Abstract
Background
Human and animal fascioliasis is emerging in many world regions, among which Andean countries constitute the largest regional hot spot and Peru the country presenting more human endemic areas. A survey was undertaken on the lymnaeid snails inhabiting the hyperendemic area of Cajamarca, where human prevalences are the highest known among the areas presenting a "valley transmission pattern", to establish which species are present, genetically characterise their populations by comparison with other human endemic areas, and discuss which ones have transmission capacity and their potential implications with human and animal infection.
Methods
Therefore, ribosomal DNA ITS-2 and ITS-1, and mitochondrial DNA 16S and cox 1 were sequenced by the dideoxy chain-termination method.
Results
Results indicate the presence of three, morphologically similar, small lymnaeid species belonging to the Galba/Fossaria group: Galba truncatula, Lymnaea neotropica and L. schirazensis. Only one combined haplotype for each species was found. The ITS-1, 16S and cox 1 haplotypes of G. truncatula are new. No new haplotypes were found in the other two species. This scenario changes previous knowledge, in which only L. viator (= L. viatrix) was mentioned. Galba truncatula appears to be the most abundant, with high population densities and evident anthropophyly including usual presence in human neighbourhood. Infection by Fasciola hepatica larval stages were molecularly confirmed in two populations of this species. The nearness between G. truncatula populations presenting liver fluke infection and both human settings and schools for children, together with the absence of populations of other lymnaeid species in the locality, suggest a direct relationship with human infection.
Conclusions
The geographical overlap of three lymnaeid species poses problems for epidemiological studies and control action. First, a problem in classifying lymnaeid specimens in both field and laboratory activities, given their transmission capacity differences: G. truncatula mainly involved in transmission to humans, L neotropica typically responsible for livestock infection, and L. schirazensis unable for transmission. Although several phenotypic characteristics may be helpful for a preliminary specimen classification, a definitive classification can only be obtained by marker sequencing. Aditionally, L. schirazensis increases the confusion, owing to its ability to mix with other Galba/Fossaria species and distort fascioliasis data such as transmission capacity and infection susceptibility. Second, a problem for epidemiological analysis, surveillance and control by methods as mathematical modelling and Remote Sensing - Geographical Information Systems. In Cajamarca, low resolution mapping may be insufficient, as already verified in Andean areas where different lymnaeid species overlap.
Similar content being viewed by others
Background
Fascioliasis is a parasitic disease transmitted by freshwater lymnaeid snails and caused by Fasciola hepatica distributed almost throughout and F. gigantica in large regions of Africa and Asia [1]. Distribution, both in space (latitudinal, longitudinal and altitudinal) and time (seasonal, yearly), of fascioliasis depends on the presence and population dynamics of the specific intermediate host or vector species in its turn linked to the presence of the appropriate water bodies and on adequate climate characteristics enabling fluke development [1, 2].
Although livestock species play an important reservoir role [3], transmission studies have shown that the metacercarial infective stage from different origins, such as sheep, cattle, pig and donkey, represent similar infectivity sources [4, 5]. On the contrary, the specificity of fasciolid species regarding given lymnaeid species [6] represent a crucial factor in establishing not only the geographical distribution of the disease in both animals and humans, but also prevalences and intensities due to more or less appropriate ecological characteristics (population dynamics, anthropophylic characteristics, type of water bodies, etc.) of the different lymnaeid intermediate host or vector species. That is why different lymnaeid species appear linked to the different transmission patterns and epidemiological scenarios of this very heterogeneous disease in humans [1, 7]. Similarly as in other vector-borne diseases, this relationship supports the use of lymnaeids as biomarkers of the disease at both local and large scales and can thus be useful for the validation of mathematical modelling and remote sensing – geographical information system (RS-GIS) tools for the control of the disease [8, 9].
In the Americas, the greatest problems are known in Andean countries. Peru appears as the country presenting a larger public health problem due to human infection by F. hepatica. Human fascioliasis has been diagnosed in inhabitants from almost all Andean areas, including from the Altiplano [10] up to inter-Andean valleys [11–13], and even urban areas surrounding the capital of Lima [14] and low altitude areas closer to the Pacific coast [15]. Many of these areas have proved to be human endemic. A rural population of almost 8 million people is estimated at risk in Peru [16]. Bolivia presents the endemic area of the Northern Altiplano with the highest human prevalences and intensities known [17–19]. In Chile, a human endemic area has been described and human cases are reported yearly [20, 21]. Available data on human infection in Venezuela [22], Ecuador [23] and Argentina [24] also suggest that the respective real situations in these three countries may be underestimated.
The increasing importance of human fascioliasis does not only rely on the recent wide emergence it shows, but also on the results obtained in studies on pathogenicity [25–28] and immunity [29, 30], according to which this disease appears to be pronouncedly more complicated and with a greater impact in long-term infection than what was believed until the 90s. The origin of the emergence of fascioliasis in recent years has been argued to be related to climate change, at least in part and in given countries [31, 32], as a consequence of the high dependence of fascioliasis transmission and freshwater lymnaeid snails on climate and environmental characteristics [8, 9, 33].
Emergence, long-term pathogenicity and immunological interactions are in the background of the decision taken by the World Health Organization (WHO) to include this disease within the so-called neglected tropical diseases (NTDs). The great concern related to the epidemiological situations in many countries led WHO to launch a worldwide initiative against this disease [34, 35]. The first step of this initiative was a pilot intervention in different countries selected according to their different epidemiological situations and transmission patterns [1, 7].
Peru was one of the countries selected for priority intervention. Within the human fascioliasis high altitude transmission pattern related to F. hepatica transmitted by lymnaeid vectors of the Galba/Fossaria group, two different subpatterns have been distinguished in Peru [1, 7]: a) the altiplanic pattern, with endemicity distributed throughout an area of homogeneous altitude and transmission throughout the whole year due to high evapotranspiration rates leading lymnaeid vectors to concentrate in permanent water bodies [2]; examples are the Northern Bolivian Altiplano and the Peruvian Altiplano of Puno; b) the valley pattern, with endemicity distributed throughout an area of heterogeneous altitude and seasonal transmission related to climate [36, 37]; Peruvian examples are the valleys of Cajamarca and Mantaro.
The present article deals with the lymnaeid snail surveys performed in the human and animal endemic areas of Cajamarca. The aim of the present study is to analyse the DNA sequences obtained from lymnaeids collected, mainly in the neighbourhood of localities where human infection is known to be high. The purpose is to establish which lymnaeid snail species are present, perform a molecular characterisation of their populations in Cajamarca by comparison with other populations of the same lymnaeid species in other human endemic areas, and finally discuss which ones have disease transmission capacity and their potential implications with the disease in humans and animals. Therefore, four DNA markers were selected: the two internal transcribed spacers of the nuclear ribosomal DNA (rDNA ITS-2 and ITS-1), and two genes of the mitochondrial DNA such as the large subunit 16S codifying for rRNAs and the cytochrome c oxidase subunit I (mtDNA cox 1) codifying gene [6, 22, 38–46]. DNA markers have also proved to be useful in Planorbidae, the other snail family of medical interest [47, 48]. Similarly as in other invertebrates [49], in lymnaeids ITS-2 and ITS-1 appear to be the best markers at species and supraspecific levels, whereas 16S and cox 1 are useful at infraspecific and population levels [42, 46].
Methods
Lymnaeid snail materials
The snail specimens studied were collected in the field, from lymnaeid populations present in geographical areas of the Departamento de Cajamarca in the northern Andean part of Peru, with human infection and/or animal fascioliasis endemicity. The majority of the snail populations studied were found close to schools whose children proved to be infected [13] or around neighbouring villages.
For an estimation of lymnaeid population densities, the method of counting the number of snails in a marked area in a unit of time was used. Marked areas were mainly a segment of a canal or, less frequently, a portion of the edge of a pool. In each locality, snails were collected by the same four people during 1 hour, with the purpose of collecting as many specimens as possible, so as to increase the probability of detecting infected snails.
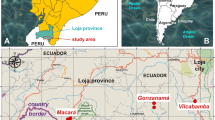
Localities and their altitudes furnishing the lymnaeid specimens sequenced are noted in Table 1 and Figure 1. Snail specimens for molecular analyses, as well as trematode larval stages found in several of them, were fixed in 70% ethanol for DNA extraction procedures.
Geographical distribution of lymnaeid sampling localities. Maps showing location of the Department of Cajamarca within Peru (A) and fascioliasis endemic area of Cajamarca studied (B): 1 = Encañada, Encañada district; 2 = Santa Rosa de Chaquil, Encañada district; 3 = Tauripampa, Llacanora district; 4 = Baños del Inca (locality A), Baños del Inca district; 5 = Baños del Inca (locality B), Baños del Inca district; 6 = Yanamarca, Jesus district; 7 = Chaquicocha, Cajabamba district; 8 = Valle de Condebamba, Cajabamba district
Molecular techniques
DNA extraction
The same procedure was performed for both lymnaeids and fluke larvae. The feet of lymnaeid snail specimens and trematode rediae found were suspended in 400 μl of lysis buffer (10 mM Tris–HCl, pH 8.0, 100 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate SDS) containing 500 μg/ml Proteinase K (Promega, Madison, WI, USA) and digested for 2 hr at 55°C with alternate shaking each 15 min. The procedure steps were performed according to methods outlined previously [6, 50]. The extraction was then performed with phenol-chloroform and DNA was precipitated with ethanol. The pellet was dried and resuspended in 30 μl sterile TE buffer (pH 8.0). This suspension was stored at –20 °C until use.
DNA sequence amplification
Each one of the four DNA markers were PCR amplified independently for each lymnaeid specimen and each PCR product was sequenced for a bona-fide haplotype characterization. The complete sequences of the rDNA spacers ITS-2 and ITS-1 were amplified using primers previously described [6, 41, 50, 51]. The complete ITS-1 of the trematode rediae was amplified by PCR according to methods outlined previously [51, 52]. The target 16S gene region was amplified by PCR using a set of universal primers [53]. Amplification procedures and thermal cycler conditions were carried out as previously described for lymnaeids [38, 42, 44]. A mitochondrial DNA cox 1 gene fragment was amplified using universal primers [54]. Amplifications were generated in a Mastercycle epgradient (Eppendorf, Hamburg, Germany) using 4–6 μl of genomic DNA for each 50 μl PCR reaction. PCR conditions were 30 cycles of 30 sec at 94°C, 30 sec at 50°C and 1 min at 72°C, preceded by 30 sec at 94°C and followed by 7 min at 72°C for ITS-2 and ITS-1, and by 40 cycles of 30 sec at 90°C, 1 min at 48°C and 1 min at 72°C, preceded by 2.5 min at 94°C and followed by 10 min at 72°C for cox 1. Ten μl of each PCR product was checked by staining with ethidium bromide on 1% Nusieve® GTG agarose (FMC) gel electrophoresis, using the Molecular Weight Marker VI (Boehringer Mannheim) at 0.1 μg DNA/μl as control.
Purification and quantification of PCR products
Primers and nucleotides were removed from PCR products by purification on Wizard™ PCR Preps DNA Purification System (Promega, Madison, WI, USA) according to the manufacturer's protocol and resuspended in 50 μl of 10 mM TE buffer (pH 7.6). The final DNA concentration was determined by measuring the absorbance at 260 and 280 nm.
DNA sequencing
The sequencing of the complete rDNA ITS-2 and ITS-1 and the fragments of the mtDNA 16S and cox 1 genes was performed on both strands by the dideoxy chain-termination method [55]. It was carried out with the Taq dye-terminator chemistry kit for ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA), using PCR primers.
Sequence alignments
Sequences were aligned using CLUSTAL-W version 1.8 and MEGA 5.0, and assembly was made with the Staden Package [56]. Subsequently, minor corrections were manually introduced for a better fit of nucleotide correspondences in insertions/deletions (indels) and/or microsatellite sequence regions. Genetic distances were measured, using parameters provided by PAUP v.4.0b10.
DNA haplotype nomenclature
The codes for the sequences obtained follow the standard nomenclature proposed for lymnaeid snails previously [1, 41, 46]. It shall be noted that haplotype codes are only definitive in the case of complete sequences (ITS-2 and ITS-1 in the present study). When dealing with fragments or incomplete sequences, haplotype codes are provisional (16S and cox 1 in the present study).
Lymnaeid sequence comparisons
The following sequences from GenBank-EMBL have been used for comparison analyses:
-
rDNA ITS-2: G. truncatula H1 [EMBL: AJ296271], H2 [EMBL: AJ243017] and H3 (= L. viatrix sensu Ueno et al., 1975; = L. cubensis sensu Ueno et al., 1975) [EMBL: AJ272051] [6, 50, 51]; L. cubensis H1 [EMBL: AM412223], H2 [EMBL: FN182200], H3 [EMBL: FN182201] and H4 [GenBank: JF514088] [22, 50]; L. viator H1 from the type locality Rio Negro, Argentina [EMBL: AM412224] [50] and H2 [GenBank: JN051366] [21]; L. neotropica H1 from the type locality of Lima, Peru [EMBL: AM412225] [50] and H2 [GenBank: JF514089] [22]; L. schirazensis H1 [GenBank: JF272601] and H2 [GenBank: JF272602] [42].
-
rDNA ITS-1: G. truncatula HA [EMBL: AJ243018], HB [AJ296270], HC (= L. viatrix sensu Ueno et al., 1975; = L. cubensis sensu Ueno et al., 1975) [EMBL: AJ272052] and HD [GenBank: JF514090] [22, 41, 50, 51]; L. cubensis HA from the type locality of Cuba [EMBL: AM412226], HB [EMBL: FN182202] and HC [EMBL: FN182203] [22, 50]; L. viator HA from the type locality of Rio Negro, Argentina [EMBL: AM412227] [50] and HB [GenBank: JN051368] [21]; L. neotropica HA from the type locality of Lima, Peru [EMBL: AM412228] [50]; L. schirazensis HA [GenBank: JF272603] and HB [GenBank: JF272604] [42].
-
mtDNA 16S rRNA gene: G. truncatula 16S-A from Spain [EMBL: HE610431] (new sequence obtained for comparison purposes); L. cubensis 16S-A from the type locality of Cuba [EMBL: FN182204] (new sequence obtained for comparison purposes); L. viator 16S-A from the type locality of Rio Negro, Argentina, and also Chile [EMBL: HE610434] (new sequence obtained for comparison purposes); L. schirazensis 16S-A [GenBank: JF272605] and 16S-B [GenBank: JF272606] [42]; and other proximal species of the Galba/Fossaria group such as Fossaria bulimoides [GenBank: AF485657] and F. obrussa [GenBank: AF485658] [57].
-
mtDNA cox 1 gene: G. truncatula cox 1a [EMBL: AM494011) [50], cox 1b [GenBank: JF461487] [22], cox1c [GenBank: JN051372] [21] and G. truncatula [GenBank: EU818799] [58]; L. cubensis cox 1a from the type locality of Cuba [EMBL: AM494009] [50] and cox 1b [GenBank: FN182205] [22]; L. viator cox1a from the type locality Rio Negro, Argentina [EMBL: AM494010] [50], cox1b [GenBank: JN051373] and cox1c [GenBank: JN051374] [21]; L. neotropica cox 1a from the type locality of Lima, Peru [EMBL: AM494008], cox 1b [EMBL: FN356741], cox 1c [GenBank: JF461485] and cox 1d [GenBank: JF461486] [22, 50, 52]; L. schirazensis cox 1a [GenBank: JF272607], cox 1b [GenBank: JF272608], cox 1c [GenBank: JF272609] and cox 1d [GenBank: JF272610] [42].
Trematode sequence comparisons
The rDNA ITS-1 sequence of the trematode larval stages was compared with the following sequences from GenBank-EMBL:
-
rDNA ITS-1: F. hepatica from Spain, France, Poland, Ireland, Iran, Japan, Korea, Vietnam, Australia, Egypt, Bolivia, Peru, Uruguay, Argentina, Chile, Venezuela, Ecuador and Mexico [GenBank: AB207139; GenBank: AB207140; GenBank: AB207141; GenBank: AB207145; GenBank: AB211236; GenBank: AB385611; EMBL: AJ243016; GenBank: EF612468; GenBank: EF612469] [1, 51].
Results
Lymnaeid populations and their densities
Populations of lymnaeid species found are noted in Table 1, including coordinates, altitudes, number of lymnaeid specimens collected and population densities. Within a high altitude range between 2,080 and 3,130 m, the very high population densities, in localities such as Santa Rosa de Chaquil, Tauripampa and Yanamarca, are worth mentioning.
Molecular characterisation of lymnaeids
Nuclear rDNA ITS-2 and ITS-1 and mtDNA 16S and cox 1 nucleotide sequence data reported in this study are available in the GenBank™, EMBL and DDBJ databases under the accession numbers noted in Table 1. Initial snail species classifications, based on previous complete morphological decriptions [42, 50], were subsequently verified by sequence comparisons, as noted in the following.
Galba truncatula
Specimens from six different populations found in the localities of Encañada, Santa Rosa de Chaquil, Tauripampa, Baños del Inca (locality B), Yanamarca and Chaquicocha, preliminarily classified as L. viatrix or Lymnaea sp., proved to be G. truncatula by ribosomal and mitochondrial DNA markers (Table 1).
rDNA ITS-2
All the specimens showed identical ITS-2 sequence, of 401 bp and a 59.10% GC content. When compared with the three ITS-2 haplotypes of G. truncatula available in EMBL (H1, H2, H3), this sequence proved to be identical to the previously described ITS-2 haplotype 1 (H1) for G. truncatula [EMBL: AJ243017].
rDNA ITS-1
Similarly, all specimens studied showed identical ITS-1 sequence, of 504 bp and a 57.74% GC content. This sequence was compared with the four ITS-1 haplotypes of G. truncatula available in EMBL (HA, HB, HC) and GenBank (HD) and proved to be different. It has consequently been ascribed to the new haplotype G.tru HE [EMBL: HE610430].
A comparison between the combined rDNA ITS-2 and ITS-1 haplotypes of G. truncatula present in Cajamarca, Peru (G.tru CH1E) and the Northern Bolivian Altiplano (G.tru CH3C) shows interesting information about the nucleotide differences shown by this lymnaeid vector in the two human fascioliasis endemic areas with the highest prevalences known (Figure 2).
Comparison of the combined rDNA ITS-2 and ITS-1 haplotypes of Galba truncatula of the human fascioliasis hyperendemic areas of Cajamarca and the Bolivian Altiplano. Nucleotide differences between the combined haplotype 1E representing the "valley transmission pattern" in Cajamarca and the combined haplotype 3C representing the "altiplanic transmission pattern" in the Northern Bolivian Altiplano. Position = numbers (to be read in vertical) refer to variable positions obtained in the concatenated ITS-1 and ITS-2 alignment made with MEGA 5.0
mtDNA 16S rRNA gene
Only one halotype was detected in the specimens from the six different populations studied. This partial sequence was 425 bp-long, presented a biased AT content of 68.70%, and was noted as G.tru-16S-B provisional haplotype [EMBL: HE610432]. Differences with haplotype 16S-A, present in Europe and in the Northern Bolivian Altilplano endemic area, are restricted to only one mutation A/T in position 345 of 16S-A/16S-B haplotype respective alignment.
mtDNA cox1
Only one haplotype was detected, being identical in all specimens analysed and including 672 bp and a 68.45% of AT content. This sequence was compared with the cox 1 haplotypes of G truncatula known so far and proved to be different. Hence, it is here noted as the new provisional haplotype G.tru-cox 1d [EMBL: HE610435]. Nucleotide and amino acid differences between the five described haplotypes for G. truncatula are listed in Figure 3.
Differences found in the mtDNA cox 1 gene sequence of the Galba truncatula populations from Cajamarca and other countries. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 5.0. Nucleotides = Nt; amino acids = Aa; Identical = .; Indel = −. Haplotype codes only provisional due to incomplete sequences of the gene: * [EMBL: AM494011]; ** [GenBank: JF461487]; *** [GenBank: JN051372]. **** sequence shorter than the partial sequence here obtained: no haplotype ascribed [GenBank: EU818799]
Lymnaea neotropica
Specimens from the lymnaeid population collected in the locality of Valle de Condebamaba, previously identified as L. viatrix, proved to be L. neotropica by ribosomal and mitochondrial DNA markers (Table 1).
rDNA ITS-2
All the specimens sequenced showed identical ITS-2 sequence, of 415 bp and a 56.87% GC content. When compared with the ITS-2 haplotypes of L. neotropica available in GenBank (H1 and H2), the Peruvian sequence proved to be identical to the previously described ITS-2 haplotype 1 (H1) for L. neotropica of its type locality [EMBL: AM412225] (Table 1).
rDNA ITS-1
All specimens presented the same ITS-1 sequence of 533 bp and a 56.66 GC content. This sequence was compared with the ITS-1 haplotype of L. neotropica available in GenBank (L.neo-HA) and proved to be identical. This haplotype was previously reported in the type locality of Rio Lurin, Lima, Peru and also in Argentina [EMBL: AM412228].
mtDNA 16S rRNA gene
Only one halotype was detected in the specimens studied from this population. This partial sequence was 425 bp-long, presented a biased AT content of 69.48%, and was described as L.neo-16S-A provisional haplotype, as it proved to be identical to the one found in the type locality of this lymnaeid species [EMBL: HE610433].
mtDNA cox1
All of the specimens sequenced showed identical cox 1 nucleotide sequence, of 672 bp and with a biased AT content of 69.5%. This sequence was compared with the four cox 1 haplotypes of L. neotropica known so far and proved to be identical to the previously described haplotype L.neo-cox 1a from the type locality of this species [EMBL: AM494008]. Nucleotide and amino acid differences between the four described haplotypes for L. neotropica are listed in Figure 4.
Differences found in the mtDNA cox 1 sequence of Lymnaea neotropica from Cajamarca and other countries. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 5.0. Nucleotides = Nt; amino acids = Aa; Identical = .; Indel = −. Haplotype codes only provisional due to incomplete sequences of the gene. * [EMBL: AM494008]; ** [GenBank: FN356741]; *** [GenBank: JF461485]; **** [GenBank: JF461486]
Lymnaea schirazensis
Snail specimens collected from the locality of Baños del Inca (locality A), preliminarily identified as L. viatrix, proved to be L. schirazensis after ribosomal and mitochondrial DNA marker sequencing (Table 1).
rDNA ITS-2
All the specimens presented the same ITS-2 sequence, of 436 bp and a 53.90% GC content. When compared to the two ITS-2 haplotypes of L. schirazensis available in GenBank, it proved to be identical to the previously described L.schi-H1 [GenBank: JF272601].
rDNA ITS-1
Similarly, all the lymnaeid individuals showed identical ITS-1 sequences, of 533 bp long and a 59.91% GC content. This haplotype was compared with two ITS-1 haplotypes of L. schirazensis available in GenBank (HA, HB) and proved to be the same as the previously described L.schi-HB [GenBank: JF272604].
mtDNA 16S rRNA gene
Only one halotype was detected in the specimens studied from this population. This partial sequence was 425 bp-long, presented a biased AT content of 69.48%, and corresponded to the provisional haplotype L.schi-16S-A [GenBank: JF272605].
The nucleotide differences between the several haplotypes described for different Galba/Fossaria species, including L. schirazensis plus G. truncatula, L. neotropica, L. viator, L. cubensis, F. bulimoides and F. obrussa are listed in Figure 5. The most proximal species are L. neotropica and L. cubensis, differing in only one polymorphic site in position 130.
Nucleotide differences found in the mtDNA 16S gene sequence of Galba/Fossaria species and populations studied from Cajamarca and other countries. Position = numbers (to be read in vertical) refer to the 50 variable positions obtained in the 426-bp-long alignment made with MEGA 5.0. Identical = .; Indel = −. Haplotype codes only provisional due to incomplete sequences of the gene. (p) = new haplotypes obtained in the present study. * [GenBank: AF485657]; ** [GenBank: JF272605]; *** [GenBank: JF272605]; **** [GenBank: AF485658]. Horizontal lines delimitate groups of proximal or similar sequences
mtDNA cox1
All of the specimens sequenced showed identical cox 1 nucleotide sequence, of 672 bp and with a biased AT content of 69.1%. This sequence was compared with the four cox 1 haplotypes of L. schirazensis known so far and proved to be identical to the previously described haplotype L.schi-cox 1d [GenBank: JF272610].
Genetic comparison of lymnaeid species found
Four DNA sequence alignments, one for each marker used, where made to show the total nucleotide differences between the respective haplotypes of the three lymnaeid species present in the fascioliasis hyperendemic area of Cajamarca.
When comparing the ITS-2 sequences, a 487-bp-long alignment was obtained, in which a total of 172 variable positions (35.3%) including mutations and indels appeared (Figure 6A). In the analysis of the ITS-1 sequences, the alignment obtained had a length of 562 bp, with a total of 187 variable positions (33.2%) including mutations and indels (Figure 6B).
Nucleotide differences found in the sequences of the rDNA ITS-2 (A) and ITS-1 (B) of the three Galba/Fossaria species present in Cajamarca. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 5.0. Alignment manually adjusted. Identical = .; Indel = −
In the comparison of the mtDNA 16S sequences, the alignment obtained was 425 bp long, including a total of only 31 variable positions (7.2%), with very few gaps (Figure 7A). In the analysis of the mtDNA cox 1 sequences, a total of 96 variable positions (14.2%) including only mutations were found in the 672-bp-long alignment. Most of these mutations were however silent, as indeed only two variable positions (0.8%) appeared in the 224-aa-long alignment of the corresponding partial protein sequences (Figure 7B).
Differences found in the sequences of the mtDNA 16S ribosomal gene (A) and mtDNA cox 1 gene (B) of the three Galba/Fossaria species present in Cajamarca. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 5.0. Nucleotides = Nt; amino acids = Aa; Identical = .; Indel = −
Molecular characterisation of trematode larval stages
The complete sequence of the rDNA ITS-1 obtained from the trematode rediae found in two specimens of G. truncatula from Santa Rosa de Chaquil and another two specimens of the same lymnaeid species from Tauripampa is 432 bp long and with a 51.85% GC content. It showed no nucleotide difference when compared with the sequence of that spacer in F. hepatica from the Northern Bolivian Altiplano and Spain [EMBL: AJ243016], and thus corresponds to the haplotype code Fh ITS1-HA.
Discussion
Lymnaeid species present and their genetic characteristcs
DNA marker sequences prove that there are three different lymnaeid species present in the fascioliasis endemic area of Cajamarca: G. truncatula L. neotropica and L. schirazensis. However, previous studies only refered to one lymnaeid species as being responsible for the disease in that area, namely L. viatrix[36, 37, 59–61]. This lymnaeid species appears repeatedly in the Peruvian literature as the main responsible for fascioliasis transmission throughout Peru [62–69]. Indeed, Peruvian L. viatrix refer to L. viator variety B elongata described long time ago from the surroundings of Lima [70, 71] that recently proved, by multiple DNA marker sequencing, to belong to a new species to which the name L. neotropica was ascribed [50]. Thus, the existence of true L. viatrix, which corresponds to L. viator variety A ventricosa [70, 71], has so far only been molecularly verified in Argentina [50] and Chile [21]. Additionally, it should be considered that the correct species name for this species is Lymnaea viator D’Orbigny, 1835, because the feminine spelling viatrix proposed time ago [72] does not fit articles 31.2.1 and 34.2.1 of the International Code of Zoological Nomenclature, as it has been recently highlighted [21].
Unfortunately, it is at present impossible to ascertain whether the populations from Cajamarca classified as L. viatrix were in fact L. neotropica. These two species are very similar and may be easily confused when only relying on traditional malacological methods [50]. Moreover, both species belong to the Galba/Fossaria group, which is well known due to the pronounced morphological similarity of the numerous species it includes and which makes specimen classification and species differentiation extremely difficult [42]. Consequently, reports on L. viatrix in the Cajamarca area may be result of confusion, not only with L. neotropica, but also with G. truncatula and L. schirazensis.
The pronouncedly less numerous nucleotide differences found in the two mtDNA markers 16S and cox 1 between the three lymnaeid species present in Cajamarca (Figure 7A,B) when compared to their two rDNA markers ITS-2 and ITS-1 (Figure 6A,B) should be highlighted. This result, together with the high number of silent mutations in cox 1 (Figure 7B), suggests a saturation in both mtDNA markers, a phenomenon already described in lymnaeids [42] and well known in several invertebrate groups [49].
Despite the numerous nucleotide differences in their DNA sequences in both rDNA and mtDNA (Figure 6 and Figure 7), the three lymnaeid species found in Cajamarca may easily be confused one another both in the field and also in the laboratory. Nevertheless, there are several details that may help for at least a preliminary classification (Table 2). However, although a morphological shell trend may help in distinguishing a given species [73], confirmation of the species to which a population belongs can so far only be obtained by DNA sequencing. Moreover, it should be taken into account that mixed populations of G. truncatula and L. schirazensis have already been described in the field [42]. Hence, care should even be taken when working experimentally in the laboratory.
The apparent monomorphic genetic characteristics of each one of the three lymnaeid species in Cajamarca should be highlighted. Each lymnaeid species appear to present only one haplotype for each molecular marker sequenced, although of course additional studies on more populations of these species throughout the endemic area in question are needed to verify this assumption. Anyway, the lack or pronouncedly reduced genetic variability of populations living at very high altitude is known in different organisms and has already been also observed in lymnaeids such as G. truncatula in the Northern Bolivian Altiplano [51, 74], an area located between 3800 and 4100 m high altitude [2]. This lack of genetic variability of G. truncatula in the Northern Bolivian Altiplano is related to the usual selfing trend followed by this lymnaeid [75]. Autofecundation has been verified to be the normal fecundation process in L. schirazensis and has also been observed in other species of the Galba/Fossaria group [42].
The aforementioned monomorphic genetic characteristics suggest an introduction of each one of the three lymnaeid species from only one source. In the case of G. truncatula, although its European origin appears evident [1, 51], the unexpected new haplotypes of ITS-1 (G.tru-HE), 16S (G.tru-16S-B) and cox 1 (G.tru-cox 1d) found in this species in Cajamarca pose a problem when attempting to elucidate the direct geographical source. In South America, the presence of G. truncatula has already been molecularly verified in the Northern Altiplano of both Bolivia [51] and Peru [10], Argentina [76, 77], Chile [21] and Venezuela [22]. In Colombia, the only report of G. truncatula published so far [78] appears to be a misclassification [42].
Lymnaea neotropica, originally described from Lima and surroundings [50], appears to be a species restricted to South America but with a very broad geographical distribution covering from Argentina in the Southern Cone [52] up to Venezuela [22]. The combined rDNA and mtDNA haplotype of this species in Cajamarca is identical to the one from the type locality. The presence of this combined haplotype in both Peruvian areas may probably be related to the livestock trade between Lima and Cajamarca which was very intense along the old rural Inca routes at a given period of the early Spanish colonisation [79].
The combined rDNA and mtDNA haplotype of L. schirazensis present in Cajamarca appears to be a mix. It shares the ITS-2 haplotype L.schi-H1 with Spain, the Dominican Republic and Venezuela, and is different from the one in Rio Lurin, close to Lima. Its ITS-1 haplotype L. schir-HB has also been found in Mexico and Ecuador and in Peru it is present in both Cajamarca and Rio Lurin. The ribosomal 16S gene of the mtDNA does unfortunately not furnish any biogeographical information, as the haplotype L.schir-16S-HA found in Cajamarca is the same as everywhere. And finally, the mtDNA cox 1 haplotype L.schir-cox 1d appears to be unique [42]. Although a trans-Andean livestock introduction route was launched between western Andean Venezuela and the Colombian Bogota and also further southward [80], the introduction of L. schirazensis with livestock and humans should have most probably occurred from northern Peru, probably by livestock transported between Quito and Lima through Cajamarca along the old rural Inca routes followed by the Spanish conquerors [42].
Implications for fascioliasis transmission and epidemiology
Fascioliasis in Cajamarca has been the focus of several studies on both livestock [36, 37, 61, 81] and humans [12, 13, 82–84]. According to results from these studies, Cajamarca is a typical representative of the "valley transmission pattern" of fascioliasis [1, 7, 81] and the area presenting the highest prevalences in children, both in average (24.4%) and local maximum (47.7%), among all human endemic areas in Peru [13]. With regard to human prevalences, Cajamarca is only surpassed by the Northern Bolivian Altiplano [2, 19]. Interestingly, only one lymnaeid species, G. truncatula, is responsible for such a human hyperendemic situation in the Northern Altiplano [51], whereas DNA sequence results here obtained demonstrate that there are three different lymnaeid species present in the hyperendemic area of Cajamarca.
Among the three lymnaeid species present in Cajamarca, Galba truncatula is considered the best F. hepatica transmitter known, with such a parasite/vector interaction as to support that this lymnaeid may be considered the original vector of this fasciolid [1]. Moreover, G. truncatula is known to give rise to very high human infection rates, both prevalences and intensities, at the very high altitude of the Northern Altiplano of both Bolivia [2, 18, 19] and Peru (10). Such high fascioliasis transmission rates have been proved to be the consequence of life cycle modifications in both F. hepatica and G. truncatula as an adaptation response to the extreme conditions of the very high altitude [51]. Several aspects suggest that G. truncatula may behave in the 2600–3100 m high altitudes of Cajamarca similarly to in the 3800–4100 m altitudes of the Northern Altiplano, namely:
a) the results of the field surveys carried out showing that populations of this species are the most abundant in the endemic area;
b) the typical anthropophylic characteristics of this lymnaeid appeared evident when proving to be the usual snail species in the neighbourhood of human communities such as Encañada, Santa Rosa de Chaquil, Tauripampa and Yanamarca (Figure 8), close to localities where schoolchildren appeared infected [13];
c) the high population densities of thois species found in the high altitude localities studied (Table 1);
d) the finding of infected specimens in two populations of this lymnaeid by means of the DNA sequencing methods (Table 1), despite the relatively few specimens analysed for infection detection (prevalences in lymnaeids are usually low).
In front of the aforementioned similarities, the genetic differences between G. truncatula populations from two different environments such as Cajamarca and the Northern Bolivian Altiplano should be emphasized (Figure 2). Whereas in Cajamarca the disease transmission follows a seasonality mainly related to the cyclic temperatures [37], in the Northern Altiplano fascioliasis transmission takes place throughout the year due to (i) the scarcely varying temperatures [8] and (ii) the link of G. truncatula populations to permanent water bodies as the consequence of the absence of sufficiently long-standing temporal water bodies due to the high evapotranspiration rates [2, 8].
Lymnaea neotropica has also been found to be linked to human infection. This species was originally described from the surroundings and other areas near Lima, Peru [50], where human infection has repeatedly been detected [14, 85]. Although none of the L. neotropica specimens sequenced in Cajamarca was infected by F. hepatica, Peruvian L. viatrix (= L. viator) variety B elongata (= L. neotropica according to [50]) has been shown to transmit fascioliasis both experimentally and in nature [62, 67, 86]. The transmission capacity of this lymnaeid vector has also been molecularly confirmed in Argentina, where it appeared linked to animal infection [52], as in Venezuela [22].
With regard to L. schirazensis, experimental infection assays of different geographical strains of this snail species from different continents with F. hepatica and F. gigantica have proved that fasciolid larval stages are not able to fully develop within this lymnaeid, which does therefore not participate in disease transmission [42]. A large multidisciplinar study has demonstrated that L. schirazensis has an outstandingly broad distribution from lowlands to highlands, including from below sea level (−23 m) in Iran up to very high altitude areas in Ecuador (3158 m). Throughout this wide altitudinal range, it has always been confused with G. truncatula in the Old World and with G. truncatula and other similar Galba/Fossaria vector species in the Americas. This hitherto overlooked species has been masking not only the geographical distribution of fascioliasis, but also fasciolid population specificity/susceptibility analyses. It shall be considered that L. schirazensis has been shown molecularly to be able to cohabit with other Galba/Fossaria group species in such proximity that a natural population of specimens of an apparently unique lymnaeid species may in fact involve specimens from two different species unnoticeably mixed. Such mixed populations have already been described in the case of G. truncatula and L. schirazensis[42].
Conclusions
Sequences of the rDNA and mtDNA markers indicate that two lymnaeid vectors, G. truncatula and L. neotropica, and a third non-transmitting lymnaeid, L. schirazensis, inhabit the fascioliasis hyperendemic area of Cajamarca. This new scenario pronouncedly changes the situation described so far, in which disease transmission was mentioned to be related to only one lymnaeid species, L. viator (= L. viatrix). All suggests that this has been the consequence of misclassifications due to the inaccuracy of traditional malacological methods for species differentiation among the Galba/Fossaria group of lymnaeids to which the three aforementioned, morphologically similar, small snail species belong [42, 50].
The nearby detection of populations of G. truncatula and L. schirazensis in Baños del Inca district, and of G. truncatula and L. neotropica in Cajabamba district, indicate that these three lymnaeid species geographically overlap inside the hyperendemic area. This fact poses two problems of increasing difficulty for epidemiological studies and control action.
First, there is a problem in classifying lymnaeid specimens in both field and laboratory activities in Cajamarca. Indeed, although several phenotypic characteristics may a priori be helpful for a preliminary specimen classification (Table 2), a definitive classification of a specimen can only be obtained by the sequencing of at least one of the molecular markers here used, ITS-2, ITS-1, 16S and cox 1. This problem becomes extremely important given the transmission capacity differences of these three lymnaeid species: G. truncatula as the vector mainly involved in disease transmission to humans, L neotropica as a typical responsible for livestock infection, and L. schirazensis as a lymnaeid unable to transmit fascioliasis. Moreover, from the ecological point of view, these three species may be found to show similar amphibious characteristics. Additionally, L. schirazensis pronouncedly increases the confusion problem, owing to its ability to mix its specimens and populations with those of other Galba/Fossaria species and distort fascioliasis data such as transmission capacity and infection susceptibility. Such L. schirazensis mixing has already been described with G. truncatula[42].
Second, the complexity of the overlap of two different lymnaeid vector species poses a serious problem for the development of methods which have shown to be useful for epidemiological analysis, surveillance and control of human fascioliasis in Andean high altitude endemic areas, such as mathematical modelling through the application of different climatic indices (Mt, Wb-bs) [8], and Remote Sensing (RS) with Geographical Information Systems (GIS) [9]. Thus, both methods appear to be useful to monitor the fascioliasis situation in the Northern Bolivian Altiplano, where only one lymnaeid species, G. truncatula, is responsible for disease transmission. However, the same methods did not appear to be sufficiently accurate when analysing fascioliasis in the central regions of Chile where human fascioliasis is endemic and livestock prevalences are the highest in the country [87]. Recent DNA sequencing results have shown that the endemic regions of Chile where the RS-GIS method did not appropriately work were in fact areas where there is an overlap of two different lymnaeid vector species with supposedly different transmission capacity [21]. The same problem may be expected in Cajamarca if similar low resolution mapping is applied.
These conclusions should be considered within future control activities in the province of Cajamarca. In this hyperendemic area, a pilot intervention to assess human treatment strategies was successfully performed in 2007 and 2008, human fascioliasis treatment activities have since then been yearly implemented, thanks to the availability of Egaten® (triclabendazole for human use; donation by Novartis Pharma AG), provided by WHO through the Ministry of Health in Lima and the Dirección Regional de Salud of Cajamarca, and a large initiative for animal control to diminish risk infection has already been approved.
References
Mas-Coma S, Valero MA, Bargues MD: Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol. 2009, 69: 41-146.
Mas-Coma S, Angles R, Esteban JG, Bargues MD, Buchon P, Franken M, Strauss W: The Northern Bolivian Altiplano: a region highly endemic for human fascioliasis. Trop Med Int Health. 1999, 4: 454-467. 10.1046/j.1365-3156.1999.00418.x.
Torgerson P, Claxton J: Epidemiology and control. Fasciolosis. Edited by: Dalton JP. 1999, CAB International Publishing, Wallingford, Oxon, UK, 113-149.
Valero MA, Mas-Coma S: Comparative infectivity of Fasciola hepatica metacercariae from isolates of the main and secondary reservoir animal host species in the Bolivian Altiplano high human endemic region. Folia Parasitol. 2000, 47: 17-22.
Valero MA, Darce NA, Panova M, Mas-Coma S: Relationships between host species and morphometric patterns in Fasciola hepatica adults and eggs from the Northern Bolivian Altiplano hyperendemic region. Vet Parasitol. 2001, 102: 85-100. 10.1016/S0304-4017(01)00499-X.
Bargues MD, Vigo M, Horak P, Dvorak J, Patzner RA, Pointier JP, Jackiewicz M, Meier-Brook C, Mas-Coma S: European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS-2 sequences. Inf Genet Evol. 2001, 1: 85-107. 10.1016/S1567-1348(01)00019-3.
Mas-Coma S: Epidemiology of fascioliasis in human endemic areas. J Helminthol. 2005, 79: 207-216. 10.1079/JOH2005296.
Fuentes MV, Valero MA, Bargues MD, Esteban JG, Angles R, Mas-Coma S: Analysis of climatic data and forecast indices for human fascioliasis at very high altitude. Ann Trop Med Parasitol. 1999, 93: 835-850. 10.1080/00034989957844.
Fuentes MV, Malone JB, Mas-Coma S: Validation of a mapping and predicting model for human fasciolosis transmission in Andean very high altitude endemic areas using remote sensing data. Acta Trop. 2001, 79: 87-95. 10.1016/S0001-706X(01)00106-1.
Esteban JG, Gonzalez C, Bargues MD, Angles R, Sanchez C, Naquira C, Mas-Coma S: High fascioliasis infection in children linked to a man-made irrigation zone in Peru. Trop Med Int Health. 2002, 7: 339-348. 10.1046/j.1365-3156.2002.00870.x.
Stork MG, Venables GS, Jennings SMF, Beesley JR, Bendezu P, Capron A: An investigation of endemic fasciolasis in Peruvian village children. J Trop Med Hyg. 1973, 76: 231-235.
Ortiz P, Cabrera M, Jave J, Claxton J, Williams D: Human fascioliasis: prevalence and treatment in a rural area of Peru. Infect Dis Rev. 2000, 2: 42-46.
Gonzalez LC, Esteban JG, Bargues MD, Valero MA, Ortiz P, Naquira C, Mas-Coma S: Hyperendemic human fascioliasis in Andean valleys: An altitudinal transect analysis in children of Cajamarca province. Peru. Acta Trop. 2011, 2011 (120): 119-129.
Blancas G, Terashima A, Maguina C, Vera L, Alvarez H, Tello R: Fasciolosis humana y compromiso gastrointestinal: estudio de 277 pacientes en el Hospital Nacional Cayetano Heredia 1970–2002. Rev Gastroenterol Perú. 2004, 24: 143-157.
Picoaga J, Lopera J, Montes J: Fasciolasis en Arequipa. Bol Peru Parasitol. 1980, 2: 1-11.
World Health Organization: Control of foodborne trematode infections. WHO Techn Rep Ser. 1995, 849: 1-157.
Hillyer GV, Soler de Galanes M, Rodriguez-Perez J, Bjorland J, Silva de Lagrava M, Guzman SR, Bryan RT: Use of the Falcon™ assay screening test-enzyme-linked immunosorbent assay (FAST-ELISA) and the enzyme-linked immunoelectrotransfer blot (EITB) to determine the prevalence of human fascioliasis in the Bolivian altiplano. AmJTrop Med Hyg. 1992, 46: 603-609.
Esteban JG, Flores A, Angles R, Strauss W, Aguirre C, Mas-Coma S: A population-based coprological study of human fascioliasis in a hyperendemic area of the Bolivian Altiplano. Trop Med Int Health. 1997, 2: 695-699. 10.1046/j.1365-3156.1997.d01-356.x.
Esteban JG, Flores A, Angles R, Mas-Coma S: High endemicity of human fascioliasis between Lake Titicaca and La Paz valley, Bolivia. Trans Roy Soc Trop Med Hyg. 1999, 93: 151-156. 10.1016/S0035-9203(99)90289-4.
Apt W, Aguilera X, Vega F, Alcaino H, Zulantay I, Apt P, Gonzalez V: Retamal, Rodriguez J, Sandoval J: Prevalencia de fascoliasis en humanos, caballos, cerdos y conejos silvestres en tres provincias de Chile. Bol Of Sanit Panam. 1993, 115: 405-414.
Artigas P, Bargues MD: Mera y Sierra R, Agramunt VH, Mas-Coma S: Characterisation of fascioliasis lymnaeid intermediate hosts from Chile by DNA sequencing, with emphasis onLymnaea viatorandGalba truncatula. Acta Trop. 2011, 120: 245-257. 10.1016/j.actatropica.2011.09.002.
Bargues MD, Gonzalez LC, Artigas P, Mas-Coma S: A new baseline for fascioliasis in Venezuela: lymnaeid vectors ascertained by DNA sequencing and analysis of their relationships with human and animal infection. Parasit Vector. 2011, 4: 200 (18pp.)-
Trueba G, Guerrero T, Fornasini M, Casariego I, Zapata S, Ontaneda S, Vasco L: Detection of Fasciola hepatica infection in a community located in the Ecuadorian Andes. AmJTrop Med Hyg. 2000, 62: 518-
Mera y Sierra R, Agramunt V, Cuervo P, Mas-Coma S: Human fascioliasis in Argentina: retrospective overview, critical analysis and baseline for future research. Parasit Vector. 2011, 4: 104-10.1186/1756-3305-4-104. 18 pp
Mas-Coma S, Bargues MD, Esteban JG: Human Fasciolosis. Fasciolosis. Edited by: Dalton JP. 1999, CAB International Publishing, Wallingford, Oxon, UK, 411-434.
Valero MA, Santana M, Morales M, Hernandez JL, Mas-Coma S: Risk of gallstone disease in advanced chronic phase of fascioliasis: an experimental study in a rat model. J Inf Dis. 2003, 188: 787-793. 10.1086/377281.
Valero MA, Navarro M, Garcia-Bodelon MA, Marcilla A, Morales M, Garcia JE, Hernandez JL, Mas-Coma S: High risk of bacterobilia in advanced experimental chronic fasciolosis. Acta Trop. 2006, 100: 17-23. 10.1016/j.actatropica.2006.09.002.
Valero MA, Girones N, Garcia-Bodelon MA, Periago MV, Chico-Calero I, Khoubbane M, Fresno M, Mas-Coma S: Anemia in advanced chronic fasciolosis. Acta Trop. 2008, 108: 35-43. 10.1016/j.actatropica.2008.08.007.
Brady MT, O’Neill SM, Dalton JP, Mills KH: Fasciola hepatica supresses a protective Th1 response against Bordetella pertussis. Infect Immunity. 1999, 67: 5372-5378.
Girones N, Valero MA, Garcia-Bodelon MA, Chico-Calero MI, Punzon C, Fresno M, Mas-Coma S: Immune supression in advanced chronic fascioliasis: an experimental study in a rat model. J inf Dis. 2007, 195: 1504-1512. 10.1086/514822.
Mas-Coma S, Valero MA, Bargues MD: Effects of climate change on animal and zoonotic helminthiases. Rev Sci Techn Off Int Epiz. 2008, 27: 443-457.
Mas-Coma S, Valero MA, Bargues MD: Climate change effects on trematodiases, with emphasis on zoonotic fascioliasis and schistosomiasis. Vet Parasitol. 2009, 163: 264-280. 10.1016/j.vetpar.2009.03.024.
Ollerenshaw CB, Smith LP: Meteorological factors and forecast of helminthic diseases. Adv Parasitol. 1969, 7: 283-232.
World Health Organization: Report of the WHO Informal Meeting on use of triclabendazole in fascioliasis control. 2007, World Health Organization, Headquarters, Geneva, Switzerland, WHO/CDS/NTD/PCT/2007.1
World Health Organization: Fact sheet on fascioliasis. Action Against Worms. 2008, World Health Organization, Headquarters; Newsletter, Geneva, Switzerland, 1-8. 10
Claxton JR, Zambrano H, Ortiz P, Amoros C, Delgado E, Escurra E, Clarkson MJ: The epidemiology of fasciolosis in the inter-Andean valley of Cajamarca, Peru. Parasitol Int. 1997, 46: 281-288. 10.1016/S1383-5769(97)00039-1.
Claxton JR, Sutherst J, Ortiz P, Clarkson MJ: The effect of cyclic temperatures on the growth of Fasciola hepatica and Lymnaea viatrix. Vet J. 1999, 157: 166-171. 10.1053/tvjl.1998.0293.
Remigio EA, Blair D: Molecular systematics of the freshwater snail family Lymnaeidae (Pulmonata: Basommatophora) utilising mitochondrial ribosomal DNA sequences. J Moll Stud. 1997, 63: 173-185. 10.1093/mollus/63.2.173.
Remigio EA, Blair D: Relationships among problematic North American stagnicoline snails (Pulmonata: Lymnaeidae) reinvestigated using nuclear ribosomal DNA internal transcribed spacer sequences. Can J Zool. 1997, 75: 1540-1545. 10.1139/z97-779.
Bargues MD, Horak P, Patzner RA, Pointier JP, Jackiewicz M, Meier-Brook C, Mas-Coma S: Insights into the relationships of Palaearctic and Nearctic lymnaeids (Mollusca: Gastropoda) by rDNA ITS-2 sequencing and phylogeny of stagnicoline intermediate host species of Fasciola hepatica. Parasite. 2003, 10: 243-255.
Bargues MD, Artigas P, Jackiewicz M, Pointier JP, Mas-Coma S: Ribosomal DNA ITS-1 sequence analysis of European stagnicoline Lymnaeidae (Gastropoda). Heldia (Münchner Malakologische Mitteilungen), München. 2006, 6 (1/2): 29-40.
Bargues MD, Artigas P, Khoubbane M, Flores R, Glöer P, Rojas-Garcia R, Ashrafi K, Falkner G, Mas-Coma S: Lymnaea schirazensis, an overlooked snail distorting fascioliasis data: Genotype, phenotype, ecology, worldwide spread, susceptibility, applicability. PLoS ONE. 2011, 6 (9): e24567-10.1371/journal.pone.0024567. 33 pp. + 3 Suppl. Tables + 5 Suppl. Figures
Bargues MD, Artigas P, Khoubbane M, Mas-Coma S: DNA sequence characterisation and phylogeography of Lymnaea cousini and related species, vectors of fascioliasis in northern Andean countries, with description of Lymnaea meridensis n. sp. (Gastropoda: Lymnaeidae). Parasit Vector. 2011, 4: 132-10.1186/1756-3305-4-132. 22 pp
Bargues MD, Mera y Sierra RL, Artigas P, Mas-Coma S: DNA multigene sequencing of topotypic specimens of the fascioliasis vector Lymnaea diaphana and phylogenetic analysis of the genus Pectinidens (Gastropoda). Mem Inst Osw Cruz. 2011, 107: 111-124 + 2 Suppl. Tables
Remigio EA, Hebert PD: Testing the utility of partial COI sequences for phylogenetic estimates of gastropod relationships. Mol Phylogenet Evol. 2003, 29: 641-647. 10.1016/S1055-7903(03)00140-4.
Bargues MD, Mas-Coma S: Reviewing lymnaeid vectors of fascioliasis by ribosomal DNA sequence analyses. J Helminthol. 2005, 79: 257-267. 10.1079/JOH2005297.
Kane RA, Stothard JR, Emery AM, Rollinson D: Molecular characterization of freshwater snails in the genus Bulinus: a role for barcodes?. Parasit Vector. 2008, 1: 15-10.1186/1756-3305-1-15.
Hauswald A, Remais J, Xiao N, Davis GM, Lu D, Bale MJ, Wilke T: Stirred, not shaken: genetic structure of the intermediate snail host Oncomelania hupensis robertsoni in an historically endemic schistosomiasis area. Parasit Vector. 2011, 4: 206-10.1186/1756-3305-4-206.
Mas-Coma S, Bargues MD: Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. 2009, 110: 112-136. 10.1016/j.actatropica.2008.10.013.
Bargues MD, Artigas P: Mera y Sierra R, Pointier JP, Mas-Coma S: Characterisation ofLymnaea cubensis,L. viatrixandL. neotropican. sp., the main vectors ofFasciola hepaticain Latin America, by analysis of their ribosomal and mitochondrial DNA. Ann Trop Med Parasitol. 2007, 101: 621-641.
Mas-Coma S, Funatsu IR, Bargues MD: Fasciola hepatica and lymnaeid snails occurring at very high altitude in South America. Parasitology. 2001, 123: S115-S127.
Mera y Sierra R, Artigas P, Cuervo P, Deis E, Sidoti L, Mas-Coma S, Bargues MD: Fascioliasis transmission by Lymnaea neotropica confirmed by nuclear rDNA and mtDNA sequencing in Argentina. Vet Parasitol. 2009, 166: 73-79. 10.1016/j.vetpar.2009.08.001.
Simon C, Franke A, Martin A: The polymerase chain reaction: DNA extraction and amplification. Molecular Techniques in Taxonomy. Edited by: Hewitt GM, Johnston AWB, Young JPW. 1991, Springer-Verlag, Berlin, 329-355.
Folmer O, Black M, Hoch W, Lutz R, Vrijenhoek R: DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotech. 1994, 3: 294-299.
Sanger F, Nicklen S, Coulson AR: DNA sequencing with chain-terminating inhibitors. Proc Natn Acad Sc USA. 1977, 74: 5463-5467. 10.1073/pnas.74.12.5463.
Staden R, Judge DP, Bonfield JK: Sequence assembly and finishing methods. Meth Bioch Anal. 2001, 43: 302-322.
Remigio EA: Molecular phylogenetic relationships in the aquatic snail genus Lymnaea, the intermediate host of the causative agent of fascioliasis: insights from broader taxon sampling. Parasitol Res. 2002, 88: 687-696. 10.1007/s00436-002-0658-8.
Albrecht C, Wolf C, Glöer P, Wilke T: Concurrent evolution of ancient sister lakes and sister species: the freshwater gastropod genus Radix in lakes Ohrid and Prespa. Hydrobiologia. 2008, 615: 157-167. 10.1007/s10750-008-9555-1.
Grados O, Ibañez N: Huésped intermediario de Fasciola hepatica en Cajamarca. Arch Peru Patol Clín. 1971, 25: 185-190.
Larrea H, Oviedo ML, Huaman MP: Observaciones morfológicas de Fossaria viatrix (Orbigny,1835) agente etiológico de la fascioliasis en el Perú. Bol Lima. 1990, 72: 33-37.
Claxton JR, Zambrano H, Ortiz P, Delgado E, Escurra E, Clarkson MJ: Strategic control of fasciolosis in the inter-Andean valley of Cajamarca, Peru. Vet Rec. 1998, 143: 42-45. 10.1136/vr.143.2.42.
Tantalean M, Huiza A, Capuñay R: Los hospederos intermediarios de Fasciola hepatica en el Perú: estudio de la infección natural y experimental de Lymnaea viator. L. diaphana y Physa venustula. Biota. 1974, 10: 243-250.
Paraense WL: Lymnaea viatrix and Lymnaea columella in the Neotropical region: a distributional outline. Mem Inst Oswaldo Cruz. 1982, 77: 181-188.
Paraense WL: Planorbidae, Lymnaeidae and Physidae of Perú (Mollusca: Basommatophora). Mem Inst Oswaldo Cruz. 2003, 98: 767-771. 10.1590/S0074-02762003000600010.
Oviedo ML, Larrea H, Vivar G, Huaman P, Pachas L: Distribución geográfica de la familia Lymnaeidae (Mollusca, Gastropoda) en el Perú y su importancia como transmisores de la fasciolasis. Bol Lima. 1993, 88: 43-50.
Larrea H, Vivar R, Huaman P, Pachas L: Clave de identificación de las especies de la familia Lymnaeidae vectores de la fasciolasis en el Perú. Bol Lima. 1993, 89: 85-96.
Larrea H, Florez M, Vivar R, Huaman P, Velasquez J: Hospederos intermediarios de Fasciola hepatica en el Perú. Rev Horizonte Méd. 2007, 7: 39-46.
Durand P, Pointier JP, Escoubeyrou K, Arenas JA, Yong M, Amarista M, Bargues MD, Mas-Coma S, Renaud F: Occurrence of a sibling species complex within Neotropical lymnaeids, snail intermediate hosts of fascioliasis. Acta Trop. 2002, 83: 233-240. 10.1016/S0001-706X(02)00128-6.
Pointier JP, Cazzaniga NJ, González-Salas C, Gutiérrez A, Arenas JA, Bargues MD, Mas-Coma S: Anatomical studies of sibling species within Neotropical lymnaeids snail intermediate hosts of fascioliasis. Mem Inst Osw Cruz. 2006, 101: 431-435. 10.1590/S0074-02762006000400015.
D’Orbigny A: Synopsis terrestrium et fluviatilium molluscorum, in suo per Americam Meridionalem itinere collectorum. Mag Zool. 1835, 5 (61–62): 1-44. Classe 5
D’Orbigny A: Voyage dans l'Amérique Méridionale. 1837, Biodiversity Heritage Library, Paris, Vol. 5, 3ème Partie: Mollusques. P. Bertrand
Paraense WL: Lymnaea viatrix: a study of topotypic specimens (Mollusca: Lymnaeidae). Rev Bras Biol. 1976, 36: 419-428.
Samadi S, Roumegoux A, Bargues MD, Mas-Coma S, Yong M, Pointier JP: Morphological studies of lymnaeid snails from the human fascioliasis endemic zone of Bolivia. J Moll Stud. 2000, 66: 31-44. 10.1093/mollus/66.1.31.
Meunier C, Tirard C, Hurtrez-Bousses S, Durand P, Bargues MD, Mas-Coma S, Pointier JP, Jourdane J, Renaud F: Lack of molluscan host diversity and the transmission of an emerging parasitic disease in Bolivia. Mol Ecol. 2001, 10: 1333-1340. 10.1046/j.1365-294X.2001.01284.x.
Trouve S, Degen L, Goudet J: Ecological components and evolution of selfing in the freshwater snail Galba truncatula. J Evol Biol. 2005, 18: 358-370.
Bargues MD: Mera y Sierra R, Gomez HG, Artigas P, Mas-Coma S: Ribosomal DNA ITS-1 sequencing ofGalba truncatula(Gastropoda: Lymnaeidae) and its potential impact on fascioliasis transmission in Mendoza, Argentina. Animal Biodiv Conserv. 2006, 29: 191-194.
Bargues MD, Mera y Sierra R, Gomez HG, Artigas P, Mas-Coma S: Caracterización molecular de Galba truncatula, vector principal de la Fascioliasis, en Argentina. Implicaciones en salud pública. Enf Emerg, Barcelona. 2007, 9: 77-82.
Longo M, Zamora H, Vasquez R, Velasquez LE: Aspectos ecologicos de Limnaea (Mollusca: Lymnaeidae) en la region de Aguas Tibias, municipio de Purace - Coconuco. Rev Asoc Col Cienc Biol. 2005, 17: 47-58.
Serrera RM: La conquista. Historia de España. Volume 8 Descubrimiento, Colonización y Emancipacion de América. Edited by: Dominguez A. 1990, Editorial Planeta SA, Barcelona, 127-185.
Oviedo y Baños Jd: Historia de la Conquista y Población de la Provincia de Venezuela. Edited by: Morón G. 1965, Ediciones Atlas, Madrid, Volume 2:82 and Volume 7:233
Valero MA, Perez-Crespo I, Khoubbane M, Artigas P, Panova P, Ortiz P, Maco P, Espinoza JR, Mas-Coma S: Fasciola hepatica phenotypic characterisation in Andean human endemic areas: valley versus altiplanic patterns analysed in liver flukes from sheep from Cajamarca and Mantaro, Peru. Inf Genet Evol. 2012, 12: 403-410. 10.1016/j.meegid.2012.01.009.
Knobloch J, Delgado E, Alvarez AG, Reymann U, Bialek R: Human fascioliasis in Cajamarca/Peru. I. Diagnostic methods and treatment with praziquantel. Trop Med Parasitol. 1985, 36: 88-90.
Espinoza JR, Maco V, Marcos L, Saez S, Neyra V, Terashima A, Samalvides F, Gotuzzo E, Chavarry E, Huaman C, Bargues MD, Valero MA, Mas-Coma S: Evaluation of Fas2-ELISA for the serological detection of Fasciola hepatica infection in humans. AmJTrop Med Hyg. 2007, 76: 977-982.
Valero MA, Perez-Crespo I, Periago MV, Khoubbane M, Mas-Coma S: Fluke egg characteristics for the diagnosis of human and animal fascioliasis by Fasciola hepatica and F. gigantica. Acta Trop. 2009, 111: 150-159. 10.1016/j.actatropica.2009.04.005.
Marcos LA, Romani L, Florencio L, Terashima A, Canales M, Nestares J, Huayanay L, Gotuzzo E: Zonas hiperendémicas y mesoendémicas de la infección por Fasciola hepatica aledañas a la ciudad de Lima: Una enfermedad emergente?. Rev Gastroenterol Perú. 2007, 27: 21-26.
Londoñe P, Chavez A, Li O, Suarez F, Pezo D: Presencia de caracoles Lymnaeidae con formas larvarias de Fasciola hepatica en altitudes sobre los 4000 msnm en la Sierra Sur del Perú. Rev Inv Vet Perú. 2009, 20: 58-65.
Fuentes MV, Malone JB: Development of a forecast system for fascioliasis in central Chile using remote sensing and climatic data in a Geographic Information System. Res Rev Parasitol. 1999, 59: 129-134.
Acknowledgements
Studies funded by Projects No. SAF2006-09278 and SAF No. 2010–20805 of the Ministry of Science and Innovation, Madrid; and by the Red de Investigación de Centros de Enfermedades Tropicales – RICET (Projects No. C03/04, No. ISCIII2005-PI050574 and No. ISCIII-RETIC RD06/0021/0017 of the Programa de Redes Temáticas de Investigación Cooperativa RETICS/FEDER), FIS, Ministry of Health, Madrid, Spain.
Field work activities performed within the worldwide initiative of WHO (Headquarters Geneva, Switzerland) against human fascioliasis. Joint coordination activities carried out within Project No. RLA5049 of the International Atomic Energy Agency (Animal Production and Health Section, Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, Department of Nuclear Sciences and Applications, IAEA Headquaters Vienna, Austria). Technical support provided by the Servicio Central de Secuenciación para la Investigación Experimental (SCSIE) of the Universidad de Valencia (Dr. A. Martínez).
Special thanks are given to Dr. Zoila Villavicencio, Dirección Regional de Salud de Cajamarca, for collaboration in coordination tasks, to Dr. María Adela Valero and postgraduate student Miss Sandra Sáez Durán (Valencia, Spain) for collaboration in field work, and to the Universidad Nacional de Cajamarca and the General Office for Technical Cooperation of this university, for kindly providing laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MDB contributed to the design of the study, participated in field collections, analysed the sequences, and helped to draft the manuscript. PA carried out the DNA sequencing processes. MK performed the phenotypic characterisation. PO participated in laboratory procedures and facilitated logistics for both experimental and field work in the endemic areas of Cajamarca. CN participated in lymnaeid collections in the endemic areas of Cajabamba. SMC designed and supervised the study, participated in field collections, performed the epidemiological analyses, and wrote the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bargues, M.D., Artigas, P., Khoubbane, M. et al. Molecular characterisation of Galba truncatula, Lymnaea neotropica and L. schirazensis from Cajamarca, Peru and their potential role in transmission of human and animal fascioliasis. Parasites Vectors 5, 174 (2012). https://doi.org/10.1186/1756-3305-5-174
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-5-174