić Z, et al. Molecular detection of Theileria annae and Hepatozoon canis in foxes (Vulpes vulpes) in Croatia. Vet Parasitol. 2010;172:333–6.
Ortuño A, Sanfeliu I, Nogueras M, Pons I, López-Claessens S, Castellà J, et al. Detection of Rickettsia massiliae/Bar29 and Rickettsia conorii in red foxes (Vulpes vulpes) and their Rhipicephalus sanguineus complex ticks. Ticks Tick Borne Dis. 2018;3:629–31.
Xie WG, Yin WJ. Surveillance and analysis of plague in Junggar desert in Xinjiang northwestern China (in Chinese). Bull Dis Control Prev. 2016;6:41–2.
Gherman CM, Sándor AD, Kalmár Z, Marinov M, Mihalca AD. First report of Borrelia burgdorferi sensu lato in two threatened carnivores: the marbled polecat, Vormela peregusna and the European mink, Mustela lutreola (Mammalia: Mustelidae). BMC Vet Res. 2012;8:137.
Liu XF, Yang MH, Liu GY, Zhao SS, Yuan WM, Xiao RH, et al. Molecular evidence of Rickettsia raoultii, “CandidatusRickettsia barbariae” and a novel Babesia genotype in marbled polecats (Vormela peregusna) at the China-Kazakhstan border. Parasit Vectors. 2018;11:450.
Jongejan F, Uilenberg G. The global importance of ticks, 2004. Parasitology. 2004;129:3–14.
Isogai E, Isogai H, Kawabata H, Masuzawa T, Yanagihara Y, Kimura K, et al. Lyme disease spirochetes in a wild fox (Vulpes vulpes schrencki) and in ticks. J Wildl Dis. 1994;30:439–44.
Jemeršić L, Dežđek D, Brnić D, Prpić J, Janicki Z, Keros T, et al. Detection and genetic characterization of tick-borne encephalitis virus (TBEV) derived from ticks removed from red foxes (Vulpes vulpes) and isolated from spleen samples of red deer (Cervus elaphus) in Croatia. Ticks Tick Borne Dis. 2014;5:7–13.
Hornok S, Sándor AD, Beck R, Farkas R, Beati L, Kontschán J, et al. Contributions to the phylogeny of Ixodes (Pholeoixodes) canisuga, I. (Ph.) kaiseri, I. (Ph.) hexagonus, and a simple pictorial key for the identification of their females. Parasit Vectors. 2017;10:545.
Guo LP, Mu LM, Xu J, Jiang SH, Wang AD, Chen CF, et al. Rickettsia raoultii in Haemaphysalis erinacei from marbled polecats China-Kazakhstan border. Parasit Vectors. 2015;8:461.
Zhang JZ, Fan MY, Wu YM, Fournier PE, Roux V, Raoult D. Genetic classification of “Rickettsia heilongjiangii” and “Rickettsia hulinii”, two Chinese spotted fever group Rickettsiae. J Clin Microbiol. 2000;9:3498–501.
Tian ZC, Liu GY, Shen H, Xie JR, Luo J, Tian MY. First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. 2012;5:19.
Zhang J, Lu G, Kelly P, Zhang Z, Wei L, Yu D, et al. First report of Rickettsia felis in China. BMC Infect Dis. 2014;14:682.
Wei QQ, Guo LP, Wang AD, Mu LM, Zhang K, Chen CF, et al. The first detection of Rickettsia aeschlimannii and Rickettsia massiliae in Rhipicephalus turanicus ticks, in northwest China. Parasit Vectors. 2015;8:631.
Ye X, Sun Y, Ju W, Wang X, Cao W, Wu M. Vector competence of the tick Ixodes sinensis (Acari: Ixodidae) for Rickettsia monacensis. Parasit Vectors. 2014;7:512.
Li H, Zhang PH, Du J, Yang ZD, Cui N, Xing B, et al. Rickettsiajaponica Infections in Humans, Xinyang, China, 2014–2017. Emerg Infect Dis. 2019;9:1719–22.
Song S, Chen C, Yang M, Zhao S, Wang B, Hornok S, et al. Diversity of Rickettsia species in border regions of northwestern China. Parasit Vectors. 2018;1:634.
Zhao S, Yang M, Liu G, Hornok S, Zhao S, Sang C, et al. Rickettsiae in the common pipistrelle Pipistrellus pipistrellus (Chiroptera: Vespertilionidae) and the bat soft tick Argas vespertilionis (Ixodida: Argasidae). Parasit Vectors. 2020;1:10.
Liu H, Li Q, Zhang X, Li Z, Wang Z, Song M, et al. Characterization of rickettsiae in ticks in northeastern China. Parasit Vectors. 2016;1:498.
Huang Y, Zhao L, Zhang Z, Liu M, Xue Z, Ma D, et al. Detection of a novel rickettsia from Leptotrombidium scutellare Mites (Acari: Trombiculidae) from Shandong of China. J Med Entomol. 2017;3:544–9.
Estrada-Peña A, Mihalca AD, Petney TN. Ticks of Europe and North Africa: a guide to species identification. Berlin: Springer; 2017.
Zhao S, Yang MH, Jiang MM, Yan B, Zhao SS, Yuan WM, et al. Rickettsia raoultii and Rickettsia sibirica in ticks from the long-tailed ground squirrel near the China–Kazakhstan border. Exp Appl Acarol. 2019;77:425–33.
Anstead CA, Chilton NB. A novel Rickettsia species detected in Vole Ticks (Ixodes angustus) from Western Canada. Appl Environ Microbiol. 2013;79:7583–9.
Anstead CA, Chilton NB. Detection of a novel Rickettsia (Alphaproteobacteria: Rickettsiales) in rotund ticks (Ixodes kingi) from Saskatchewan Canada. Ticks Tick Borne Dis. 2013;4:202–6.
Sheng JL, Jiang MM, Yang MH, Bo XW, Zhao SS, Zhang YY, et al. Tick distribution in border regions of Northwestern China. Ticks Tick Borne Dis. 2019;10:665–9.
Zipcodezoo. ZipcodeDev Team (2018). http://zipcodezoo.com/
Klompen JS, Black WC, Keirans JE, Oliver JH. Evolution of ticks. Annu Rev Entomol. 1996;41:141–61.
Hofmann-Lehmann R, Wagmann N, Meli ML, Riond B, Novacco M, Joekel D, et al. Detection of “Candidatus Neoehrlichia mikurensis” and other Anaplasmataceae and Rickettsiaceae in Canidae in Switzerland and Mediterranean countries. Schweiz Arch Tierheilkd. 2016;158:691–700.
Spitalská E, Stefanidesová K, Kocianová E, Boldiš V. Rickettsia slovaca and Rickettsia raoultii in Dermacentor marginatus and Dermacentor reticulatus ticks from Slovak Republic. Exp Appl Acarol. 2012;57:189–97.
Jiang J, Blair PJ, Felices V, Moron C, Cespedes M, Anaya E, et al. Phylogenetic analysis of a novel molecular isolate of spotted fever group Rickettsiae from northern Peru: Candidatus Rickettsia andeanae. Ann N Y Acad Sci. 2005;1063:337–42.
Cheng C, Fu W, Ju W, Yang L, Xu N, Wang YM, et al. Diversity of spotted fever group Rickettsia infection in hard ticks from Suifenhe Chinese–Russian border. Ticks Tick Borne Dis. 2016;5:715–9.
Boldis V, Kocianová E, Strus J, Tusek-Znidaric M, Sparagano OA, Stefanidesová K, et al. Rickettsial agents in Slovakian ticks (Acarina, Ixodidae) and their ability to grow in Vero and L929 cell lines. Ann N Y Acad Sci. 2008;1149:281–5.
Merino FJ, Nebreda T, Serrano JL, Fernández-Soto P, Encinas A, Pérez-Sánchez R. Tick species and tick-borne infections identified in population from a rural area of Spain. Epidemiol Infect. 2005;5:943–9.
Chisu V, Foxi C, Masala G. First molecular detection of the human pathogen Rickettsia raoultii and other spotted fever group rickettsiae in Ixodid ticks from wild and domestic mammals. Parasitol Res. 2018;11:3421–9.
Shpynov S, Fournier PE, Rudakov N, Tankibaev M, Tarasevich I, Raoult D. Detection of a rickettsia closely related to Rickettsia aeschlimannii, "Rickettsia heilongjiangensis," Rickettsia sp. strain RpA4, and Ehrlichia muris in ticks collected in Russia and Kazakhstan. J Clin Microbiol. 2004;5:2221–3.
Paddock CD, Fournier PE, Sumner JW, Goddard J, Elshenawy Y, Metcalfe MG, et al. Isolation of Rickettsia parkeri and identification of a novel spotted fever group Rickettsia sp. from Gulf Coast ticks (Amblyomma maculatum) in the United States. Appl Environ Microbiol. 2010;9:2689–96.
de Sousa R, Barata C, Vitorino L, Santos-Silva M, Carrapato C, Torgal J, et al. Rickettsia sibirica isolation from a patient and detection in ticks. Portugal Emerg Infect Dis. 2006;7:1103–8.
Mahara F. Japanese spotted fever: report of 31 cases and review of the literature. Emerg Infect Dis. 1997;2:105–11.
Yu X, Jin Y, Fan M, Xu G, Liu Q, Raoult D. Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from China. J Clin Microbiol. 1993;1:83–8.
Eremeeva ME, Balayeva NM, Ignatovich VF, Raoult D. Proteinic and genomic identification of spotted fever group rickettsiae isolated in the former USSR. J Clin Microbiol. 1993;10:2625–33.
Chen M, Fan MY, Bi DZ, Zhang JZ, Huang YP. Detection of Rickettsia sibirica in ticks and small mammals collected in three different regions of China. Acta Virol. 1998;1:61–4.
Acknowledgements
The authors thank the contributions by the staff at the School of Medicine and School of Animal Science and Technology, Shihezi University.
Funding
This work was supported by the National Natural Science Foundation of China (81960379 and 31960709), International Scientific and Technological Cooperation in Bingtuan (2020BC008) and International Cooperation Projects of Xinjiang Uygur Autonomous Region (2020E01008).
Author information
Authors and Affiliations
Contributions
GL, SZ, WT and YW conceived and designed the study, and wrote the manuscript. WY, LM, SW, YZ, ZL, WH and XG performed the experiments and analyzed the data. SH contributed to study design and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
This study was approved by the Animal Ethics Committee of Shihezi University (Approval No. AECSU2015-11).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Sampling data of 12 red foxes (Vulpes vulpes), a marbled polecat (Vormela peregusna) and their ticks.
Additional file 2.
Nested PCR protocol for the detection of rickettsiae in 13 wild carnivores as well as from their ticks, northern Xinjiang, China.
Additional file 3.
The morphological characteristics of ticks from 12 red foxes and a marble polecat in northwestern China.
Additional file 4.
Phylogenetic tree of the 17-kDa-ompA-gltA-sca1 concatenated sequences of Rickettsia raoultii from Ixodes canisuga and Dermacentor marginatus ticks.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, G., Zhao, S., Tan, W. et al. Rickettsiae in red fox (Vulpes vulpes), marbled polecat (Vormela peregusna) and their ticks in northwestern China. Parasites Vectors 14, 204 (2021). https://doi.org/10.1186/s13071-021-04718-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-04718-1