Abstract
Background
In South America, fascioliasis stands out due to the human endemic areas in many countries. In Argentina, human endemic areas have recently been detected. Lymnaeid vectors were studied in two human endemic localities of Catamarca province: Locality A beside Taton and Rio Grande villages; Locality B close to Recreo town.
Methods
Lymnaeids were characterised by the complete sequences of rDNA ITS-2 and ITS-1 and fragments of the mtDNA 16S and cox1. Shell morphometry was studied with the aid of a computer image analysis system. Climate analyses were made by nearest neighbour interpolation from FAO data. Koeppen & Budyko climate classifications were used. De Martonne aridity index and Gorczynski continentality index were obtained. Lymnaeid distribution was assessed in environmental studies.
Results
DNA sequences demonstrated the presence of Lymnaea neotropica and L. viator in Locality A and of L. neotropica in Locality B. Two and four new haplotypes were found in L. neotropica and L. viator, respectively. For interspecific differentiation, ITS-1 and 16S showed the highest and lowest resolution, respectively. For intraspecific analyses, cox1 was the best marker and ITS-1 the worst. Shell intraspecific variability overlapped in both species, except maximum length which was greater in L. viator. The desertic-arid conditions surrounding Locality A, the semiaridity-aridity surrounding Locality B, and the very low yearly precipitation in both localities, are very different from the typical fascioliasis transmission foci. Lymnaeids are confined to lateral river side floodings and small man-made irrigation systems. Water availability only depends on the rivers flowing from neighbouring mountains. All disease transmission factors are concentrated in small areas where humans and animals go for water supply, vegetable cultures and livestock farming.
Conclusions
The unusually high number of DNA haplotypes and the extreme climate unsuitable for F. hepatica and lymnaeid development, demonstrate that the transmission foci are isolated. Seasonal transmission may depend on the timely overlap of appropriate temperature and river water availability. Lymnaeids and F. hepatica have probably reached these localities by livestock introduction. DNA differences regarding other populations of L. neotropica and L. viator in Argentina suggest an introduction independent from the spreading movements which allowed these two lymnaeids to expand throughout the country.
Similar content being viewed by others
Background
Fascioliasis is a foodborne trematodiasis caused by species of the genus Fasciola. This disease affects livestock almost everywhere and humans in many countries of Europe, Africa, Asia, the Americas and Oceania [1, 2], where there is tradition of eating uncooked vegetables carrying infective metacercariae [3]. Fascioliasis is emerging in many countries, with progressive detection of new human fascioliasis endemic areas and an increasing number of human case reports. This emergence phenomenon has partly been related to climate change [4] and also global change aspects [5] such as anthropogenic modifications of the environment [6], travelling [7] and import/export of livestock [2]. These changes appear to be related to the high dependence of both fasciolid larval stages and their freshwater lymnaeid snail vectors on climatic and environmental characteristics [8–10]. The increasing importance of human fascioliasis also relies on pathogenicity and immunity. Thus, this disease appears to be pronouncedly complicated including diagnosis difficulties [11] and a great morbidity impact on children in long-term infections, such as in human fascioliasis endemic areas [12–15]. The clinical complexity of the symptoms and syndromes due to the capacity of the flukes to affect vital organs other than the liver, giving rise to important sequelae and even death, add concern about human fascioliasis [16].
Fascioliasis is transmitted by freshwater lymnaeid snails which show marked specificity according to Fasciola spp. [17]. Fasciola hepatica, distributed throughout all continents, is mainly transmitted by species of the “fossarine” or Galba/Fossaria group [18, 19]. Fasciola gigantica, present in large regions of Africa and Asia, is mainly transmitted by species of the Radix group [20]. Another vector species is Pseudosuccinea columella, a species with global distribution, able to transmit both Fasciola spp. and above all related to animal infection [21]. Other lymnaeid groups include species which may only act as secondary or sporadic vectors and without epidemiological importance except under special circumstances [22, 23].
Although livestock species play an important reservoir role, transmission studies have shown that the metacercarial infective stage from different origins, such as sheep, cattle, pig and donkey, represent similar infectivity sources [24, 25]. On the contrary, the susceptibility of a given lymnaeid species to fasciolid infection represents a crucial factor in establishing not only the geographical distribution of the disease in both animals and humans, but also prevalences and intensities due to more or less appropriate ecological characteristics (population dynamics, anthropophylic characteristics, type of water bodies, etc.) of the different lymnaeid vector species. This is why different lymnaeid species appear linked to the different transmission patterns [26] and epidemiological scenarios [27] of this very heterogeneous disease in humans [2]. The continental differences in lymnaeid faunas also explain that in the Americas fascioliasis is only caused by F. hepatica, due to the absence of lymnaeids of the genus Radix which act as vectors of F. gigantica [20]. Similarly as in other vector-borne diseases, this relationship supports the use of lymnaeids as biomarkers of the disease at both local and large scales [28]. Distribution, both in space (latitudinal, longitudinal and altitudinal) and time (seasonal, yearly), of fascioliasis depends on the presence and population dynamics of the specific mollusc species, which in turn is linked to the presence of the appropriate water bodies and on adequate climate characteristics enabling fluke development.
Within the several human fascioliasis regions known [2], the Americas stand out due to the human endemic areas described in many countries. High prevalences and intensities of F. hepatica in humans have been reported in Andean countries such as Bolivia [29–33], Peru [34–36] and Chile [37, 38]. Human endemic areas have also been described, although with lower prevalences and intensities, in Central America [39] and recently also in North America [40]. Several human cases have also been reported from altitude areas of Ecuador [41], Colombia [21] and Venezuela [28]. In Uruguay and Brazil, human reports only concern sporadic and isolated cases. In the Caribbean region, human fascioliasis mainly poses problems in Cuba, where patients are continuously diagnosed [42, 43] even in high numbers [44], and Haiti [45]. Puerto Rico may still be considered an area of risk for human infections considering the epidemiological situation in the past [46].
In Argentina, physiography, climate, animal prevalence of F. hepatica and lymnaeid species composition are similar to those of countries where human fascioliasis endemic areas are present, such as Bolivia, Peru and Chile. A retrospective analysis highlights that human fascioliasis may have been overlooked in the past and its real epidemiological situation in high risk rural, mainly altitudinal areas, may currently be underestimated. Moreover, a long delay in diagnosis (almost 3.5 years on average) and high lithiasis proportion suggest that many patients are frequently overlooked and pose a question mark about fascioliasis detection in the country [47]. The recent detection of lymnaeid vector species such as G. truncatula [48, 49] and L. neotropica [19, 50], well-known to be responsible for high prevalences and intensities of human fascioliasis in neighbouring and closely located countries such as Bolivia and Peru [51], adds concern to this question.
In Argentina, several outbreaks presenting typical food-borne characteristics appear related to the most common risk factor: ingestion of watercress naturally growing along the river- and stream-beds picked during recreational, weekend or vacation activities. Many of these field excursions are undertaken by a family or as a group activity. This explains why family outbreaks have been noted to be common, whereas isolated cases seem to be rare in the country [52]. Outbreaks described in eleven families involving a total of 63 people [52–58] including a maximum of up to 15 family members affected at once [52], are good examples.
However, different epidemiological situations were recently detected in two areas of this country as a result of two serological surveys performed with an ELISA test for the detection of anti-recombinant procathepsine L1 of F. hepatica (Fh-rproCL1) antibodies [59, 60]. In the area of Arroyo El Juncal, La Toma, province of San Luis, between 500 and 600 m altitude, the prevalence was 11.90 % in a total of 42 human subjects (2 infected out of 23 males and 3 out of 19 females) serologically analysed [61]. This corresponds to a human hyperendemic situation according to the WHO classification [27]. In the same El Juncal area, 5.26 % out of 19 livestock species proved to be infected after coprological analysis (only in cattle; sheep and horses were not infected) and a surprisingly high prevalence (61.76 % out of 34 snails) was detected by sequencing a 447 bp long fragment of the mtDNA cox1 gene of F. hepatica [61] in snails identified as Lymnaea viator (= L. viatrix; for nomenclature see [38]).
In the area of the villages of Taton and Rio Grande, Department of Tinogasta, province of Catamarca, at an altitude of 1,630 m, the situation appears to be worse. A total of 54 subjects proved to be positive out of 148 serum samples analysed (17 positive out of 61 males and 37 out of 87 females) [62]. This means a very high prevalence of 36.5 %, at the level of the highest hyperendemic situations known in the Northern Altiplano of Bolivia [30–33] and Peru [34], as well as the valley of Cajamarca also in Peru [36]. Indeed, a public health problem posed by fascioliasis throughout the Argentinian province of Catamarca was already suspected in preliminary studies which allowed for the detection of ten serologically positive patients (without eggs in stools) in the locality of Recreo in 2002 [63].
The present study aims to (i) identify the lymnaeid vector species involved in both endemic areas of Taton-Rio Grande and Recreo by means of molecular tools; (ii) characterise these lymnaeids by their shell morphometry and genital anatomy; and (iii) analyse their habitats and respective relationships with climatic factors to assess the transmission pattern of fascioliasis in the endemic areas. For the molecular characterisation, both nuclear ribosomal DNA and mitochondrial DNA markers are sequenced and subjected to phylogenetic analyses. Snails are studied by traditional malacological methods to ascertain whether morphology may allow species differentiation. In the Americas, except P. columella, all lymnaeid species involved in the transmission belong to the Galba/Fossaria group. Species differentiation within Galba/Fossaria is crucial given (i) their different disease transmission capacities to humans; (ii) the difficulties in species differentiation due to their pronounced morphological similarities; and (iii) the fact that there is a snail species of this group which, although usually present in high transmission areas, does not transmit [18]. For the climatic assessment, a deep analysis by different climate classifications and indices is made additionally for the study of the climatic factors related to fascioliasis transmission, given the very peculiar, isolation characteristics of the transmission foci [64, 65].
Methods
Lymnaeid snails
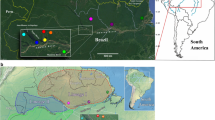
The snail specimens studied were collected in the field, from lymnaeid populations present in two geographical areas of the Province of Catamarca (Fig. 1):
-
Locality A: two neighbouring fertile plains where livestock go for drinking (27°29′52.62″S, 67°35′53.51″W; altitude 1,630 m), only 300 m far from the river, in the way to the small villages of Taton (1860 m altitude) and Rio Grande (2825 m) (Departament of Tinogasta). The zone comprises two villages, Taton of about 150 inhabitants living in dwellings spread throughout 2 km at the two banks of the river coming down from the smaller Rio Grande of 40 inhabitants and several more or less scattered dwellings distributed on the hills (called “Puestos”); human infection by F. hepatica has been reported from inhabitants of both villages [62];
-
Locality B: an artificial dyke or dam (28°49′08.83″S, 65°32′20.00″W; altitude 944 m) beside the village of Ipizca (Department of Ancasti) and close to the town of Recreo (Department of La Paz) where human infection has also been reported [63].
Maps showing the transmission foci studied where the lymnaeid snails were collected. a Location of Argentina in South America. b Location of the province of Catamarca inside the country (names of provinces only in those where human infection has been diagnosed). c Location of the sampling Locality A (white circle) beside the human fascioliasis endemic villages of Taton-Rio Grande and Tinogasta (red circles), in the Department of Tinogasta, and sampling Locality B (white circle) beside the villages of Ipizca and Ancasti and close to the human fascioliasis endemic town of Recreo (red circles), in the Department of La Paz
For molecular analyses, live snail specimens were fixed in 70 % ethanol for DNA extraction procedures. A total of 50 lymnaeid specimens from Locality A and six specimens from Locality B were collected and used for molecular and morphometric studies.
Molecular techniques
DNA markers
The molecular characterisation of the snails has been made by DNA sequencing of the complete nuclear ribosomal DNA (rDNA) spacer markers ITS-2 and ITS-1 and fragments of the mitochondrial DNA (mtDNA) gene markers 16S rDNA and cox1. The usefulness of these markers has been already proven for invertebrates in general [66] and for the classification of the lymnaeid species and the comparative assessment of the intraspecific variability of their populations in many countries of Latin America [17–21, 28], also including Argentina [48–50].
DNA extraction
DNA extraction was performed individually from the head-foot tissue of each snail with phenol-chloroform and DNA was precipitated with ethanol. The snail specimens were suspended in 400 μl of lysis buffer (10 mM Tris–HCl, pH 8.0, 100 mM EDTA, 100 mM NaCl, 1 % sodium dodecyl sulfate SDS) containing 500 μg/ml Proteinase K (Promega, Madison, WI, USA) and digested for 2 h at 55 °C with alternate shaking each 15 min. The procedure steps were performed according to methods outlined previously [19, 20]. The pellet was dried and resuspended in 30 μl sterile TE buffer (pH 8.0); this suspension was stored at −20 °C until use.
Sequencing of rDNA and mtDNA markers
The four DNA markers were PCR amplified independently for each specimen and each PCR product was sequenced for a bona-fide haplotype characterisation. The complete rDNA spacers ITS-2 and ITS-1 were amplified using previously described primers [2, 19, 20, 48, 49]. The target fragments of the 16S and cox1 genes were amplified by PCR using a set of universal primers [67, 68]. Amplification procedures and thermal cycler conditions for each one of the DNA markers were carried out in a Mastercycle epgradient (Eppendorf, Hamburg, Germany), as previously described [2, 18].
PCR products were purified using the Ultra Clean™ PCR Clean-up DNA Purification System (MoBio, Solana Beach, CA, USA) according to the manufacturer’s protocol and resuspended in 50 μl of 10 mM TE buffer (pH 7.6). The final DNA concentration was determined by measuring the absorbance at 260 and 280 nm on an Eppendorf BioPhotometer (Hamburg, Germany).
Sequencing was performed on both strands by the dideoxy chain-termination method and carried out with the Taq dye-terminator chemistry kit on an Applied Biosystems 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using the PCR primers.
Sequence analyses
Sequences were aligned using CLUSTALW2 [69] in MEGA 6.0.6 [70] using default settings. Minor corrections for a better fit of nucleotide or indel correspondences were made in the cases of the ITS spacers. Homologies were performed using the BLASTN programme from the National Center for Biotechnology information website (http://www.ncbi.nlm.nih.gov/BLAST). As the systematics of the South American lymnaeids belonging to the Galba/Fossaria group is controversial and specimen identification cannot be made with the use of morphological criteria alone [19, 50], sequence comparisons were made using all ribosomal and mitochondrial sequence data for molluscs available in the GenBank database.
Phylogenetic inference
For phylogenetic analyses, ITS-1 and ITS-2 combined haplotypes were used, given that the Galba/Fossaria group includes species originating in both the Old and New Worlds. The division of the Gondwana is estimated back to around 100 million years [71]. This long evolutionary period indicates the inappropriateness of using mtDNA markers for the phylogenetic assessment, due to the well-known problem of mtDNA nucleotide saturation distorting the phylogenetic information [66]. Details of combined haplotypes and GenBank codes of the sequences of lymnaeid species used in the phylogenetic analyses, others than the new ones obtained for L. neotropica and L. viator in the present study, may be found in a previous analysis [18].
The phylogenetic analyses were performed with maximum likelihood (ML) and distance-based (Neighbour Joining) (NJ) methods implemented in MEGA 6.0 [70] and PAUP 4.0 b10 [72], respectively. The best-fitting substitution model was determined with jModeltest version 0.1.1 [73] based on the corrected Akaike’s information criterion (AICc) [74], which led to the selection of the Kimura 2-parameter model with a gamma-distributed rate heterogeneity among sites, with five rate categories (K2 + G). The initial tree for the heuristic search was automatically obtained by applying NJ and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach.
To provide an assessment of the reliability of the nodes in the tree, a bootstrap analysis using 1,000 replicates was performed with two types of search, Neighbour-Joining and “Fast” stepwise-addition in PAUP. Moreover, a Bayesian approach was applied to reconstruct the phylogeny of the concatenated dataset in MrBayes 3.1.2 [75], using the same evolutionary model as above. Posterior probabilities were estimated by a 1,000,000 generations (four chains) and trees were sampled every 100 generations. The first 1,000 trees sampled were ruled out (“burn-in”), and clade posterior probabilities (PP) were computed from the remaining trees. The intergenic region sequence (GenBank AY030361) [76] including both ITS spacers of the planorbid Biomphalaria pfeifferi, was used as the outgroup.
DNA haplotype nomenclature
The haplotype (H) terminology used for the sequences obtained follows the previously described standard nomenclature proposed for lymnaeid snails [2, 17, 48]. It should be noted that haplotype codes are only definitive in the case of complete sequences; when dealing with fragments or incomplete sequences, haplotype codes are provisional.
Morphometric study
Only shells from simultaneously molecularly assessed lymnaeid specimens were used (L. neotropica: n = 34; L. viator: n = 16) for the morphometric study and comparative analyses. The comparative morphometric characterisation of the lymnaeid shell may be a feature of interest [18, 19, 77], as it may help health officers in their field work in the case that different lymnaeid species coexisting in the same endemic area can be distinguished morphologically.
Snail shells were measured according to traditional malacological methods [77, 78], using a computerised image-analysis system (CIAS) [79, 80]. This system is based on a DXC-930P colour video camera (Sony DXC-930P, Tokyo) fitted to a stereomicroscope, supplied with image analysis software (ImagePro® Plus 4.5; Media Cybernetics Inc., Silver Spring, MD). Statistical analysis was conducted using IBM Statistics version 22 (SPSS, Armonk, NY, USA), by applying t-test to compare species samples for each variable (P < 0.05) and principal components analysis PCA (using varimax rotation) on the shell measurement dataset including all variables (data ln-transformed). Shell characteristics measured on 34 L. neotropica and 16 L. viator specimens were: length (SL); maximum width (SW); aperture length (AL); aperture width (AW); last spire length (LSL); spiral angle (SSA); SL/SW ratio; SL/AL ratio; and SL/LSL ratio.
Moreover, the soft parts of the snails were dissected under a stereomicroscope and the anatomy of the reproductive system analysed, to assess the morphology and measurements of the penial structures.
Environment and climate analyses
Field studies to look for the presence of lymnaeid snails were undertaken in freshwater bodies in close proximity to the dwellings, schools and villages where infected human subjects were detected. The analysis of aspects of the ecology of the lymnaeids, such as the characteristics of their natural habitat and environment and the local climatic factors become crucial to understand the human endemic areas in question, regarding the transmission pattern [2, 26], the epidemiological scenario [2, 27] and the best strategies for diagnosis and surveys [11].
The yearly variation of the climatic factors was obtained by interpolation furnished by the closest ten surrounding meteorological stations (four and six for the transmission foci of localities A and B, respectively) from a series database of 50 years (1950–2000). The data from the agroclimatic database of the FAO Agromet Group (FAOCLIM) were used. The interpolation method applied to assess the climatic characteristics of the two aforementioned local areas in question is by nearest neighbour by using the New LocClim 1.10 software from FAO with a temporal resolution of days. This software allows for altitude and horizontal corrections. Climatic factors analysed included the maximum, mean and minimum temperatures, precipitation, potential evapotranspiration (PET), humid period (vegetation growing season), moist period (vegetation growing season), and dry period. With regard to the vegetation period, the growing season is the period during a year when precipitation exceeds half the potential evapotranspiration and is defined by Prec/PET > 0.5 [81], although for these two foci both PET > 0.5 and PET > 0.45 were analysed due to their extreme arid conditions.
The most widely applied climatic classifications were used and different appropriate climatic indices were obtained. The Koeppen climate classification is based on the concept that native vegetation is the best expression of climate [64]. According to the Koeppen classification, climate zone boundaries are selected with vegetation distribution in mind. It combines average annual and monthly temperatures and precipitation, and the seasonality of precipitation [64]. The Budyko climatic classification system is a method of categorizing climates based on the ratio of energy received to energy required to evaporate local precipitation [64]. The principles of Budyko’s classification take into account radiation characteristics, which are closely and directly correlated with the temperature in the warm season, and circulation factors, which indirectly correlated with quantitative characteristics of precipitation and moistening regime [64].
Among the variety of existing indices to quantify aridity/moisture and continentality, the aridity index of De Martonne and the continentality index of Gorczynski were obtained. The aridity index of De Martonne provides a simple way to express the ratio of precipitation to evaporation [65]. Since evaporation is rarely observed, it is a common tradition to approximate it. De Martonne’s index is defined as the ratio of the summed annual precipitation in mm and the annual mean temperature in °C +10. The continentality index of Gorczynski is a way to estimate the influence of the ocean on the local climate [65]. Gorczynski’s index depends linearly on the annual temperature amplitude (difference of monthly mean temperature of warmest and coldest month) [65].
Results
Molecular characterisation of lymnaeids
ITS-2, ITS-1, 16S and cox1 sequences reported in this study are available in the databases under the accession numbers listed in Table 1. The results revealed two lymnaeid species: L. neotropica and L. viator. Out of the 50 specimens analysed, 34 (68.0 %) were molecularly identified as L. neotropica and 16 (32.0 %) as L. viator. The latter species was detected sharing the same ecotope with L. neotropica in the Locality A whereas only L. neotropica was found in Locality B.
Lymnaea neotropica
Nuclear markers
rDNA ITS-2
All specimens from both localities had identical sequences (417 bp long; GC content 56.83 %). No nucleotide difference was found when comparing with the ITS-2 of the L. neotropica haplotype L.neo-H1 (GenBank AM412225) from the type-locality in Peru. The haplotype L.neo-H2 from Venezuela (GenBank JF514089) differed from the Argentinian sequence in the lack of one microsatellite repeat (AT) at positions 402–403 and in one mutation at position 350 of the sequence alignment of both ITS-2 haplotypes. Other Argentinian isolates of L. neotropica available in the GenBank database (KJ425597, 415 bp; KJ425598, 415 bp; and KJ425596, 414 bp) showed only some differences (deletions) at the 3′ end of the ITS-2, although these sequences are incomplete.
rDNA ITS-1
A new haplotype for L. neotropica was detected in the populations from localities A and B. All specimens presented the same ITS-1 of 535 bp (GC content 56.45 %). This new haplotype is here annotated as L.neo-HB. Differences with haplotype L.neo-HA from the type-locality are two insertions in the “poli-A” region at the 3′ end (positions 512 to 529), a region of 16 or 18 consecutive “A” in L.neo-HA and L.neo-HB, respectively. BLASTN results showed similarity of 98 % with other L. neotropica sequences obtained in Argentina available in the GenBank database (KJ425594, 528 bp; KJ425590, 528 bp; KJ425591, 527 bp; KJ425595, 525 bp; and KJ425593, 523 bp), and curiously, with one of L. viator (JF960165, 533 bp). Differences (indels) were again found at the 3′ end of the ITS-1 spacer of these incomplete sequences.
Mitochondrial markers
mtDNA 16S
Only one haplotype was detected. This partial sequence was 435 bp long, with a biased AT content of 69.65 %, and proved to be identical to L.neo-16SA from the type-locality also reported in the human fascioliasis endemic area of Cajamarca in Peru (GenBank HE610433). In a 439 bp long alignment of this haplotype (L.neo-16SA) with other L. neotropica isolates (sequences 429–431 bp long) available in GenBank, 12 variable positions were detected, most being generated by the isolate NtS1 from Argentina (GenBank JN872471), whose mutations (most of them always “A”) should be revised (Fig. 2a).
Nucleotide differences found in mtDNA sequence of Lymnaea neotropica populations studied and other haplotypes and isolates of the same species. a 16S. b cox1. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 6.0.6; . = identical; − = indel; ? = undetermined amino acid; * = present paper. Haplotype codes only provisional due to incomplete sequences of the gene
mtDNA cox1
All specimens from localities A and B showed identical cox1 sequences of 672 bp (AT content 69.79 %). This sequence was compared with the four cox1 haplotypes described for L. neotropica so far and proved to be different in only one mutation at position 9 of the alignment with the previously described haplotype L.neo-cox1a from the type-locality (GenBank AM494008). Similarly, one mutation at position 375 was the only difference with haplotype L.neo-cox1b, previously reported in Mendoza, Argentina. Nucleotide and amino acid differences with all L. neotropica haplotypes (672 bp long) and other isolates (655 bp long) showing highly similar sequences according to BLASTN results, are detailed in Fig. 2b.
Lymnaea viator
Nuclear markers
rDNA ITS-2
All specimens had identical ITS-2 sequences (436 bp long; GC content 53.67 %). When compared with the available ITS-2 haplotypes of L. viator, the most similar to this Argentinian haplotype was L.via-H2 from Chile (GenBank JN051366), being only shorter in length due to 8 polymorphic sites corresponding to 8 indels related to a different number of repeats in the microsatellites GCTT and GCTC. These microsatellites were (GCTT)2, (GCTC)1 in Locality A, Argentina and (GCTT)4 and (GCTC)1, respectively, in Casa Blanca, Chile. The code H3 has been ascribed to this new haplotype (Table 1). Other differences with the haplotype L.via-H1 and other L. viator isolates showing incomplete 3′ end ITS-2 sequences are listed in Fig. 3a.
Polymorphic sites found in rDNA sequence of Lymnaea viator populations studied and other haplotypes and isolates of the same species. a ITS-2. b ITS-1. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 6.0.6; .= identical; - = deletions (in microsatellite area); * = present paper; ** = incomplete sequences at the 3′ or 5′ end. Shaded area corresponds to microsatellite positions
rDNA ITS-1
All individuals had identical ITS-1 sequences (570 bp; GC content 54.03 %). This sequence was similar to the haplotype L.via-HB from Chile (GenBank JN051368), differing in only five polymorphic sites corresponding to one mutation and one tetranucleotide repeat (ATTG). The code HC was ascribed to this new haplotype. Detailed differences with L. viator haplotypes HA from Argentina and HB from Chile, as well as with other L. viator isolates showing incomplete 5′ end ITS-1 sequences are listed in Fig. 3b.
Mitochondrial markers
mtDNA 16S
A new haplotype has been detected in Locality A, for which the new code L.via-16SB is assigned. This partial sequence was 438 bp long (AT content 69.41 %). This haplotype is similar to one found in the type-locality, L.via-16SA (GenBank HE610434), which differs in only two insertion/deletions (indels). Worth noting are the very few differences detected in this fragment between specimens of L. neotropica and L. viator studied in Argentina (only five variable positions: four mutations and one indel). Therefore, a comparative sequence analysis was made including L. neotropica and L. viator haplotypes and other 16S haplotypes and isolates of the same species available in the GenBank database showing highly similar sequences according to BLASTN (Fig. 4).
Nucleotide differences found in the mitochondrial rDNA 16S of Lymnaea neotropica and L. viator studied and other 16S haplotypes and isolates of the same species. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 6.0.6; . = identical; − = indel; * = present paper. Haplotype codes only provisional due to incomplete sequences of the gene
mtDNA cox1
Only one nucleotide sequence was found, identical in all specimens (672 bp; AT content 69.94 %). This sequence proved to be new and the new haplotype code L.via-cox1d is here ascribed to it. When compared with previously described haplotypes of L. viator cox1a, cox1b, and cox1c, nucleotide differences ranged between 6 and 11. The haplotype L.via-cox1a from Rio Negro, Argentina (GenBank AM494010) appears to be the most similar. BLASTN results provided 100 % similarity only with a shorter cox1 fragment of L. viatrix (isolate VxC2; 654 bp) from Mendoza, Argentina (GenBank JN872450). Nucleotide and amino acid differences with L. viator haplotypes and other isolates are detailed in Fig. 5.
Nucleotide and amino acid differences found in the mtDNA cox1 of Lymnaea viator studied and other cox1 haplotypes and isolates of the same species. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 6.0.6; . = identical; ? = undetermined amino acid; − = not sequenced; * = present paper. Shaded area corresponds to shorter fragments. Haplotype codes only provisional due to incomplete sequences of the gene
Comparative sequence analysis of L. neotropica vs L. viator
There were 60 variable positions (29 mutations and 31 indels) in a 464 bp long ITS-2 alignment of both species and the respective haplotypes and isolates. Additionally, this marker provides microsatellites for population characterisation (Fig. 3a). For ITS-1, there were 119 variable positions (93 mutations and 26 indels) in a 580 bp long alignment. Total character differences between species were clear, and consistent, showing a range of 57 to 59 (average 57.90), with a p-distance of 0.1163 on average (or a total of 80 parsimony informative positions in their respective alignment) for species distinction. A pairwise distance matrix between ITS-2 and ITS-1 sequences of haplotypes and isolates of L. neotropica and L. viator is included in Tables 2 and 3.
Regarding mtDNA, cox1 was excellent for species and populations distinction when used as nucleotide sequence, but not when translated to protein due to amino acid sequence identity of some isolates of L. neotropica and L. viator. Contrarily to the other three markers used, 16S is the only one that may create confusion if used alone for molecular differentiation of the two species. Causes for this conclusion are: the short length of the fragment used and the very low number of variable positions obtained (25, of which 19 mutations and 6 indels, in a 442 bp long alignment, 5.65 %) which do not generate a consistent separation between both species and haplotypes (Fig. 4). Total character differences were used to construct a pairwise distance matrix for 16S and cox1 sequences between haplotypes and isolates of L. neotropica and L. viator (Tables 4 and 5).
Useful information provided by the four molecular markers applied for the distinction between L. neotropica and L. viator is detailed in Table 6. For the differentiation between these two species, ITS-1 is the marker with a higher resolution and 16S as the worst. For intraspecific analyses, cox1 appears to be the best marker and ITS-1 the worst. The low resolution of the 16S gene fragment for species distinction is worth mentioning. For instance, 16S distance estimation using as substitution model total nucleotide differences (ts + tv) between L. neotropica and L. viator ranged between 4 and 14 vs 0–9 and 0–10 within L. neotropica and L. viator sequences, respectively (Table 6).
Phylogenetic results
ML, NJ and Bayesian phylogenetic methods yielded similar tree topologies for the combined dataset used (ITS-1 + ITS-2; 1417 bp).
The ML tree obtained (−Ln = 11,597.42) (Fig. 6) including the new haplotypes, together with published GenBank sequences of related species, showed that L. neotropica and L. viator, together with Lymnaea cubensis, had strong support as sister species and constituted a monophyletic clade inside the Galba/Fossaria group. Other members of this group, i.e. G. truncatula, L. humilis, L. cousini, L. meridensis and L. schirazensis clustered together in another monophyletic clade. Pseudosuccinea columella appeared basal to the two large groupings, namely stagnicolines (including both Palaearctic and Nearctic species) and the Galba/Fossaria clade which comprised the main host species of F. hepatica. Radix spp. clustered independently in a branch basal to all lymnaeids in this analysis.
Phylogenetic tree (Log-Likelihood = −11,597.42) of lymnaeid species studied based on maximum-likelihood (ML) estimates (Ts/Tv = 1.1445; Gamma = 0.6637). Scale-bar indicates the number of substitutions per nucleotide position. Node support values with 1,000 bootstrap replicates based on a/b/c: a: bootstrap with Neighbour-Joining search (only values > 70 shown); b: bootstrap with “Fast” stepwise-addition search (only values > 70 shown); c: Bayesian posterior probability (BPP) (only values > 95 shown)
Morphometric study of the lymnaeid species
The coexistence of L. neotropica and L. viator in Locality A furnished the possiblity for a comparative shell morphology study to assess whether these species may be morphometrically distinguished within the same endemic area.
The shell of both species is brownish to light brown, thin-walled, elongated conical, usually with four regular convex whorls (up to 5.5 whorls in longer specimens). The spire is more or less pointed or short with a rather obtuse apex. Growth lines and umbilicus are slightly pronounced. The spiral sculpture is very faint and not easily seen. The aperture is oval and almost round, with a thin peristome. The shape of the shell of L. neotropica and L. viator (Fig. 7) is very similar, both with a similar whorl number, although the specimens of L. viator may sometimes appear somewhat more slender (compare in Fig. 7).
The comparative data for shell measurements and shell ratios (Additional file 1: Table S1) show that the shell length/shell maximum width ratio overlaps in the two species and hence the variability of this parameter cannot be used for their differentiation. A similar problem appears evident in relation to all other shell parameters, except for the shell length which not only reaches an apparently higher length maximum (10.85 vs 9.18 mm) but also a greater shell length mean (9.21 vs 7.49 mm) in L. viator. The results of the statistical comparisons by means of t-test are included in Additional file 1: Table S1. Only the shell spiral angle and the three shell ratios furnished data exhibiting statistical significant differences between the two species. The principal components analysis furnished a graph in which data of the two species overlap (Fig. 8).
At anatomical level, only the preputium/penis-sheath length ratio allowed for a species differentiation. This ratio proved to be of 1.14 in L. neotropica and 2.78 in L. viator.
Climatic characteristics and snail habitats
The results of the climatic analyses of the localities A and B, according to the different climate classification systems and the different climatic indices used, are listed in Table 7. The results of the analyses by the climate classifications of Koeppen and Budyko, as well as by the aridity index of De Martonne, allow to highlight the extreme desertic-arid environmental characteristics surrounding Locality A (Fig. 9) and the slightly less extreme conditions of semiaridity-aridity of those surrounding Locality B. The influence of the ocean on the local climate estimated by the continentality index of Gorczynski indicates small differences between the two localities situated at different altitudes and west and east of the same mountain chain.
Transmission focus of Locality A. a Geographical image showing Locality A where lymnaeid snails were collected, on the way from the small village of Medanitos to the small villages of Taton and Rio Grande (image from Google Earth taken at 90 km altitude; image Landsat, US Department of State Geographer, 2016); note desert surroundings of sand dunes of the Fiambala Valley and high mountain chains at the east and west of the desert. b Aspect of sand dunes surrounding Locality A. c: Overview of Locality A with sand dunes and eastern mountain chain in the background
In Locality A, the maximum, mean and minimum daily temperatures show a marked monoseasonality, with a peak in January and a decrease up to the minimum values in June-July (Fig. 10a). Mean temperature values are only lower than the minimum temperature threshold of 10 °C known for F. hepatica during the period of June-July.
Yearly variation of monthly values of climatic factors during the period 1950–2000. a, b Locality A (Taton-Rio Grande). c, d Locality B (Ipizca). Left column graphs (a, c) correspond to yearly variation of temperatures (data in °C): red squares and line = mean maximum temperature (MMT); green triangles and line = mean environmental temperature (MET); blue diamonds and line = mean minimum temperature (MmT). Right column graphs (b, d) correspond to yearly variation of precipitation and potential evapotranspiration (data in mm/day) to show differences between the rainy and dry periods: blue line = precipitation; red line = potential evapotranspiration (PET); pink line = 0.45 × PET; dark blue areas = humid period (vegetation growing season - no one appears); green areas = moist period (vegetation growing season); yellow areas = dry period
In Locality B, the maximum, mean and minimum daily temperatures show a similar seasonality, although with somewhat higher temperatures (Fig. 10c). The mean temperature values are higher than the minimum temperature threshold of 10 °C known for F. hepatica throughout the whole year.
The analysis of the yearly variation of precipitation and potential evapotranspiration shows the very low availability of rainfall in both localities, with a small peak in January-February (slightly higher in Locality B) and almost inexistent precipitation during the period from April-May to October-November (Fig. 10b, d). Worth emphasising is the extension of dryness throughout the year in both localities, so that only in Locality B a very short moist period appears from mid-January to beginning of March when PET > 0.45 is applied (none when using PET > 0.5) (Fig. 10d).
These results indicate that the transmission foci of localities A and B where lymnaeid snails were collected are indeed isolated environments presenting water availability depending only on the rivers flowing from the neighbouring mountain chain. These biotopes appropriate for lymnaeid snail survival are confined to lateral river side floodings and small man-made irrigation systems in both Locality A (Additional file 2: Figure S1 a-g) and Locality B (Additional file 2: Figure S1 h-j). In Locality A, lymnaeid populations may reach high densities on mud and neighbouring superficial water borders in the adequate season (Additional file 3: Figure S2).
Discussion
DNA characterisation of lymnaeid vectors
In L. neotropica, the two populations of localities A and B were characterised by having identical sequences for the four DNA markers used, including two new haplotypes for ITS-1 and cox1 (Table 1). The new ITS-1 (L.neo-HB) haplotype is worth nothing, given the uniformity so far detected at the level of the nuclear rDNA spacers in this species throughout. The only previously known ITS-1 haplotype (L.neo-HA) has been reported from Lima (type-locality) [19] and Cajabamba [51], both in Peru, and also in Perdriel, Departamento de Lujan de Cuyo, Mendoza, Argentina [50]. The other new haplotype in a mtDNA marker (L.neo-cox1e) differs from the cox1 haplotypes described in L. neotropica in other countries such as Peru, Venezuela and also Argentina. In the latter country, this new L.neo-cox1e haplotype only shows identity with the cox1 sequence of the isolate NtC2 (GenBank: JN872453) previously reported from the same lymnaeid species in Mendoza [82], although unfortunately the fragment available from that isolate is shorter (655 bp vs 672 bp).
The haplotypes found in the other two markers in L. neotropica, one in rDNA ITS-2 (L.neo-H1) and the other in mtDNA 16S (L.neo-16SA), were already reported from another area of Argentina, namely in Perdriel [50]. These two ITS-2 and 16S haplotypes were also detected in Peru, in the type-locality besides Lima [19] and also in Cajamarca [51].
In L. viator, four new haplotypes were found in ITS-2, ITS-1, 16S and cox1 in Locality A (Table 1). Detecting a new haplotype in every marker studied in a single lymnaeid population is really surprising. Previously described haplotypes and isolate sequences of the same DNA markers of L. viator have been cited from different localities of Chile, Argentina and Uruguay (see Figs. 3, 4 and 5).
The analysis of sequences of L. neotropica and L. viator available in the GenBank database shows that several sequences are duplicated. For instance, at ITS-2 level, two sequences of L. viator from Rio Negro, Argentina (HQ283265 and JN614465), are identical to the sequence of L. viator ventricosa H1 (AM412224) from the type-locality (Rio Negro) [19]. Similarly, another ITS-1 sequence (HQ283254) is identical to the sequence of L. viator ventricosa HA (AM412227) from the same type-locality (Rio Negro) [19]. Similar situations are also found in mtDNA sequences, e.g. a 16S sequence of L. neotropica (JN872473) which is identical to that of the haplotype L.neo-16SA (HE610433) from the type-locality of the species [51]. Also another L. viator cox1 sequence (JN614397) from Rio Negro is identical to that of L.via-cox1a (AM494010) from the same type-locality [19]. Additionally, two sequences deposited in the databases as belonging to L. viator ITS-1 (JF960165 and JF960166) actually belong to L. neotropica, although species ascription was corrected in the publication [83].
Phylogenetic analysis
Although many attempts to divide the Galba/Fossaria group of lymnaeids into different genera have been made since long ago, proposals have never been unanimously accepted [84–89]. Moreover, molecular analyses on genetic distances by appropriately considering the different weights of the different nucleotide differences and phylogenetic assessments by different tree reconstruction methods offer different results which do not allow for a clear taxonomic arrangement of the Galba/Fossaria species so far. Such a lack of molecular systematics consistency appears using either mitochondrial DNA markers [90] or nuclear ribosomal DNA markers [20], as well as both markers together [18].
The phylogenetic tree obtained in the present study reinforces the arguments against including the American native species L. neotropica, L. viator and L. cubensis within the genus Galba, which is defined by the type-species G. truncatula of Euro-Asian origin. Therefore, we have kept the two lymnaeid species found in the Catamarca province within the genus Lymnaea (sensu lato) throughout.
Morphometric characterisation of L. neotropica and L. viator
In lymnaeids, the interspecific morphological and anatomic uniformity exhibited by numerous species gives usually rise to serious difficulties in specimen identification, sometimes even impeding it [17–19]. Moreover, intraspecific variation of shell shape is particularly well marked within lymnaeids depending on environmental conditions. Such specimen identification problems are well known in species of the Galba/Fossaria group [18].
Within the Galba/Fossaria group, there are species which show high transmission capacity and others non-susceptible to F. hepatica, such as L. schirazensis [18]. Moreover, there are Galba/Fossaria species clearly related to human fascioliasis endemic areas because of their anthropophilic ecology and behaviour, whereas other species do usually inhabit biotopes frequented by livestock [38, 51]. There is therefore an applied interest in species differentiation when dealing with an endemic area and also their geographical distribution throughout a country [28].
The morphometric study performed to assess whether L. neotropica and L. viator could be morphologically distinguished in Locality A has shown an overlap in all shell features, except for the maximum length that may be greater in L. viator (Additional file 1: Table S1). However, no definitive conclusion on this potentially differing feature depending on growth can be reached through measurements obtained from specimens collected in nature. Unfortunately, the single absolute measurement of the shell which showed a statistical capacity for species differentiation was the shell spiral angle. Despite its usefulness for shell description, this angle is known to provide a useless marker for species differentiation because (i) there is no exact way to measure it [78] and (ii) it shows large intraspecific and even intrapopulational variability in lymnaeids [84]. The statistical analysis suggested that the three shell ratios might be helpful for species differentiation. However, regarding SL/SW, the existence of slender shells together with somewhat stouter ones was already emphasised in the type-locality of L. viator [91]. Concerning SL/AL and SL/LSL, ranges overlap completely and means are almost equal for both species (see Additional file 1: Table S1). Even using the combination of all shell parameters, the identification of a specimen is subject to a high risk of confusion, as shown by the results of the principal components analysis (see Fig. 8). The only species difference was the preputium/penis-sheath length ratio, close to 1 in L. neotropica vs close to 3 in L. viator. Unfortunately, obtaining this ratio needs a time-consuming anatomical dissection and malacological expertise which may not be available among public health responsibles working in the field. Moreover, a variability in this ratio of 1.1–3.9 in L. neotropica has been found throughout its geographical distribution [19, 82, 91, 92].
Additionally, the two species appear to coexist in the same water collections in the aforementioned locality, reiterating a similar situation of overlap of these two species in many places of the Argentinian province of Mendoza [93]. A similar phenomenon leading to confusion of the lymnaeid species due to the impossibility of distinguishing between two species and the repercussions for the disease transmission has recently been evoked in the case of G. truncatula and L. schirazensis [51].
Climatic and environmental characterisation of the lymnaeid habitats
The extreme desertic-arid environmental characteristics surrounding Locality A and the slightly less extreme conditions of semiaridity-aridity of those surrounding Locality B, as well as the very low yearly precipitation in both localities, are surprising and very different from the typical environmental characteristics surrounding transmission foci of fascioliasis. In the two localities studied, lymnaeid snail habitats are confined to lateral river side floodings and small man-made irrigation systems.
In the aforementioned extreme situations, water availability only depends on water from the rivers and hence almost exclusively from rainfall, snow melting and deglaciation in the mountains where the rivers originate. A similar fascioliasis transmission dependence on river water coming from rainfall (monzoon) in the neighbouring mountains (Himalaya) in a markedly very large dry area (Punjab province) has recently been highlighted in Pakistan [6]. In the Argentinian areas studied here, the data indicate a concentration of all disease transmission factors in small areas where humans and animals go for water supply, vegetable cultures and livestock farming. Such a concentrated transmission focus such as in Locality A reminds the epidemiological characteristics of the transmission foci of schistosomiasis in oases of the Sahara Desert in Africa [94].
In both localities studied, a seasonal transmission may be expected depending on the timely overlap of the appropriate temperature and water availability from the rivers. Such a seasonality is known in endemic areas following the “valley pattern” of fascioliasis transmission known in other Andean countries [26, 95]. Lymnaea neotropica has already been linked to human fascioliasis hyperendemic areas following the “valley pattern”, as in the valley of Cajamarca, Peru [51]. Lymnaea viator was so far related to areas where human infection concerning a sporadic case or a few patients has been reported in Argentina [61, 96], because recent knowledge suggests that its link to human endemic areas in Chile may involve a confusion with G. truncatula [38].
Conclusions
The finding of an unusual high number of DNA haplotypes in both lymnaeid vector species and the extreme climatic factors (almost total lack of rainfall and moisture) unsuitable for F. hepatica and freshwater lymnaeid snail development, demonstrate that the transmission foci are completely isolated. Consequently, it may be concluded that both the lymnaeid species and F. hepatica have reached both localities by means of livestock introduction. Moreover, DNA differences from other populations of L. neotropica and L. viator previously studied in Argentina, suggest that this introduction phenomenon was independent from the spreading movements which allowed these two lymnaeid species to expand throughout the country.
Abbreviations
- 16S rDNA:
-
ribosomal RNA coding large subunit gene (16S) of the mitochondrial genome
- COX1:
-
cytochrome c oxidase subunit 1, amino acid sequence
- cox1:
-
cytochrome c oxidase subunit 1, nucleotide sequence
- FAOCLIM:
-
agroclimatic database of the FAO Agromet Group
- H:
-
haplotype
- indel:
-
insertion/deletion
- ITS-1:
-
first internal transcribed spacer
- ITS-2:
-
second internal transcribed spacer
- MET:
-
mean environmental temperature
- MMT:
-
mean maximum temperature
- MmT:
-
mean minimum temperature
- mtDNA:
-
mitochondrial DNA
- PET:
-
potential evapotranspiration
- rDNA:
-
nuclear ribosomal DNA
- ts:
-
transition
- tv:
-
transversion
References
Chen MG, Mott KE. Progress in assessment of morbidity due to Fasciola hepatica infection: a review of recent literature. Trop Dis Bull. 1990;87:R1–38.
Mas-Coma S, Valero MA, Bargues MD. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol. 2009;69:41–146.
Ashrafi K, Valero MA, Massoud J, Sobhani AR, Solaymani-Mohammadi S, Conde P, et al. Plant-borne human contamination by fascioliasis. Am J Trop Med Hyg. 2006;75:295–302.
Mas-Coma S, Valero MA, Bargues MD. Effects of climate change on animal and zoonotic helminthiases. Rev Sci Techn Off Int Epiz. 2008;27:443–57.
Boissier J, Mone H, Mitta G, Bargues MD, Molyneux D, Mas-Coma S. Schistosomiasis reaches Europe. Lancet Infect Dis. 2015;15:757–8.
Afshan K, Fortes-Lima CA, Artigas P, Valero MA, Qayyum M, Mas-Coma S. Impact of climate change and man-made irrigation systems on the transmission risk, long-term trend and seasonality of human and animal fascioliasis in Pakistan. Geospat Health. 2014;8:317–34.
Ashrafi K, Bargues MD, O’Neill S, Mas-Coma S. Fascioliasis, a worldwide parasitic disease of importance in travel medicine. Travel Med Infect Dis. 2014;12:636–49.
Fuentes MV, Valero MA, Bargues MD, Esteban JG, Angles R, Mas-Coma S. Analysis of climatic data and forecast indices for human fascioliasis at very high altitude. Ann Trop Med Parasitol. 1999;93:835–50.
Fuentes MV, Malone JB, Mas-Coma S. Validation of a mapping and predicting model for human fasciolosis transmission in Andean very high altitude endemic areas using remote sensing data. Acta Trop. 2001;79:87–95.
Mas-Coma S, Valero MA, Bargues MD. Climate change effects on trematodiases, with emphasis on zoonotic fascioliasis and schistosomiasis. Vet Parasitol. 2009;163:264–80.
Mas-Coma S, Bargues MD, Valero MA. Diagnosis of human fascioliasis by stool and blood techniques: Update for the present global scenario. Parasitology. 2014;141:1918–46.
Valero MA, Santana M, Morales M, Hernandez JL, Mas-Coma S. Risk of gallstone disease in advanced chronic phase of fascioliasis: an experimental study in a rat model. J Inf Dis. 2003;188:787–93.
Valero MA, Navarro M, Garcia-Bodelon MA, Marcilla A, Morales M, Garcia JE, et al. High risk of bacterobilia in advanced experimental chronic fasciolosis. Acta Trop. 2006;100:17–23.
Valero MA, Girones N, Garcia-Bodelon MA, Periago MV, Chico-Calero I, Khoubbane M, et al. Anemia in advanced chronic fasciolosis. Acta Trop. 2008;108:35–43.
Girones N, Valero MA, Garcia-Bodelon MA, Chico-Calero MI, Punzon C, Fresno M, et al. Immune supression in advanced chronic fascioliasis: an experimental study in a rat model. J inf Dis. 2007;195:1504–12.
Mas-Coma S, Agramunt VH, Valero MA. Neurological and ocular fascioliasis in humans. Adv Parasitol. 2014;84:27–149.
Bargues MD, Mas-Coma S. Reviewing lymnaeid vectors of fascioliasis by ribosomal DNA sequence analyses. J Helminthol. 2005;79:257–67.
Bargues MD, Artigas P, Khoubbane M, Flores R, Glöer P, Rojas-Garcia R, et al. Lymnaea schirazensis, an overlooked snail distorting fascioliasis data: Genotype, phenotype, ecology, worldwide spread, susceptibility, applicability. PLoS One. 2011;6:e24567.
Bargues MD, Artigas P, Mera y Sierra R, Pointier JP, Mas-Coma S. Characterisation of Lymnaea cubensis, L. viatrix and L. neotropica n. sp., the main vectors of Fasciola hepatica in Latin America, by analysis of their ribosomal and mitochondrial DNA. Ann Trop Med Parasitol. 2007;101:621–41.
Bargues MD, Vigo M, Horak P, Dvorak J, Patzner RA, Pointier JP, et al. European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS-2 sequences. Infect Genet Evol. 2001;1:85–107.
Bargues MD, Artigas P, Khoubbane M, Mas-Coma S. DNA sequence characterisation and phylogeography of Lymnaea cousini and related species, vectors of fascioliasis in northern Andean countries, with description of Lymnaea meridensis n. sp. (Gastropoda: Lymnaeidae). Parasit Vectors. 2011;4:132.
Bargues MD, Horak P, Patzner RA, Pointier JP, Jackiewicz M, Meier-Brook C, et al. Insights into the relationships of Palaearctic and Nearctic lymnaeids (Mollusca: Gastropoda) by rDNA ITS-2 sequencing and phylogeny of stagnicoline intermediate host species of Fasciola hepatica. Parasite. 2003;10:243–55.
Rondelaud D, Vignoles P, Dreyfuss G. Adaptation of preadult Lymnaea fuscus and Radix balthica to Fasciola hepatica through the infection of several successive generations of snails. Parasit Vectors. 2014;7:296.
Valero MA, Mas-Coma S. Comparative infectivity of Fasciola hepatica metacercariae from isolates of the main and secondary reservoir animal host species in the Bolivian Altiplano high human endemic region. Folia Parasitol. 2000;47:17–22.
Valero MA, Darce NA, Panova M, Mas-Coma S. Relationships between host species and morphometric patterns in Fasciola hepatica adults and eggs from the Northern Bolivian Altiplano hyperendemic region. Vet Parasitol. 2001;102:85–100.
Mas-Coma S. Epidemiology of fascioliasis in human endemic areas. J Helminthol. 2005;79:207–16.
Mas-Coma S, Esteban JG, Bargues MD. Epidemiology of human fascioliasis: a review and proposed new classification. Bull WHO. 1999;77:340–6.
Bargues MD, Gonzalez LC, Artigas P, Mas-Coma S. A new baseline for fascioliasis in Venezuela: lymnaeid vectors ascertained by DNA sequencing and analysis of their relationships with human and animal infection. Parasit Vectors. 2011;4:200.
Hillyer GV, Soler de Galanes M, Rodriguez-Perez J, Bjorland J, Silva de Lagrava M, Guzman SR, et al. Use of the Falcon™ assay screening test-enzyme-linked immunosorbent assay (FAST-ELISA) and the enzyme-linked immunoelectrotransfer blot (EITB) to determine the prevalence of human fascioliasis in the Bolivian altiplano. Am J Trop Med Hyg. 1992;46:603–9.
Esteban JG, Flores A, Angles R, Strauss W, Aguirre C, Mas-Coma S. A population-based coprological study of human fascioliasis in a hyperendemic area of the Bolivian Altiplano. Trop Med Int Health. 1997;2:695–9.
Esteban JG, Flores A, Aguirre C, Strauss W, Angles R, Mas-Coma S. Presence of very high prevalence and intensity of infection with Fasciola hepatica among Aymara children from the Northern Bolivian Altiplano. Acta Trop. 1997;66:1–14.
Esteban JG, Flores A, Angles R, Mas-Coma S. High endemicity of human fascioliasis between Lake Titicaca and La Paz valley, Bolivia. Trans Roy Soc Trop Med Hyg. 1999;93:151–6.
Mas-Coma S, Angles R, Esteban JG, Bargues MD, Buchon P, Franken M, et al. The Northern Bolivian Altiplano: a region highly endemic for human fascioliasis. Trop Med Int Health. 1999;4:454–67.
Esteban JG, Gonzalez C, Bargues MD, Angles R, Sanchez C, Naquira C, et al. High fascioliasis infection in children linked to a man-made irrigation zone in Peru. Trop Med Int Health. 2002;7:339–48.
Espinoza JR, Maco V, Marcos L, Saez S, Neyra V, Terashima A, et al. Evaluation of Fas2-ELISA for the serological detection of Fasciola hepatica infection in humans. Am J Trop Med Hyg. 2007;76:977–82.
Gonzalez LC, Esteban JG, Bargues MD, Valero MA, Ortiz P, Naquira C, et al. Hyperendemic human fascioliasis in Andean valleys: An altitudinal transect analysis in children of Cajamarca province, Peru. Acta Trop. 2011;120:119–29.
Apt W, Aguilera X, Vega F, Alcaino H, Zulantay I, Apt P. Prevalencia de fascioliasis en humanos, caballos, cerdos y conejos silvestres en tres provincias de Chile. Bol Of Sanit Panam. 1993;115:405–14.
Artigas P, Bargues MD. Mera y Sierra R, Agramunt VH, Mas-Coma S. Characterisation of fascioliasis lymnaeid intermediate hosts from Chile by DNA sequencing, with emphasis on Lymnaea viator and Galba truncatula. Acta Trop. 2011;120:245–57.
Arroyo R, Mora J, Molina S, Troper L, Amador A. Fascioliasis hepática humana en Costa Rica. Rev Costarricense Cienc Méd. 1981;2:35–57.
Zumaquero-Rios JL, Sarracent-Perez J, Rojas-Garcia R, Rojas-Rivero L, Martinez-Tovilla Y, Valero MA, et al. Fascioliasis and intestinal parasitoses affecting schoolchildren in Atlixco, Puebla State, Mexico: Epidemiology and treatment with nitazoxanide. PLoS Negl Trop Dis. 2013;7:e2553.
Trueba G, Guerrero T, Fornasini M, Casariego I, Zapata S, Ontaneda S, et al. Detection of Fasciola hepatica infection in a community located in the Ecuadorian Andes. Am J Trop Med Hyg. 2000;62:518.
Millan JN, Mull R, Freise S, Richter J. The efficacy and tolerability of triclabendazole in Cuban patients with latent and chronic Fasciola hepatica infection. Am J Trop Med Hyg. 2000;63:264–9.
Diaz Fernandez R, Garces Martinez M, Millan Alvarez LM, Perez Lastre J, Millan Marcelo JC. Comportamiento clínico-terapéutico de Fasciola hepatica en una serie de 87 pacientes. Rev Cub Med Trop. 2011;63:268–74.
Gonzales Santana B, Dalton JP, Vasquez Camargo F, Parkinson M, Ndao M. The diagnosis of human fascioliasis by enzyme-linked immunosorbent assay (ELISA) using recombinant cathepsin L protease. PLoS Negl Trop Dis. 2013;7:e2414.
Agnamey P, Fortes-Lopes E, Raccurt CP, Boncy J, Totet A. Cross-sectional serological survey of hunman fascioliasis in Haiti. J Parasitol Res. 2012;2012:751951. doi:10.1155/2012/751951.
Hillyer GV. Fascioliasis in Puerto Rico: A review. Bol Asoc Méd Puerto Rico. 1981;73:94–101.
Mera y Sierra R, Agramunt V, Cuervo P, Mas-Coma S. Human fascioliasis in Argentina: retrospective overview, critical analysis and baseline for future research. Parasit Vectors. 2011;4:104.
Bargues MD. Mera y Sierra R, Gomez HG, Artigas P, Mas-Coma S. Ribosomal DNA ITS-1 sequencing of Galba truncatula (Gastropoda: Lymnaeidae) and its potential impact on fascioliasis transmission in Mendoza, Argentina. Animal Biodiv Conserv. 2006;29:191–4.
Bargues MD, Mera y Sierra R, Gomez HG, Artigas P, Mas-Coma S. Caracterización molecular de Galba truncatula, vector principal de la Fascioliasis, en Argentina. Implicaciones en salud pública. Enf Emerg Barcelona. 2007;9:77–82.
Mera y Sierra R, Artigas P, Cuervo P, Deis E, Sidoti L, Mas-Coma S, et al. Fascioliasis transmission by Lymnaea neotropica confirmed by nuclear rDNA and mtDNA sequencing in Argentina. Vet Parasitol. 2009;166:73–9.
Bargues MD, Artigas P, Khoubbane M, Ortiz P, Naquira C, Mas-Coma S. Molecular characterisation of Galba truncatula, Lymnaea neotropica and L. schirazensis from Cajamarca, Peru and their potential role in transmission of human and animal fascioliasis. Parasit Vectors. 2012;5:174.
Nieto Sosa L, Barnes A, Jimenez G, Cicaerlli G. Fasciolosis: evaluación clínica y terapéutica. In: Resúmenes de las Comunicaciones del III Congreso Latinoamericano de Zoonosis y VI Congreso Argentino de Zoonosis, Asociación Argentina de Zoonosis, Universidad Católica Argentina, Buenos Aires, Argentina; 2008. p. 35.
Bacigalupo J. Fasciola hepatica, su ciclo evolutivo en la República Argentina. An Fac Vet Montevideo. 1942;4:7–134.
Ossola A, Siciliano C, Garcia FJ. Distomatosis hepática, tres epidemias familiares en Córdoba, Argentina. Rev Asoc Bioq Arg. 1972;37:100–6.
Peiretti JA, Morales JR. Epidemia familiar de distomatosis por Fasciola hepatica. Prensa Méd Arg. 1973;60:262–4.
Siciliano C, Chalub E, Sosa L, Barnes A, Shicton G. Distomatosis hepática, nuevos casos en Córdoba, Argentina. Prensa Méd Arg. 1989;76:106–9.
Minoprio JE, Donna RA, Borremans CG, Passardi AH, Glatstein de Wolf T, Olindo AL, et al. Epidemia familiar de distomatosis humana por Fasciola hepatica. Localización errática (cutánea). Prensa Méd Arg. 1995;82:869–77.
Salomon MC, De Jong L, Tonelli RL, Borremans CG, Bertello D, Jofre CA, et al. Confirmación etiológica de un brote de distomatosis hepática en Mendoza. In: Resúmenes de las Comunicaciones de las XII Jornadas Argentinas de Microbiología, Asociación Argentina de Microbiología - Filial Cuyo, Universidad Nacional de Cuyo, Mendoza, Argentina; 2006. p. 108.
Carnevale S, Rodriguez MI, Guarnera EA, Carmona C, Tanos T, Angel SO. Immunodiagnosis of fasciolosis using recombinant procathepsin L cysteine proteinase. Diagn Microbiol Infect Dis. 2001;41:43–9.
Carnevale S, Rodriguez MI, Santillan G, Labbe JH, Cabrera MG, Belegarde EJ, et al. Immunodiagnosis of human fascioliasis by an Enzyme-Linked Immunosorbent Assay (ELISA) and a Micro-ELISA. Clin Diagn Lab Immunol. 2001;8:174–7.
Carnevale S, Cabrera MG, Cucher MA, di Risio CA, Malandrini JB, Kamenetzky L, et al. Direct, immunological and molecular techniques for a fasciolosis survey in a rural area of San Luis, Argentina. J Parasit Dis. 2013;37:251–9.
Malandrini JB, Carnevale S, Velazquez J, Soria CC. Diagnóstico de Fasciola hepatica con la técnica de ELISA en el Departamento de Tinogasta. Ciencia. 2009;4:143–51.
Malandrini JB, Soria CC, Foresi I, Correa S. La Investigación Científica en la Facultad de Ciencias de la Salud, en los Umbrales del Siglo XXI. In: Fasciolosis en Catamarca. Primeros resultados. Catamarca: Editorial Universitaria; 2002. p. 287–93.
Oliver JE, Wilson L. The Encyclopedia of Climatology. Encyclopedia of Earth Sciences Series. In: Oliver JE, Fairbridge RW, editors. Climatic classification. New York: Van Nostrand Reinhold, Springer-Verlag; 1987. p. 221–37.
Grieser J, Gommes R, Cofield S, Bernardi M. Data sources for FAO worldmaps of Koeppen climatologies and climatic net primary production. FAO, Environment, Climate Change and Bioenergy Division. 2006. http://www.fao.org/nr/climpag/globgrids/kc_commondata_en.asp. Accessed 6 May 2015.
Mas-Coma S, Bargues MD. Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. 2009;110:112–36.
Simon C, Franke A, Martin A. The polymerase chain reaction: DNA extraction and amplification. In: Hewitt GM, Johnston AB, Youn JPW, editors. Molecular Techniques in Taxonomy. Berlin: Springer; 1991. p. 329–55.
Folmer O, Black M, Hoch W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotech. 1994;3:294–9.
Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, et al. ClustalW and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Bargues MD, Marcilla A, Ramsey J, Dujardin JP, Schofield CJ, Mas-Coma S. Nuclear rDNA-based molecular clock of the evolution of Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mem Inst Oswaldo Cruz. 2000;95:567–73.
Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other Methods). Version 4. Computer program distributed by the Smithsonian Institution. Sunderland, Massachusetts: Sinauer Associates; 2002.
Posada D. ModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6.
Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
De Jong RJ, Morgan JAT, Paraense WL, Pointier JP, Amarista M, Ayeh-Kumi PFK, et al. Evolutionary relationships and biogeography of Biomphalaria (Gastropoda: Planorbidae) with implications regarding its role as host of the human bloodfluke, Schistosoma mansoni. Mol Biol Evol. 2001;18:2225–39.
Samadi S, Roumegoux A, Bargues MD, Mas-Coma S, Yong M, Pointier JP. Morphological studies of lymnaeid snails from the human fascioliasis endemic zone of Bolivia. J Molluscan Stud. 2000;66:31–44.
Oviedo JA, Bargues MD, Mas-Coma S. Lymnaeid snails in the human fascioliasis high endemic zone of the Northern Bolivian Altiplano. Res Rev Parasitol. 1995;55:35–43.
Valero MA, Panova M, Mas-Coma S. Phenotypic analysis of adults and eggs of Fasciola hepatica by computer image analysis system. J Helminthol. 2005;79:217–25.
Valero MA, Perez-Crespo I, Periago MV, Khoubbane M, Mas-Coma S. Fluke egg characteristics for the diagnosis of human and animal fascioliasis by Fasciola hepatica and F. gigantica. Acta Trop. 2009;111:150–9.
FAO. Report on the agro-ecological zones project, Vol 1. Results for Africa. FAO, Rome: World Soil Resources Report 48/1. 1978.
Standley CJ, Prepelitchi L, Pietrokovsky SM, Issia L, Stothard JR, Wisnivesky-Colli C. Molecular characterization of cryptic and sympatric lymnaeid species from the Galba/Fossaria group in Mendoza Province, Northern Patagonia, Argentina. Parasit Vectors. 2013;6:304.
Sanabria R, Mouzet R, Pankrac J, Djuikwo Teukeng FF, Courtioux B, Novobilsky A, et al. Lymnaea neotropica and Lymnaea viatrix, potential intermediate hosts for Fascioloides magna. J Helminthol. 2013;87:494–500.
Hubendick B Recent Lymnaeidae. Their variation, morphology, taxonomy, nomenclature, and distribution. Kungliga Svenska Vetenskapsakademiens Handlingar, Fjärde Serien, Royal Swedish Academy of Sciences, Stockholm, 1951;3:1–223 + 5 pl.
Burch JB. North American freshwater snails. Identification keys, generic synonymy, supplemental notes, glossary, references, index. Walkerana. Trans POETS Soc. 1982;1:216–365.
Burch JB. North American freshwater snails. Introduction, systematics, nomenclature, identification, morphology, habitats, distribution. Walkerana. Trans POETS Soc. 1988;2:1–80.
Kruglov ND, Starobogatov YI. The volume of the subgenus Galba and of the other Lymnaea (Gastropoda, Pulmonata) subgenera, similar to it. Zool Zhurnal. 1985;64:24–35.
Glöer P. Meier-Brook C. Süsswassermollusken. Ein Bestimmungsschlüssel für die Bundesrepublik Deutschland, 13th Edition. Hamburg: Deutscher Jugendbund für Naturbeobachtung; 2003. p. 1–134.
Kruglov ND. Molluscs of the Family Lymnaeidae (Gastropoa, Pulmonata) in Europe and Northern Asia. Smolensk: SGPU Publishing; 2005. p. 1–507.
Remigio EA, Blair D. Molecular systematics of the freshwater snail family Lymnaeidae (Pulmonata: Basommatophora) utilising mitochondrial ribosomal DNA sequences. J Molluscan Stud. 1997;63:173–85.
Paraense WL. Lymnaea viatrix: a study of topotypic specimens (Mollusca: Lymnaeidae). Rev Brasil Biol. 1976;36:419–28.
Paraense WL. Lymnaea viatrix and Lymnaea columella in the neotropical region: a distributional outline. Mem Inst Oswaldo Cruz. 1982;77:181–8.
Mera y Sierra R, Cuervo P, Sidoti L, Artigas P, Bargues MD, Mas-Coma S. And then there were three: Lymnaeid vectors of fascioliasis in highly endemic province of Argentina. In: Abstracts in CD-ROM of the XII International Congress of Parasitology. Melbourne: ICOPA XII; 2010. p. 1141.
Doumenge JP, Mott KE, Cheung C, Villenave D, Chapuis O, Perrin MF, et al. Atlas of the Global Distribution of Schistosomiasis. CRGET-CNRS, Talence; OMS/WHO, Genève. Talence: Presses Universitaires de Bordeaux; 1987.
Valero MA, Perez-Crespo I, Khoubbane M, Artigas P, Panova M, Ortiz P, et al. Fasciola hepatica phenotypic characterisation in Andean human endemic areas: valley versus altiplanic patterns analysed in liver flukes from sheep from Cajamarca and Mantaro, Peru. Infect Genet Evol. 2012;12:403–10.
Rubel D, Prepelitchi L, Kleiman F, Carnevale S, Wisnivesky-Colli C. Estudio del foco en un caso de fasciolosis humana en Neuquén. Med B Aires. 2005;65:207–12.
Acknowledgements
The studies were funded by Project No. SAF2010-20805 of the Ministry of Economy and Competitiveness, Madrid, Spain; by the Red de Investigación de Centros de Enfermedades Tropicales - RICET (Project No. RD12/0018/0013 of the VI PN de I + D + I 2008–2011, ISCIII-Subdirección General de Redes y Centros de Investigación Cooperativa RETICS), Ministry of Health and Consumption, Madrid; and by Project No. 2012/042 of the PROMETEO Program, Generalitat Valenciana, Valencia, Spain. This study is part of the worldwide initiative of WHO (Headquarters Geneva, Switzerland) against human fascioliasis. Technical support was provided by the Servicio Central de Secuenciación para la Investigación Experimental (SCSIE) of the Universidad de Valencia (Dr. A. Martínez).
Authors’ contributions
MDB, JBM and SMC conceived the study, its design and coordination. MDB, PA and LM carried out the molecular studies and genetic analyses. MK and SMC carried out the morphometric studies. JBM, CCS, JNV and SC carried out the environment analyses in field studies. PA and SMC carried out the climate analyses. CCS, JNV and SC made substantial contributions to acquisition and interpretation of data. MDB, JBM and SMC drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Lymnaeid shell measurements for natural populations of morphologically similar species, Lymnaea viator and Lymnaea neotropica from the present study and Galba truncatula, including t-test statistical comparison of variables of L. viator vs L. neotropica in Locality A. (PDF 67 kb)
Additional file 2: Figure S1.
Freshwater habitats of lymnaeids. a-g Locality A: in villages of Taton and Rio Grande. h-j Locality B: in the neighbourhood of Ipizca. b Note general aridity. c Sand dunes in the background. b, d Closeness to human dwellings. g Lymnaeids present on mud. i Small artificial dyke on the river. (PDF 448 kb)
Additional file 3: Figure S2.
High density of lymnaeids on mud and freshwater border in Locality A (village of Taton). (PDF 415 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bargues, M.D., Malandrini, J.B., Artigas, P. et al. Human fascioliasis endemic areas in Argentina: multigene characterisation of the lymnaeid vectors and climatic-environmental assessment of the transmission pattern. Parasites Vectors 9, 306 (2016). https://doi.org/10.1186/s13071-016-1589-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1589-z