Abstract
Background
Chlorophyte microalgae have a cell wall containing a large quantity of cellulose Iα with a triclinic unit cell hydrogen-bonding pattern that is more susceptible to hydrolysis than that of the cellulose Iβ polymorphic form that is predominant in higher plants. This study addressed the enzymatic hydrolysis of untreated Chlorella homosphaera biomass using selected enzyme preparations, aiming to identify the relevant activity profile for the microalgae cellulose hydrolysis. Enzymes from Acremonium cellulolyticus, which secretes a complete pool of cellulases plus β-glucosidase; Trichoderma reesei, which secretes a complete pool of cellulases with low β-glucosidase; Aspergillus awamori, which secretes endoglucanases and β-glucosidase; blends of T. reesei-A. awamori or A. awamori-A. cellulolyticus enzymes; and a purified A. awamori β-glucosidase were evaluated.
Results
The highest initial glucan hydrolysis rate of 140.3 mg/g/h was observed for A. awamori enzymes with high β-glucosidase, low endoglucanase, and negligible cellobiohydrolase activities. The initial hydrolysis rates when using A. cellulolyticus or T. reesei enzymes were significantly lower, whereas the results for the T. reesei-A. awamori and A. awamori-A. cellulolyticus blends were similar to that for the A. awamori enzymes. Thus, the hydrolysis of C. homosphaera cellulose was performed exclusively by the endoglucanase and β-glucosidase activities. X-ray diffraction data showing negligible cellulose crystallinity for untreated C. homosphaera biomass corroborate these findings. The A. awamori-A. cellulolyticus blend showed the highest initial polysaccharide hydrolysis rate of 185.6 mg/g/h, as measured by glucose equivalent, in addition to the highest predicted maximum glucan hydrolysis yield of 47% of total glucose (w/w). T. reesei enzymes showed the lowest predicted maximum glucan hydrolysis yield of 25% (w/w), whereas the maximum yields of approximately 31% were observed for the other enzyme preparations. The hydrolysis yields were proportional to the enzyme β-glucosidase load, indicating that the endoglucanase load was not rate-limiting.
Conclusions
High rates of enzymatic hydrolysis were achieved for untreated C. homosphaera biomass with enzymes containing endoglucanase and β-glucosidase activities and devoid of cellobiohydrolase activity. These findings simplify the complexity of the enzyme pools required for the enzymatic hydrolysis of microalgal biomass decreasing the enzyme cost for the production of microalgae-derived glucose syrups.
Similar content being viewed by others
Background
In the pursuit of a new, renewable, and environmentally friendly feedstock supply, the use of seed plants, energy crops, grasses, and agricultural wastes has raised serious concerns regarding the use of land and water for food versus feedstock for chemicals and fuel production. Alternatively, attention has been turned to the use of algae, in particular microalgae, as feedstock for the production of renewable chemicals and biofuels, as they do not compete for arable land [1] despite some skeptical views [2,3]. Microalgae are versatile photosynthetic organisms that can adapt to and tolerate a variety of environmental conditions and can be cultivated in non-freshwater sources including salt and wastewater [4,5] besides being able to use flue gases as a source of carbon, sulfur, and nitrogen [6]. The calorific values of microalgae can be increased by growing them in tubular bioreactors [7] or in cheaper, low nitrogen media [8] because nitrogen starvation triggers lipid and carbohydrate accumulation [9]. Several products can be obtained from the same algal biomass using an integrated biorefinery concept whereby different biomass processing methods can be sequentially employed, including lipid extraction, algae biomass polysaccharide hydrolysis for sugar syrup production and subsequent fermentation [10], followed by biodigestion or pyrolysis for the production of biodiesel, bioethanol, methane, and syngas [11-13]. Furthermore, milder conditions for microalgae, with the aim of biomass polysaccharide hydrolysis, are necessary for the production of sugar syrups in comparison with lignocellulosic biomass processing [14,15].
The carbohydrates in green algae, primarily cellulose, come chiefly from the cell wall components although depending on the growth conditions in which starch accumulates in the chloroplasts [16]. Early studies on the algal cell wall [17] classified the algae species into three groups according to their cell wall constituents. Group 1 includes chlorophyte algae, in which native cellulose is the major component of the cell walls and is usually highly crystalline. These algae belong to the Cladophorales order. Group 2 includes the chlorophyte algae, which have cell walls that contain a large quantity of mercerized-like cellulose, which is presumably a derivative of native cellulose. This cellulose has a low degree of crystallinity, and the chains are randomly oriented. Most algae fall into this category. Group 3 contains algae for which the walls contain a well-oriented and highly crystalline skeletal substance, which is neither native nor mercerized cellulose, as the major constituent [17]. Three types of cellulose were identified in the algal system as follows: the Iα-rich, broad microfibril, 0.6 nm-oriented type; the Iβ-dominant flat-ribbon, 0.53 nm-oriented type; and the Iβ-dominant, small, random-oriented type. The first type appears to occur in more primitive organisms than the other types. The three types of algal cellulose correlate well with the arrangements of cellulose synthesizing complexes, i.e., a multiple-row linear type, a consolidated rosette type, and an isolated rosette type, respectively [18,19]. Linear-type terminal complexes are found in algae belonging to the Chlorophyta division whereas the rosette terminal complex is found in algae belonging to the Charophyta division and land plants [18-21]. Cellulose Iα differs from Iβ not only in the hydrogen-bonding pattern, and consequently in the crystalline unit cell [22,23], but also in its stability and susceptibility to hydrolysis. Cellulose Iα, which possesses a triclinic unit cell, is metastable and converts into the monoclinic form (Iβ) after annealing in dilute alkali at 260°C [24]; the theoretical density of the monoclinic unit cell is slightly greater than that of the triclinic unit cell [25]. These differences in the stability, hydrogen-bonding pattern, and distribution of the two crystalline phases in the microfibril may render cellulose Iα more prone to enzymatic hydrolysis, as indicated by FTIR and electron diffraction data [25,26]. The X-ray diffraction patterns of cellulose in plants and algae differ in the presence of well-resolved and narrow peaks, especially at 2θ’s of 14° and 16°, which are not common for native cellulose obtained from higher plants [27].
Chlorella homosphaera is a microalga that belongs to the Chlorophyta division. The microalgae from this genus divide by autosporulation and therefore possess lytic activity [28-31] that leads to the enzymatic dissolution of their inner cell wall polysaccharides. The Chlorella cell wall is formed by an inner and outer layer and possesses unusual polysaccharides other than cellulose and hemicellulose [32,33]. The inner layer is composed of a matrix and a rigid fibrillar structure, shows high cellulose content, and is susceptible to degradation by cellulolytic enzymes [34]. The outer cell wall, which is resistant to treatment with a number of enzymes, presents two types of ultrastructure, one of which is a trilaminar structure with several resistant components including the biopolymer algaenan, a non-hydrolyzable aliphatic biopolymer composed of long-chain, even-carbon-numbered, ω9-unsaturated ω-hydroxy fatty acid monomers that vary in their chain lengths from 30 to 34 carbon atoms. These monomers are intermolecularly ester-linked to form linear chains in which the unsaturated carbons act as the starting position of ether cross-linking [35].
This study evaluates the enzymatic hydrolysis of polysaccharides from untreated C. homosphaera biomass with enzyme preparations that were produced by the fungi Trichoderma reesei, Aspergillus awamori, and Acremonium cellulolyticus, in addition to the mixtures of enzymes produced by T. reesei and A. awamori or A. cellulolyticus and A. awamori. The focus was to identify the enzyme activities necessary for the efficient hydrolysis of C. homosphaera cell wall polysaccharides because each enzyme preparation showed a different enzyme activity profile and load concerning its endoglucanase, cellobiohydrolase, and β-glucosidase content, aside from the differences regarding their xylanolytic enzyme profiles. This work also reports on the initial hydrolysis rates, the effect of the β-glucosidase load on the hydrolysis rates, and the final hydrolysis yields. The XRD crystallinity profile of the C. homosphaera cell wall from untreated algal biomass is also presented.
Results and discussion
C. homosphaera biomass sugar composition
Data for the total sugar content and sugar composition of C. homosphaera biomass dry weight for the biomass total hydrolysis by 1 mol/L H2SO4 followed by high-performance anion exchange chromatography and pulse amperometric detection (HPAEC) analysis is presented in Table 1. C. homosphaera biomass possessed a carbohydrate content of 59.7% with a glucose contribution of 53.9%. Considering that amyloglucosidase hydrolysable starch (AHS) contributed to 7% (w/w) of the total glucose, non-AHS polysaccharides corresponded to 46.1% of the C. homosphaera biomass. As such, glucose constituted 90.4% of all carbohydrates present in C. homosphaera (85.5% non-AHS polysaccharides) besides the minor contributions of galactose (5.4%), arabinose (2.6%), mannose (1.3%), and xylose (0.5%), which makes this microalga a glucose-rich biomass. This high carbohydrate content is caused by the growth conditions used in this study; after nitrate depletion, within the fifth day of cultivation, the microalga was kept under nitrogen starvation for 15 days before harvest. Nitrogen starvation reportedly triggers the accumulation of carbohydrates, primarily starch, and lipids in both Chlorella [9,16,36] and Chlamydomonas reinhardtii [37,38], increasing their calorific value [8]. The low AHS content of C. homosphaera late stationary grown phase cells found in this study suggests that reserve carbohydrates, which may have accumulated during the exponential phase of growth as suggested by the Lugol’s solution coloration used for cell counting (data not shown), were converted into structural carbohydrates and incorporated into the algal cell wall [37-39] primarily as a glucan type, most likely cellulose. Accordingly, cellulose was the primary polysaccharide present in Chlorella fusca [30,31] and Coelastrum sphaericum [35]; however, a similar low starch content was found in the chlorophyte Chlorococcum humicola [40].
These results resemble those previously reported for the cell wall sugar composition of some Chlorophytes [41-43], for which glucose, galactose, mannose, arabinose, xylose, rhamnose, and sometimes fucose were detected. A similar composition was also found in the TFA-hydrolyzed microalgal biomass of C. reinhardtii [37] and mucilage sheaths of C. sorokiniana, although sucrose was among the saccharides that were found [44]. Interestingly, glucose and mannose are the monosaccharides that are reportedly found in the rigid fibrillar structure of the inner layer of the cell wall, and all the others were found only in the matrix structure of the inner layer, with glucose as the primary saccharide [42,43]. Because we estimated the sugar composition of the C. homosphaera total biomass and not that of the purified cell walls, we expected that non-cell wall polysaccharides such as starch would contribute to the total glucose composition, as already noted; nevertheless, the chlorophyte microalgae Scenesdesmus obliquus possesses a cell wall that is enriched with glucose [45].
Biomass enzymatic hydrolysis
Hydrolysis experiments were performed using enzymes that were produced by selected fungal strains according to their different profiles in relation to cellulase, β-glucosidase, and xylanase activities, as follows: Acremonium cellulase (Meiji Seika Co., Japan), a preparation possessing a complex set of biomass-hydrolyzing enzyme activities; exoglucanase (4.3 filter paper unit (FPU)/mL) and endoglucanase (67.1 IU/mL); xylanase (208.0 IU/mL) and β-glucosidase (29.2 IU/mL); a T. reesei RUT C-30 preparation containing exoglucanase (1.2 FPU/mL), endoglucanase (37.3 IU/mL), xylanase (228.0 IU/mL), and low β-glucosidase content (1.4 IU/mL); the A. awamori 2B.361 U2/1 enzyme was devoid of exoglucanase but exhibited xylanase (117.0 IU/mL), β-glucosidase (8.4 IU/mL), and low endoglucanase (1.9 IU endo-β-1,4 glucanase (CMCase)/mL).
The A. awamori-T. reesei blend contained exoglucanase (5.3 IU FPU/mL), endoglucanase (20.5 IU/mL), xylanase (202.0 IU/mL), and β-glucosidase (69.5 IU/mL) activities; the supernatants of individual fungal cultures were concentrated by ultrafiltration using a 30-kDa membrane before blending. The A. awamori-A. cellulolyticus blend possessed exoglucanase (2.0 FPU/mL), endoglucanase (32.5 IU/mL), xylanase (159.7 IU/mL), and β-glucosidase (18.2 IU/mL) activities.
Sugar analyses
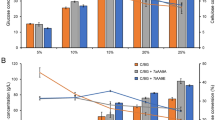
Table 2 shows the sugar composition and yield released from the C. homosphaera biomass after 48 h of enzymatic hydrolysis. The overall analysis of the effectiveness of the different enzyme preparations in hydrolyzing the algae cell wall polysaccharides indicates that the A. awamori-A. cellulolyticus enzyme blend released the highest amounts of glucose and galactose, besides releasing mannose, and was the only preparation that was able to release the pentoses xylose and arabinose and thus able to attack pentose-containing polysaccharides, besides glucan and galactose and/or mannose-containing polysaccharides. The A. awamori-T. reesei blend was able to hydrolyze glucan aside from the galactose and/or mannose-containing polysaccharides, and all the other enzyme preparations were solely able to hydrolyze glucan and galactose-containing polysaccharides, which were noteworthy in that the A. cellulolyticus enzymes released the largest amount of galactose. Partially purified β-glucosidase was solely able to release limited amounts of glucose in comparison with the non-enzymatic control experiments; the small detected galactose release can be considered non-enzymatic when taking into account the data for the non-enzymatic galactose release. No cellobiose was detected in the hydrolysate for all enzyme preparations. Because A. awamori can also produce amylases [46], one could argue that the high glucose content found in the hydrolysates of A. awamori and the T. reesei-A. awamori blend might result from starch hydrolysis. To test this hypothesis, 50 mg/mL starch solution was incubated under the same conditions used for algae biomass hydrolysis for each enzyme preparation; however, all preparations were unable to hydrolyze starch (data not shown). Because the C. homosphaera starch content was not converted into glucose, all the measured glucose was solely released from the cell wall polysaccharides.
Glucose and mannose are reportedly found in the rigid fibrillar structure of the inner layer of the C. homosphaera cell wall, and galactose, arabinose, and xylose are present in the matrix of the cell wall inner layer [42,43]. Thus, the absence of mannose in all enzyme preparations but its presence in the A. awamori-A. cellulolyticus and A. awamori-T. reesei blends might indicate that these enzyme preparations are able to hydrolyze only the glucan and galactose-containing polysaccharides in the matrix of the inner cell wall. However, the presence of mannose in the hydrolysates resulting from the use of A. awamori-A. cellulolyticus and A. awamori-T. reesei blends indicate that these preparations were able to attack the rigid fibrillar structure of the inner cell wall layer. Moreover, because xylose and arabinose, in addition to glucose, galactose, and mannose, were detected in the hydrolysates resulting from the use of the A. awamori-A. cellulolyticus blend, this preparation was also able to attack different glycosidic bonds or a different structural polysaccharide. The use of enzyme blends showed the synergistic effect of the enzymes produced by these two fungi. The importance of hemicellulases in the polysaccharide hydrolysis in the algae inner layer cell wall was not assessed in this study. However, even when considering that substances other than glucose monosaccharides represent less than 10% of the total algae biomass sugar, key glucosidic bonds involving galactose, arabinose, mannose, and xylose may hinder high-yield glucan hydrolysis. In this context, it is noteworthy that the best performing enzyme preparation, i.e., the A. awamori-A. cellulolyticus blend that resulted in the highest glucose hydrolysis yield, also released the highest amount of galactose, besides releasing mannose, arabinose, and xylose, most likely because of the action of the hemicellulose-degrading enzyme complex.
Nevertheless, interesting results were obtained in this study with regard to the selective hydrolysis of structures in the algae inner cell wall, and conclusive data regarding the specificity of the selected enzyme pool towards the structural polysaccharides could only be gathered from hydrolysis experiments that were performed with purified components of the cell wall. Those results would be compared to sequentially TFA and H2SO4-hydrolyzed cell wall components because TFA hydrolyzes only the matrix of the inner cell wall and H2SO4 hydrolyzes both the matrix and the inner cell wall fibrillar structure [47]. This question will be a matter of further investigation.
These results are consistent with the time course of algae biomass hydrolysis shown in Figures 1 and 2. The glucose and total reducing sugar yields curves are very similar when the biomass was hydrolyzed with the enzymes of T. reesei, A. awamori, or a mixture of both. However, when the enzymes of A. cellulolyticus or the A. awamori-A. cellulolyticus blend were used, the amount of released glucose was noticeably lower than that of the total reduced sugar, suggesting that non-glucan-type polysaccharides were hydrolyzed.
A time course for the release of total reducing sugars from the C. homosphaera biomass (50 mg d.w./mL) in 50 mM citrate buffer, pH 4.8, at 50° C. The biomass was hydrolyzed using the enzyme preparations from T. reesei (filled circles), A. cellulolyticus (open triangles), A. awamori (inverted filled triangles), the T. reesei-A. awamori blend (open circles), the A. cellulolyticus-A. awamori blend (open squares), and partially purified β-glucosidase (diamonds). Biomass suspended in buffer without enzyme preparation was used as a control (filled squares). The data were fitted into an exponential function as described in the ‘Materials and methods’ section. Standard deviation was less than 10% of the mean value, and bars were omitted for clarity.
The time course for the release of glucose from C. homosphaera biomass (50 mg d.w./mL) in 50 mM citrate buffer, pH 4.8, at 50° C . The biomass was hydrolyzed using the enzyme preparations from T. reesei (filled circles), A. cellulolyticus (open triangles), A. awamori (inverted filled triangles), the T. reesei-A. awamori blend (open circles), the A. cellulolyticus-A. awamori blend (open squares), and partially purified β-glucosidase (diamonds). Biomass suspended in buffer without enzyme preparation was used as the control (filled squares). The data were fitted into an exponential function as described in the ‘Materials and methods’ section. Standard deviation was less than 10% of the mean value and bars were omitted for clarity.
C. homosphaera biomass enzymatic hydrolysis
Figures 1 and 2 present time course profiles for the release yield of total reducing sugars and of glucose, in terms of total sugar and total glucose, respectively, on the enzymatic hydrolysis of C. homosphaera biomass. In Figure 1, curves for the A. cellulolyticus, A. awamori, A. cellulolyticus-A. awamori blend, partially purified β-glucosidase, and the control were best fitted into a mono-exponential model, and the curves for T. reesei and the T. reesei-A. awamori blend were best fitted into a double exponential model. In Figure 2, the curve for T. reesei was best fitted into a double exponential model, and the others were best fitted into a mono-exponential model. Figure 1 shows that the initial rates and final yields for the release of reducing sugars for the A. cellulolyticus-A. awamori blend were the highest followed by A. awamori and T. reesei-A. awamori blend enzyme preparations, which were comparable to one another, plateauing at a 10-h reaction time. The enzymes from A. cellulolyticus exhibited a lower initial rate, plateauing with comparable hydrolysis yields at an 18-h reaction time. Enzymes from T. reesei showed the smallest initial rate and did not reach the yield plateau within the time frame of the hydrolysis reaction. The partially purified β-glucosidase, which is devoid of cellobiohydrolase, endoglucanase, and xylanase activities, showed a discrete ability to hydrolyze the C. homosphaera biomass with a low initial rate and maximum yield, indicating that the β-glucosidase excreted by A. awamori was able to attack the algae biomass glucan in addition to its well-known action on cellobiose and short chain oligosaccharides [48]. However, the results showed that the synergistic action of endoglucanases and β-glucosidase is essential to attain higher hydrolysis yields. The C. homosphaera biomass also showed a small amount of sugar release unrelated to the enzymatic action (control) for up to 10 h of incubation, possibly because of the reaction conditions, including the pH and temperature, or because of the residual action of native autolytic enzymes. It is well established that chlorophytes reproduce by autospore formation, in which daughter cells are enclosed inside the mother cell wall; autolytic enzymes are used to degrade the mother cell wall and release the daughter cells, which then may use the hydrolyzed monosaccharides as carbon and energy sources [29-31,34]. Figure 2 shows the algae biomass hydrolysis pattern, including the initial rates and final yields for glucose release for all enzyme preparations. The general enzyme preparation performances were quite similar, as expected considering that glucan is the most abundant component of the C. homosphaera cell wall, except for the fact that the initial rate of glucose release for the A. awamori-A. cellulolyticus blend was considerably smaller than the release of total reducing sugar, which was consistent with the results shown in Table 2.
Table 3 summarizes the data for the predicted kinetic parameters regarding initial hydrolysis rates and maximum hydrolysis yields. The data are related to the total sugar (hexose equivalents) and total glucose (glucose) hydrolysis yields. The initial hydrolysis rate of A. awamori preparation, when considering hexose equivalents, was 114.3 mg hexose/g total sugar/h, and the rate for the T. reesei-A. awamori blend was 115.8 mg hexose/g total sugar/h. Enzymes from A. cellulolyticus showed a lower initial velocity of 58.8 mg hexose/g total sugar/h); nevertheless, the maximum predicted hydrolysis yield of 336.2 mg/g was similar to that for A. awamori (311.7 mg/g) and for the T. reesei-A. awamori blends (335.4 mg/g). Enzymes from T. reesei showed the smallest initial rate (27.5 mg hexose/g sugar/h); because the hydrolysis plateau was not reached within the time frame of the experiment and the predicted maximum value by the model was unreliable, the mean experimental result (±standard deviation) at 48 h of incubation was used as the data for the hydrolysis yield of 256.8 mg/g. The highest initial rate and maximum yield were attained with the A. awamori-A. cellulolyticus blend with an initial rate and a final yield that were considerably superior to all the other enzymes preparations (185.6 mg/g/h and 494.6 mg/g, respectively), suggesting that the enzymes of these fungi act synergistically in hydrolyzing the polysaccharides that each enzyme preparation cannot hydrolyze by itself as already shown by the HPAEC-PAD. The partially purified β-glucosidase showed a low initial rate and maximum yield and the biomass incubated without enzymes released some sugar at a very low initial rate and yield.
Following the same trend, the enzyme pool of A. awamori and the blends of A. awamori-T. reesei and A. awamori-A. cellulolyticus took approximately 2 h to accomplish 50% of the maximum hydrolysis yield (t½). A. cellulolyticus enzymes took 4 h and T. reesei enzymes took 13 h to achieve 50% of the 48 h hydrolytic value (Table 3). The predicted kinetic parameters for the enzymatic hydrolysis results when using all enzyme preparations, and taking into account the glucose concentration measurements, were similar to those observed for the measurement of the total reducing sugars, as previously discussed, except for the A. awamori-A. cellulolyticus blend, which showed a similar initial rate for glucose release when compared with the enzyme preparations of A. awamori and the A. awamori-T. reesei blend. Because the glucose yield was considerably higher (470.8 mg glucose/g sugar), the t½ was larger for the A. awamori-A. cellulolyticus blend (Figures 1 and 2 and Table 3). The initial velocities found for T. reesei and A. cellulolyticus enzymes are close to those previously reported by Harun and Danquah [40] for Chlorococum humicola when the same enzyme load was applied. Nevertheless, those found for A. awamori and A. awamori-T. reesei were 1.5 times higher and those for the A. awamori-A. cellulolyticus blend were 2.4 higher. Moreover, with the exception of the T. reesei enzyme pool, we used the Marquardt non-linear regression method, which is a much more reliable procedure for estimating the initial velocity (Table 3). These results suggest that the A. awamori enzyme pools efficiently hydrolyze C. homosphaera biomass, and the results observed for the A. awamori-T. reesei blend resulted predominantly from the contribution of the A. awamori enzyme pool. The A. awamori-A. cellulolyticus blend hydrolyzes the C. homosphaera biomass synergistically to attain a high rate and yield for hydrolyses.
The maximum hydrolytic yield found for all enzyme pools were either similar [40] or lower, sometimes considerably lower (near 50%), than those previously reported [16]. However, in reports in which a higher hydrolytic yield was found, the algae biomass showed a fourfold higher starch content [16], and the material was subjected to severe treatment beforehand, such as sonication, disc milling, or hydrothermal, acid, or alkali pre-treatment before enzymatic hydrolysis [14,16,38,40,45], which may have disrupted the structure of the outer cell wall. In the present work, the room temperature dried algal biomass was only manually ground, which is a very mild procedure for disrupting the algae cell wall; therefore, the enzymatic hydrolysis results reported herein are quite promising.
The maximum yield was near 50 mg of reducing sugar/g total sugar, indicating that approximately 50% of the algae polysaccharides were not accessed by either of the studied enzyme preparations. Considering the structure of the whole cell wall and that the outer cell wall is resistant to treatments with a number of enzymes, we could speculate that only the inner cell wall layer polysaccharides from either the rigid fibrillar structure or the matrix were selectively or entirely hydrolyzed by the studied enzyme preparations. As such, the residual non-hydrolyzed polysaccharide fraction, from the higher yield hydrolysis experiments, would be a component of the outer cell wall. In reports in which the starch content was similar to ours, a comparable maximum yield was found [40]. Other studies about algae biomass pretreatments are under way, in our laboratory, to increase the maximum hydrolysis yields.
The hydrolytic efficiency of the A. awamori, T. reesei, and A. cellulolyticus enzyme pools and blends
Because the enzyme pool of A. awamori has mostly β-glucosidase activity in addition to endoglucanase activity and negligible cellobiohydrolase activity, a lower initial hydrolysis rate and a lower maximum hydrolysis yield were expected in comparison with the complete cellulases plus β-glucosidase preparations of A. cellulolyticus and the blend of T. reesei and A. awamori enzymes. In fact, we would expect a pattern of hydrolysis similar to the partially purified β-glucosidase, but with a higher initial rate and yield because it possesses endoglucanase activity and the partially purified β-glucosidase does not. However, a similar rate and hydrolytic efficiency were observed for the A. awamori enzyme preparation and the A. awamori-T. reesei blend in comparison with the A. cellulolyticus enzymes, indicating that exoglucanase (CBH) activity is not necessary for the hydrolysis of this microalgae biomass.
The use of the A. awamori-A. cellulolyticus enzyme blend resulted in a hydrolysis reaction medium with the highest β-glucosidase load (90.3 IU/g d.w. biomass) followed by the A. cellulolyticus enzymes (49.0 IU/g) and by both the A. awamori enzymes and the T. reesei-A. awamori enzyme blend (22.3 IU/g). The T. reesei enzymes showed the lowest β-glucosidase load (8.7 IU/g). With the exception of A. cellulolyticus, the initial hydrolysis rates and final yields correlated positively with the β-glucosidase. To understand further the importance of β-glucosidase activity, we conducted two sets of hydrolysis assays with T. reesei, A. awamori, A. cellulolyticus, and a blend of A. awamori-T. reesei enzyme preparations. In the first set, the assays were performed with the same β-glucosidase load for 3 h because more striking differences regarding the initial hydrolysis yields were observed within this time interval. A control experiment, with the same load of a partially purified β-glucosidase and devoid of endo- and exoglucanase activities, was conducted. Figure 3 shows that after 3 h of hydrolysis, no significant differences among the means of hydrolyzed hexose equivalents or glucose were observed for any enzyme preparation. However, the A. cellulolyticus enzymes showed lower mean values, mostly for glucose, which were significant when compared with the A. awamori enzyme, confirming the previous results shown in Figures 1 and 2, where despite having a higher β-glucosidase load, it showed a lower initial rate but a similar final hydrolysis yield.
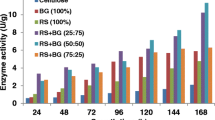
The hydrolysis yield of C. homosphaera biomass after 3 h of enzymatic treatment. C. homosphaera biomass (50 mg/mL) was incubated in 50 mM citrate buffer pH 4.8 at 50°C with the indicated enzyme preparation at a final β-glucosidase activity of 23.3 IU/g d.w. biomass load. Biomass without enzyme was incubated under the same conditions as the control. Letters a, b, c, and d indicate differences in the means at 0.05 level.
The hydrolysis results from the control experiments were surprisingly high, accounting for over 40% and 50% of the total hexose and glucose released, respectively (Figure 3). The data were even higher than that presented in Figures 1 and 2, which could be related to differences in the uneven manual grinding of the algae biomass, affecting the particle sizes as we observed in preliminary assays in our laboratory (data not shown). The C. homosphaera biomass also showed a small amount of biomass hydrolysis not caused by enzymatic action as previously found (Figures 1 and 2). The different enzyme preparations contained different endoglucanase loads (Figure 3), which might have played an important role in the hydrolysis results, although no correlation between the endoglucanase load and initial rates and final yields was found. For this reason, all enzyme preparations in the second set of assays were set to a fixed endoglucanase load (1.5 IU/g dry biomass) and increasing β-glucosidase loads as follows: 7.5, 15.0, and 22.5 IU/g dry biomass, which were achieved through purified β-glucosidase supplementation. Figure 4 shows that after 36 h of incubation, all enzymes showed an increase in the hydrolysis yield, especially when the β-glucosidase load increased from 7.5 to the 15 IU/g that was significant for all enzyme preparations. The T. reesei preparation showed no significant increase for the hydrolysis yield of reducing sugar when the β-glucosidase load increased from 15.0 to 22.5 IU/g; however, a significant increase in the hydrolysis yield of glucose was found. The same pattern was observed for the A. cellulolyticus preparation. However, no significant increase in the glucose hydrolysis yield was found for the A. awamori enzyme and the A. awamori-T. reesei blend; nevertheless, the reducing sugar yield increase was significant. An analysis among the groups showed that the hydrolysis yields for A. awamori enzyme preparations were the highest even when compared with experiments presenting a higher β-glucosidase load. Moreover, the A. awamori preparation was more responsive to the increase in the β-glucosidase load (Figure 4), indicating a unique catalytic feature for the A. awamori enzymes.
The hydrolysis yield of C. homosphaera biomass after 36 h of enzymatic treatment. C. homosphaera biomass (50 mg/mL) was incubated in 50 mM citrate buffer pH 4.8 at 50°C with 1.5 IU/g of endoglucanase load and β-glucosidase loads of 7.5, 15.0, and 22.5 IU/g biomass loads by adding partially purified β-glucosidase.
Data for the predicted kinetics parameters regarding the initial hydrolysis rates and maximum yields of each enzyme preparation with the three β-glucosidase load (Additional file 1: Figure S1 and Additional file 2: Figure S2) are shown in Table 4. The predicted maximum hydrolysis yield for all enzyme preparations followed the same pattern presented in Figure 4. The analysis within the group for the initial hydrolysis rates indicated that T. reesei and A. cellulolyticus preparations showed a significant increase when the β-glucosidase load was increased from 7.5 to 15 IU/g. A small increase was also found when the β-glucosidase load was increased from 15 to 22.5 IU/g, but the model failed to predict any significant difference in the initial rate values. A. awamori and A. awamori-T. reesei blend preparations showed no significant increase after the increment of the β-glucosidase load. An analysis among the groups showed that the A. awamori preparation showed the highest initial rates of reducing sugar release that were significant when compared with their counterparts, which showed no significant difference among them. The predicted initial rates for glucose release in the A. awamori preparation showed a large standard error and are not reliable, but the predicted means suggest higher initial rates when compared with the others.
HPAEC-PAD analyses showed that the released monosaccharides consisted of only glucose and galactose for all hydrolysis conditions, i.e., 1.5 IU endoglucanase load/g (d.w.) biomass and 7.5, 15.0, and 22.5 IU β-glucosidase load/g (d.w.) biomass. The enzyme preparation of A. awamori showed the highest sugar hydrolysis yield, and the increment in the β-glucosidase load increased the amount of sugar that was hydrolyzed. These results are consistent with those presented in Figure 4.
Altogether, these results clearly show that for the same endoglucanase load, the hydrolysis yield of C. homosphaera biomass is positively affected by the β-glucosidase load. Moreover, even in the absence of A. awamori enzymes, the β-glucosidase load affects the initial rate of hydrolysis. This finding also shows that a very small endoglucanase load is needed to attain high hydrolysis yields within just 12 h of incubation. The higher rates and yields observed for A. awamori enzymes suggest that the enzyme prepared from this fungus have a higher ability to hydrolyze C. homosphaera biomass.
C. homosphaera cell wall studies
The fact that the A. awamori enzymes, which are devoid of exoglucanase activity, hydrolyzed C. homosphaera biomass at high rates and that the β-glucosidase activity load influences the hydrolysis yields and sometimes the initial rate, raised a question regarding the nature of the microalgal cell wall glucan. Chlorophytes are known to be enriched with cellulose type Iα, which is more susceptible to degradation than cellulose type Iβ, which is predominant in Charophyta and land plants [22,25,26,49]. Although the XRD of microalgae cellulose is generally similar to that of land plants, and except for some peaks at 14° and 16° that are not found in land plants [27], our results showed otherwise. Indeed, Figure 5 shows the X-ray diffractometry of C. homosphaera biomass and, for comparison, that of pure crystalline cellulose. The figure shows that, contrary to the data for crystalline cellulose, C. homosphaera biomass has a very low crystallinity, such that it was not possible to determine its degree of crystallinity. However, it was possible to identify a clear peak near 15°, which is typical for microalgae [27]. Remarkably, the XRD of C. homosphaera closely resembles that of the ball-milled XRD of microcrystalline cellulose after enzymatic hydrolysis, as found by Teixeira et al. [50], who also showed that cellobiohydrolase activity is not required to efficiently hydrolyze amorphous cellulose. These results suggest that the cell wall polysaccharides of C. homosphaera is mostly amorphous and, as such, C. homosphaera would be categorized as belonging to group 2, as established by Nicolai and Preston [17]. However, the low crystallinity may also be an artifact of biomass preparation; under stress, chlorophytes secrete a mucilaginous substance that was observed during the centrifugation process. In addition, the presence of algaenan in the cell wall may create structural changes and amorphous zones during the drying process, as suggested by Mihranyan [27]. However, regardless of whether the centrifugation and drying processes altered the cell wall polysaccharide structure, the dried C. homosphaera biomass showed such a low crystallinity that the enzyme mixtures devoid of exoglucanase activity were able to efficiently hydrolyze 40.5% of the algal glucan. In terms of biomass processing and enzyme use, this feature poses a great advantage for the biorefinery concept because it simplifies raw material processing and enzyme blend complexity, thereby reducing processing costs.
Figure 6A shows the scanning electron microscopy of C. homosphaera cells and cell fragments that were dried and hand-milled, and Figure 6B shows the C. homosphaera cells after enzymatic hydrolysis when using the A. awamori-T. reesei enzyme blend. The images suggest that the enzyme action resulted in a more homogenous material and that an amorphous and sticky material that surrounded the undigested algae was removed by enzymatic hydrolysis.
Scanning electron microscopy of ground C. homosphaera biomass (A) before enzymatic hydrolysis and (B) after 48 h of enzymatic hydrolysis using the A. awamori-T. reesei blend. Samples were adhered to carbon tape, sputter-coated with 28 nm gold using an Emitech/K550 model and observed via SEM as described in the Materials and methods section. Magnifications of × 1,200 (A) and × 6,000 (B) were chosen to show the action of the enzyme blend more clearly on C. homosphaera biomass. The scale bars represent 50 μm (A) and 10 μm (B).
Conclusions
Enzyme mixtures of A. awamori, which have β-endoglucanase and β-glucosidase activities and are devoid of cellobiohydrolase activity, efficiently hydrolyze untreated C. homosphaera biomass, reaching 42.3% of the cell wall glucan because of its very low crystallinity, and the A. awamori-A. cellulolyticus enzyme blend attain even higher hydrolysis rates and yields of approximately 50%. This finding is most likely explained by the synergistic action of both enzyme preparations that allowed the hydrolysis of different polysaccharides that would be otherwise inaccessible. The β-glucosidase load was a determinant in the hydrolysis yield and in some cases for the initial hydrolysis rate of all the enzyme preparations.
Materials and methods
C. homosphaera cultivation
C. homosphaera cells were grown in inorganic W.C. medium [51] with continual aeration under white fluorescent light, with a 12-h photoperiod and a photosynthetic active radiation of 60 μmol/m2/s in average irradiance. The cells were cultivated in five carboys of 5 L each for 21 days and collected at the stationary phase of growth by centrifugation, then dried cold, ground manually on a mortar, and stored in the freezer until use, as previously described [52]. Under this growth condition, an average of 300 mg/L of dry weight biomass was obtained at the end of cultivation. Cell growth was followed by cell counting and by light scattering at 750 nm, to avoid the absorption of any cell chromophore, using a previously calibrated dry weight curve. Each growth phase was determined by plotting the logarithm of the cell concentration against the cultivation time.
Fungal strains and enzyme preparations
Laboratory enzyme preparations were produced by A. awamori 2B.361 U2/1 and T. reesei Rut-C30 and deposited in the fungal culture collection of the National Institute of Quality Control in Health (INCQS 40259-40251, respectively) of the Oswaldo Cruz Foundation (http://www.incqs.fiocruz.br). Both fungi were propagated on potato dextrose agar (PDA) plates at 30°C for 7 days, until dense sporulation was observed. The spores were collected by adding 2 mL of sterilized distilled water to the plate, followed by gentle scraping. A standardized spore suspension presenting 107 spores/mL in 20% (v/v) glycerol was maintained at −20°C. For enzyme production, T. reesei Rut C30 was cultivated in a liquid medium containing the following (in g/L): 30.0 lactose, 6.0 yeast extract, 0.3 urea, 0.6% (v/v) corn steep liquor, plus the following salts (in g/L): 1.4 (NH4)2SO4, 2.0KH2PO4, 0.3 CaCl2, and 0.3 MgSO4.7H2O, and the following trace elements (mg/L): 5.0 FeSO4.7H2O, 20 CoCl2, 1.6 MnSO4, and 1.4 ZnSO4, with an initial pH of 6.0. A. awamori was cultivated in a liquid medium containing the following (in g/L): 12.0 yeast extract, 30.0 wheat bran, 1.2 NaNO3, 3.0 KH2PO4, 6.0 K2HPO4, 0.2 MgSO4.7H2O, and 0.05 CaCl2, with an initial pH of 7.0 [53]. Enzyme production by both fungi took place in 1,000-mL Erlenmeyer flasks containing 300 mL of growth medium inoculated with 1% (v/v) of the spore suspension. The cultures were incubated in a rotary shaker (Innova 4340, New Brunswick, Edison, NJ, USA) at 30°C and 200 rpm. The commercial enzyme Acremonium cellulase from A. cellulolyticus was kindly provided by Meiji Seika Pharma Co., Japan.
A. awamori β-glucosidase was partially purified as follows. Fungal culture supernatant was concentrated by ultrafiltration using a 100-kDa membrane (Amicon Filtration System-Stirred Cells). A 10 mL volume of the retentate, which contained 227 mg of protein and a total β-glucosidase activity of 5672 IU, was subsequently fractionated by gel filtration on a Sephadex G-75 Column (3.0 × 62.5 cm) and pre-equilibrated with 50 mM sodium acetate buffer (pH 5.0) containing 0.15 M NaCl. The sample was eluted using a flow rate of 20 mL/h, and 5.0 mL aliquots were collected and screened for β-glucosidase activity and protein concentration by measuring the absorbance at 280 nm. Fractions presenting β-glucosidase activity were pooled and concentrated by ultrafiltration with a 30-kDa membrane. This preparation was shown to be free of FPase, CMCase, and xylanase activities. The protein concentration was also measured according to Bradford [54]. β-glucosidase preparations from gel filtration and ion-exchange chromatography were analyzed by SDS-PAGE, Native PAGE, and zymogram with an 8% polyacrylamide gel [55].
The following enzyme preparations were used to hydrolyze the C. homosphaera biomass: enzymes from T. reesei RUT-C30, A. awamori, A. cellulolyticus, and blends of T. reesei-A. awamori and A. awamori-A. cellulolyticus-excreted enzymes. The T. reesei-A. awamori blend was made by taking into account the FPase and β-glucosidase of each individual enzyme preparation, which were mixed in the correct proportion to achieve the final FPase: β-glucosidase activities ratio of 1:2. Moreover, hydrolysis experiments were performed with a purified A. awamori β-glucosidase preparation to evaluate the effect of this enzyme alone on the initial biomass hydrolysis rate. The aforementioned fungal enzymes were chosen because of their complementary activities as follows: (i) A. awamori 2B.361 U2/1, which produces high levels of glucoamylase and β-glucosidase, but lacks exoglucanase activity (8.4 IU/mL of β-glucosidase, 1.9 IU/mL of CMCase, and no FPase activity); (ii) T. reesei RUT C-30, which produces high levels of cellulases but low levels of β-glucosidase (1.4 IU/mL of β-glucosidase, 37.3 IU/mL of CMCase, and 1.2 IU/mL of FPase); (iii) Acremonium cellulase (Meiji Seika Co., Japan), which contains high CBH levels in addition to a complex set of biomass-hydrolyzing enzyme activities (29.2 IU/mL of β-glucosidase, 67.1 IU/mL of CMCase, and 4.3 IU/mL of FPase); and (iv) a blend of A. awamori and T. reesei after the concentration of each enzyme by ultrafiltration with a 30 kDa membrane (20.5 IU/mL of β-glucosidase, 69.5 IU/mL of CMCase, and 5.5 IU/mL of FPase).
FPAse, CMCase, and β-glucosidase activity determinations
The activities of A. awamori 2B.361 U2/1 and T. reesei Rut-C30 laboratory enzymes and the A. cellulolyticus commercial enzyme were determined according to standard IUPAC procedures and expressed as international units (IU), as described by Ghose (1987) [56]. Several dilutions of the enzyme pools were prepared by diluting them with 50 mM citrate buffer, pH 4.8. Enzyme activities were estimated by incubating the diluted enzyme preparations with the relevant substrate in a water bath at 50°C for the time indicated. FPAse was determined by incubating exactly 6 cm2 of Whatman 1 filter paper for exactly 1 h. The reaction was stopped by adding 3,4-dinitrosalicylic acid (DNS) and centrifuging to remove paper debris; the reducing sugar content was then determined. The endo-β1,4-glucanase activity was determined by incubating a fresh 2% (w/v) carboxymethyl cellulose CMC 7 L2 solution in 50 mM citrate buffer, pH 4.8, for exactly 30 min, followed by the reducing sugar determination. The β-glucosidase activity was determined by incubating a fresh 15 mM cellobiose solution in 50 mM citrate buffer, pH 4.8, for exactly 30 min. The enzymatic reaction was stopped by incubating in a boiling water bath for 5 min; the glucose content was then determined. As controls, the reducing sugar contents of the paper and of the enzyme dilutions, in addition to the glucose contents of the enzyme dilutions, were determined. The enzyme activities were estimated using the calculations described in [56].
C. homosphaera biomass enzymatic hydrolysis
The experiments were performed in 50-mL glass Erlenmeyer flasks that were stoppered with glass to minimize evaporation, with a 30-mL reaction mixture containing 50 mg/mL cell mass powder suspended in 50 mM citrate buffer, pH 4.8, and an enzyme load of 10 FPU (filter paper activity) units/g dry biomass. The measured β-glucosidase loads for the T. reesei and A. cellulolyticus preparations and the A. awamori-A. cellulolyticus blend were 8.7, 49.0, and 90.3 IU/g biomass, respectively. Because the A. awamori enzymes exhibited no FPAse, the enzyme load was equalized with the β-glucosidase load of the T. reesei-A. awamori blend, for 22.3 IU/g dry biomass. The mixtures were incubated at 50°C in a rotatory shaker (Innova 4340, New Brunswick, Edison, NJ, USA), and sampling was performed at 0, 2, 4, 6, 8, 12, 24, 28, 32, 36, and 48 h, followed by incubation for 5 min in a boiling water bath to halt enzyme action. Sugar analyses were performed in the supernatants of centrifuged samples.
To investigate the influence of the β-glucosidase activity on the initial enzymatic biomass hydrolysis and final yield, two sets of assays were designed. In the first, the biomass powder was incubated for 3 h under the same conditions as described above, with an enzyme load of 22.3 IU β-glucosidase/g dry biomass for all the enzymes. This time frame was chosen because the kinetics for the hydrolysis experiments using the evaluated enzyme preparations (A. cellulolyticus, T. reesei, A. awamori, and the A. awamori-T. reesei blend) with the same FPAse load but different β-glucosidase loads showed significant rate differences up to 3 h of hydrolysis, i.e., where the most β-glucosidase hydrolysis dependency was observed. For the controls, a sample of biomass powder was submitted to hydrolysis with partially purified A. awamori β-glucosidase and another biomass sample was incubated without any enzymes under the same conditions. As in the first set, the reaction medium of each enzyme preparation had a different endoglucanase load, and in the second set of the assay, each reaction medium of the evaluated enzyme preparations was set to have an endoglucanase load of 1.5 IU endoglucanase/g dry biomass. The β-glucosidase load was then adjusted by adding partially purified β-glucosidase to final load values of 7.5, 15.0, and 22.5 I.U/g dry biomass. All enzymatic assays were performed in triplicate.
AHS determination
The AHS in C. homosphaera biomass was estimated on the basis of previously described procedures [57,58]. Soluble saccharides, lipids, and pigments were extracted by incubating 50 mg of biomass powder with 3 mL of 80% ethanol (v/v) for 30 min in a water bath at 70°C. The supernatant was discarded, and the procedure was repeated three times. The pellet was washed twice in 50 mM citrate buffer, pH 4.8, and resuspended in 0.9 mL of the same buffer; 0.1 mL of Novozyme amyloglucosidase AMG 300 L solution was then added to give a final enzyme activity load of 40 IU. The suspension was incubated for 24 h at 50°C, then centrifuged; the supernatant glucose content was then estimated. Sodium azide was added to prevent contamination.
Nitrate determination
The nitrate remaining in the culture medium was determined by UV spectrophotometry as described in [59]. A 10 mL aliquot of cell suspension was withdrawn from culture cell flasks under sterile conditions. After determining the cell suspension turbidity at 750 nm, the cells were centrifuged and the clear supernatant was used to determine the nitrate content. For each 1 mL of supernatant, 20 μL of 1 mol/L HCl solution was added to prevent any interference from hydroxide and carbonate ions, and the absorbances at 220 and 275 nm were measured, assuring that the absorbance at 275 nm was less than 10% of the absorbance at 220 nm. The reading at 275 nm was used to discount the interference of organic materials that might be present in the sample. The net absorbance was calculated using the following equation: Net Abs = Abs(220) − 2 × Abs (275). The nitrate concentration was determined by means of an analytical curve of the net absorbance against the nitrate concentration, using a 20 μg/mL NaNO3 solution as a standard. All glassware was thoroughly rinsed with hot 1 mol/L HCl solution and distilled water before use, to eliminate any trace of carbonate and detergent.
Sugar determination
The C. homosphaera total biomass sugar content was determined on the basis of the procedure described in [41]. A total of 4 mg of dry cell powder was incubated with 2 mL of 1 mol/L H2SO4 at 100°C for 6 h. The samples were then neutralized with BaCO3 and centrifuged, and the supernatants were used for total sugar determination by the phenol sulfuric method [60] with an analytical curve using a 0.2 mg/mL glucose solution as a standard. For the control, the same procedure was performed with pure glucose powder. The reducing sugar levels in hydrolysates were estimated by the 3,5-dinitrosalicylic method [61] by means of an analytical curve with a 2.0 mg/mL glucose solution as a standard. The glucose concentration was estimated using a YSI 2730 glucose analyzer (Yellow Springs Incorporated, Ohio, USA). The sugar composition was determined by HPAEC-PAD analysis.
HPAEC-PAD analysis
A sugar composition determination was performed in an Ion Chromatography System 5000 (ICS-5000, Dionex Ltd., Canada) equipped with Chromeleon 6.8 (Dionex Ltd., Canada) software. The column system consisted of a CarboPac PA1 (4 × 50 mm, Thermo Scientific Ltd., USA) pre-column and a CarboPac PA1 (4 × 250 mm, Thermo Scientific Ltd., USA) analytical column. The chromatographic conditions were as follows: the samples were automatically injected at 15°C with an injection volume of 10 μL, a furnace temperature of 30°C, and a pressure of 1,050 to 1,150 psi. The detection was performed through a pulse amperometric detector specific for monosaccharides at 30°C. The mobile phase was a step gradient (0% to 85% to 0%) of 300 μmol/mL NaOH solution (reagent grade type I) and ultra-pure 0.2 μm filtered degassed water (with a resistivity of at least 18 MΩ) at a flow rate of 1.0 mL/min. The postcolumn solvent was a 400 μmoL/mL NaOH solution at a flow rate of 0.3 mL/min. The total running time was 50 min, and all solvents were kept under a N2 atmosphere.
Analytical curves (2.0 to 250.0 mg/L) were constructed for each monosaccharide to be determined using standards chosen based on previously reported Chlorella cell wall sugar compositions [42]. The sugar content was estimated by measuring the peak area.
Curve fit analysis
The hydrolysis time curves were fitted in a double exponential curve, which was rejected when it did not converge or when the predicted parameters had P values greater than 0.05, which resulted in a large standard error. Under these circumstances, a mono-exponential model was used. Fittings were performed using a non-linear regression (Levengerg-Marquardt Algorithm) with SigmaPlot 10.0 software (Systat Software Inc., San Jose, CA, USA) for Windows. A fitting analysis was used to estimate the initial hydrolytic activity rates, the maximum hydrolysis yield, and the time needed to reach 50% hydrolysis for each enzymatic preparation (t½).
Statistical analysis
The results obtained from the assays to test the dependency of biomass hydrolysis yields on β-glucosidase activity were analyzed by one-way ANOVA with MINITAB 15.0 software for Windows (Minitab Inc., PA, USA). The differences among the means were verified using the Fisher test. The means were considered to be significantly different when P ≤ 0.05.
Cell wall X-ray diffractometry
The crystallinity of the C. homosphaera dried biomass was evaluated by X-ray diffraction with a Rigaku MiniFlex diffractometer and filtered copper Kα radiation (λ = 0.1542 nm) using a monochromator at a 30 KV voltage and 15 mA electric current, with a speed of approximately 2°/min and scanning at an angle (2θ) from 2° to 60°. For comparison, a diffractogram of cellulose microcrystalline (Avicel PH101, Sigma-Aldrich) was also performed. Each sample’s crystallinity index (CrI) was calculated according to the Segal method [62].
SEM
Scanning electron microscopy (SEM-FEI/Inspect S50 model) was used to investigate the microalgae biomass morphology before and after 48 h of enzymatic treatment with the A. awamori-T. reesei enzyme blends. Samples were adhered to carbon tape, sputter-coated with 28 nm gold with an Emitech/K550 model and observed via SEM with an acceleration voltage of 20 KV and a working distance of approximately 19 mm. Several images were obtained from different areas of the samples (at least 20 images per sample) to guarantee the reproducibility of the results.
Abbreviations
- SEM:
-
scanning electron microscopy
- CrI:
-
crystallinity index
- FTIR:
-
Fourier transform infrared spectroscopy
- AHS:
-
amyloglucosidase hydrolyzable starch
- XRD:
-
X-ray diffraction
- FPAse:
-
filter paper activity
- FPU:
-
filter paper unit
- CMCase:
-
endo-β-1,4 glucanase
- INCQS:
-
Instituto Nacional de Controle de Qualidade em Saúde
- IU:
-
international unit
- IUPAC:
-
International union of pure and applied Chemistry
- TFA:
-
trifluoracetic acid
- EG:
-
endoglucanase
- CBH:
-
cellobiohydrolase
- HPAEC-PAD:
-
high-performance anion exchange chromatography and pulse amperometric detection
References
John RP, Anisha GS, Nampoothiri KM, Pandey A. Micro and macroalgal biomass: a renewable source for bioethanol. Bioresour Technol. 2011;102:186–93.
Van Beilen JB. Why microalgal biofuels won’t save the internal combustion machine. Biofuels, Bioprod Biorefining. 2010;4:41–52.
Walker DA. Biofuels - for better or worse? Ann Appl Biol. 2010;156:319–27.
Chinnasamy S, Bhatnagar A, Hunt RW, Das KC. Microalgae cultivation in a wastewater dominated by carpet mill effluents for biofuel applications. Bioresour Technol. 2010;101:3097–105.
Ellis JT, Hengge NN, Sims RC, Miller CD. Acetone, butanol, and ethanol production from wastewater algae. Bioresour Technol. 2012;111:491–5.
Pittman J. The potential of sustainable algal biofuel production using wastewater resources. Bioresour Technol. 2011;102:17–25.
Scragg A, Illman A, Carden A, Shales S. Growth of microalgae with increased calorific values in a tubular bioreactor. Biomass Bioenergy. 2002;23:67–73.
Illman A, Scragg A, Shales S. Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme Microb Technol. 2000;27:631–5.
Ho S-H, Chen C-Y, Chang J-S. Effect of light intensity and nitrogen starvation on CO2 fixation and lipid/carbohydrate production of an indigenous microalga Scenedesmus obliquus CNW-N. Bioresour Technol. 2012;113:244–52.
Harun R, Danquah MK, Forde GM. Microalgal biomass as a fermentation feedstock for bioethanol production. J Chem Technol Biotechnol. 2010;85:199–203.
Amin S. Review on biofuel oil and gas production processes from microalgae. Energy Convers Manag. 2009;50:1834–40.
Costa JAV, De Morais MG. The role of biochemical engineering in the production of biofuels from microalgae. Bioresour Technol. 2011;102:2–9.
Demirbas MF. Biofuels from algae for sustainable development. Appl Energy. 2011;88:3473–80.
Harun R, Danquah MK. Influence of acid pre-treatment on microalgal biomass for bioethanol production. Process Biochem. 2011;46:304–9.
Al-Zuhair S, Ramachandran KB, Farid M, Aroua MK, Vadlani P, Ramakrishnan S, et al. Enzymes in biofuels production. Enzyme Res. 2011;2011:2.
Ho S-H, Huang S-W, Chen C-Y, Hasunuma T, Kondo A, Chang J-S. Bioethanol production using carbohydrate-rich microalgae biomass as feedstock. Bioresour Technol. 2013;135:191–8.
Nicolai E, Preston RD. Cell-wall Studies in the chlorophyceae. I. A General survey of submicroscopic structure in filamentous species. Proc R Soc B Biol Sci. 1952;140:244–74.
Koyama M, Sugiyama J, Itoh T. Systematic survey on crystalline features of algal celluloses. Cellulose. 1997;4:147–60.
Wada M, Sugiyama J, Okano T. The crystalline phase (Iα/Iβ) system of native celluloses in relation to plant phylogenesis. Mokuzai Gakkaishi. 1995;41:186–92.
Michell AJ. Second-derivative FTIR spectra of native celluloses. Carbohydr Res. 1990;197:53–60.
Roberts AW, Roberts EM, Delmer DP. Cellulose synthase (CesA) Genes in the green alga Mesotaenium caldariorum. Eukaryot Cell. 2002;1:847–55.
Atalla RH, Vanderhart DL. Native cellulose: a composite of two distinct crystalline forms. Science. 1984;223:283–5.
VanderHart DL, Atalla RH. Studies of microstructure in native celluloses using solid-state carbon-13 NMR. Macromolecules. 1984;17:1465–72.
Sugiyama J, Vuong R, Chanzy H. Electron diffraction study on the two crystalline phases occurring in native cellulose from an algal cell wall. Macromolecules. 1991;24:4168–75.
Hayashi N, Sugiyama J, Okano T, Ishihara M. The enzymatic susceptibility of cellulose microfibrils of the algal-bacterial type and the cotton-ramie type. Carbohydr Res. 1997;305:261–9.
Hayashi N, Sugiyama J, Okano T, Ishihara M. Selective degradation of the cellulose Iα component in Cladophora cellulose with Trichoderma viride cellulase. Carbohydr Res. 1997;305:109–16.
Mihranyan A. Cellulose from cladophorales green algae: from environmental problem to high-tech composite materials. J Appl Polym Sci. 2011;119:2449–60.
Burczyk J, Terminska-Pabis K, Smietana B. Cell wall neutral sugar composition of chlorococcalean algae forming and not forming acetolysis resistant biopolymer. Phytochemistry. 1995;38:837–41.
Burczyk J, Loos E. Cell wall-bound enzymatic activities in Chlorella and Scenedesmus. J Plant Physiol. 1995;146:748–50.
Loos E, Meindl D. Cell wall-lytic activity in Chlorella fusca. Planta. 1984;160:357–62.
Loos E, Meindl D. Cell-wall-bound lytic activity in Chlorella fusca: function and characterization of an endo-mannanase. Planta. 1985;166:557–62.
Ogawa K, Yamaura M, Ikeda Y, Kondo S. New aldobiuronic acid, 3-O-alpha-D-glucopyranuronosyl-L-rhamnopyranose, from an acidic polysaccharide of Chlorella vulgaris. Biosci Biotechnol Biochem. 1998;62:2030–1.
Ogawa K, Ikeda Y, Kondo S. A new trisaccharide, α-d-glucopyranuronosyl-(1 → 3)-α-l-rhamnopyranosyl-(1 → 2)-α-l-rhamnopyranose from Chlorella vulgaris. Carbohydr Res. 1999;321:128–31.
Burczyk J, Smietana B, Terminska-Pabis K, Zych M, Kowalowski P. Comparison of nitrogen content amino acid composition and glucosamine content of cell walls of various chlorococcalean algae. Phytochemistry. 1999;51:491–7.
Rodríguez MC, Noseda MD, Cerezo AS. The fibrillar polysaccharides and their linkage to algaenan in the trilaminar layer of the cell all of Coelastrum sphaericum (Chlorophyceae). J Phycol. 1999;35:1025–31.
Dragone G, Fernandes BD, Abreu AP, Vicente AA, Teixeira JA. Nutrient limitation as a strategy for increasing starch accumulation in microalgae. Appl Energy. 2011;88:3331–5.
Choi SP, Nguyen MT, Sim SJ. Enzymatic pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. Bioresour Technol. 2010;101:5330–6.
Nguyen MT, Choi SP, Lee J, Lee JH, Sim SJ. Hydrothermal acid pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. J Microbiol Biotechnol. 2009;19:161–6.
Ogbonna JC, Tanaka H. Night biomass loss and changes in biochemical composition of cells during light/dark cyclic culture of Chlorella pyrenoidosa. J Ferment Bioeng. 1996;82:558–64.
Harun R, Danquah MK. Enzymatic hydrolysis of microalgal biomass for bioethanol production. Chem Eng J. 2011;168:1079–84.
Northcote DH, Goulding KJ, Horne RW. The chemical composition and structure of the cell wall of Chlorella pyrenoidosa. Biochem J. 1958;70:391–7.
Takeda H. Sugar composition of the cell wall and the taxonomy of Chlorella (Chlorophyceae). J Phycol. 1991;27:224–32.
Takeda H. Chemical composition of cell walls as a taxonomical marker. J Plant Res. 1993;106:195–200.
Watanabe K, Imase M, Sasaki K, Ohmura N, Saiki H, Tanaka H. Composition of the sheath produced by the green alga Chlorella sorokiniana. Lett Appl Microbiol. 2006;42:538–43.
Miranda JR, Passarinho PC, Gouveia L. Pre-treatment optimization of Scenedesmus obliquus microalga for bioethanol production. Bioresour Technol. 2012;104:342–8.
Bon E, Webb C. Glucoamylase production and nitrogen nutrition in Aspergillus awamori. Appl Biochem Biotechnol. 1993;39–40:349–69.
Takeda H. Classification of Chlorella strains by cell wall sugar composition. Phytochemistry. 1988;27:3823–6.
Pei J, Pang Q, Zhao L, Fan S, Shi H. Thermoanaerobacterium thermosaccharolyticum β-glucosidase: a glucose-tolerant enzyme with high specific activity for cellobiose. Biotechnol Biofuels. 2012;5:31.
Corgié SC, Smith HM, Walker LP. Enzymatic transformations of cellulose assessed by quantitative high-throughput fourier transform infrared spectroscopy (QHT-FTIR). Biotechnol Bioeng. 2011;108:1509–20.
Teixeira RSS, da Silva AS, Kim H, Ishikawa K, Endo T, Lee S, et al. Use of cellobiohydrolase-free cellulase blends for the hydrolysis of microcrystalline cellulose and sugarcane bagasse pretreated by either ball milling or ionic liquid [Emim] [Ac]. Bioresour Technol. 2013;149:551–5.
Guillard RR, Lorenzen CJ. Yellow-green algae with chlorophyllide. J Phycol. 1972;8:10–4.
Rodrigues MA, da Silva Bon EP. Evaluation of Chlorella (Chlorophyta) as source of fermentable sugars via cell wall enzymatic hydrolysis. Enzyme Res. 2011;2011:5.
Gottschalk LMF, Paredes RS, Teixeira RSS, Silva AS, Bon EPS. Efficient production of lignocellulolytic enzymes xylanase, β-xylosidase, ferulic acid esterase and β-glucosidase by the mutant strain Aspergillus awamori 2B.361 U2/1. Braz J Microbiol. 2013;44:569–76.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Kwon K-S, Lee J, Kang HG, Hah YC. Detection of {beta}-glucosidase activity in polyacrylamide gels with esculin as substrate. Appl Envir Microbiol. 1994;60:4584–6.
Ghose TK. Measurements of cellulase activities. Pure Appl Chem. 1987;59:257–68.
Chow PS, Landhäusser SM. A method for routine measurements of total sugar and starch content in woody plant tissues. Tree Physiol. 2004;24:1136.
Ranwala AP, Miller WB. Analysis of nonstructural carbohydrates in storage organs of 30 ornamental geophytes by high-performance anion-exchange chromatography with pulsed amperometric detection. New Phytol. 2008;180:421–33.
Federation WE. Standard Methods for the Examination of Water and Wastewater. APHA American Public Health Association, Washington. 1999.
DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–6.
Sumner JB. Dinitrosalicylic acid: a reagent for the estimation of sugar in normal and diabetic urine. J Biol Chem. 1924;62:285–9.
Segal L, Creely JJ, Martin AE, Conrado CM. An empirical method for estimating the degree of crystallinity of native cellulose using the X-ray diffractometer. Text Res J. 1959;29:764–86.
Acknowledgements
The authors are grateful to FINEP from the Brazilian Ministry of Science, Technology and Innovation for financial support (Grant No. 01.09.0566.001421/08). The authors express gratitude to the undergraduate students Mr. Michel Quintal Nunes and Mr. João Carlos Lopes do Nascimento Junior for the cultivation of C. homosphaera and for their technical assistance throughout this work. The authors are also grateful to Daniel Santos Pereira, Maria Alice Santos Cerullo, and Marcello Martins Torquato de Carvalho for performing the chromatographic analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MAR performed the cultivation of C. homosphaera and the preparation of the algae biomass, study coordination, and data analysis, trained and supervised the experimental work performed by undergraduate students for tasks related to routine experimental work, and was involved in the manuscript preparation, revision, and discussion. VSFL was involved in the determination and analysis of X-ray diffraction data for C. homosphaera biomass and the scanning electron microscopy analysis of C. homosphaera biomass before and after enzymatic hydrolysis, manuscript preparation, revision, and discussion. RSST participated in the A. awamori and T. reesei RUT C-30 strain maintenance, propagation, and cultivation for enzyme production, enzyme activity determination, partial purification of β-glucosidase from A. awamori, trained and supervised the experimental work performed by undergraduate students on tasks related to routine experimental work, and was involved in the manuscript preparation, revision, and discussion. EPSB participated in the study outline and supervision, critical analysis of the results, manuscript revision, and final approval. All authors have read and approved the final manuscript.
Additional files
Additional file 1: Figure S1.
The time course for the release of total reducing sugars from C. homosphaera biomass (50 mg d.w./mL) in 50 mM citrate buffer, pH 4.8, at 50°C. The biomass was hydrolyzed using enzyme preparations with a 1.5 IU/g endoglucanase load from T. reesei (A), A. cellulolyticus (B), A. awamori (C), and a T. reesei-A. awamori blend (D) at final β-glucosidase loads of 7.5 (filled circle), 15.0 (empty circle), and 22.5 IU/g (triangles). The data were fitted into an exponential function as described in the Materials and methods section. Standard deviation was less than 10% of the mean value and bars were omitted for clarity.
Additional file 2: Figure S2.
A time course for the release of glucose from C. homosphaera biomass (50 mg d.w./mL) in 50 mM citrate buffer, pH 4.8, at 50°C. The biomass was hydrolyzed using the enzyme preparations with a 1.5 IU/g endoglucanase load from T. reesei (A), A. cellulolyticus (B), A. awamori (C), and the T. reesei-A. awamori blend (D) at final β-glucosidase loads of 7.5 (filled circle), 15.0 (empty circle), and 22.5 IU/g (triangles). The data were fitted into an exponential function as described in the Materials and methods section. Standard deviation was less than 10% of the mean value and bars were omitted for clarity.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Rodrigues, M.A., Teixeira, R.S.S., Ferreira-Leitão, V.S. et al. Untreated Chlorella homosphaera biomass allows for high rates of cell wall glucan enzymatic hydrolysis when using exoglucanase-free cellulases. Biotechnol Biofuels 8, 25 (2015). https://doi.org/10.1186/s13068-015-0215-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-015-0215-1